Abstract
The attractant chemotaxis response of Escherichia coli to norepinephrine requires that it be converted to 3,4-dihydroxymandelic acid (DHMA) by the monoamine oxidase TynA and the aromatic aldehyde dehydrogenase FeaB. DHMA is sensed by the serine chemoreceptor Tsr, and the attractant response requires that at least one subunit of the periplasmic domain of the Tsr homodimer (pTsr) has an intact serine-binding site. DHMA that is generated in vivo by E. coli is expected to be a racemic mixture of the (R) and (S) enantiomers, so it has been unclear whether one or both chiral forms are active. Here, we used a combination of state-of-the-art tools in molecular docking and simulations, including an in-house simulation-based docking protocol, to investigate the binding properties of (R)-DHMA and (S)-DHMA to E. coli pTsr. Our studies computationally predicted that (R)-DHMA should promote a stronger attractant response than (S)-DHMA because of a consistently greater-magnitude piston-like pushdown of the pTsr α-helix 4 toward the membrane upon binding of (R)-DHMA than upon binding of (S)-DHMA. This displacement is caused primarily by interaction of DHMA with Tsr residue Thr156, which has been shown by genetic studies to be critical for the attractant response to L-serine and DHMA. These findings led us to separate the two chiral species and test their effectiveness as chemoattractants. Both the tethered cell and motility migration coefficient assays validated the prediction that (R)-DHMA is a stronger attractant than (S)-DHMA. Our study demonstrates that refined computational docking and simulation studies combined with experiments can be used to investigate situations in which subtle differences between ligands may lead to diverse chemotactic responses.
Significance
3,4-Dihydroxymandelic acid (DHMA) is a Tsr chemoattractant that binds to the periplasmic domain of the E. coli Tsr chemoreceptor (pTsr). DHMA produced in E. coli is presumably a racemic mixture of its two enantiomers, (R)-DHMA and (S)-DHMA. We used a combination of computational studies, primarily based on docking and molecular dynamics simulations, to predict that the two enantiomers will bind in different orientations and interact differently with key pTsr residues. The binding of (R)-DHMA is predicted to cause a greater downward piston-like movement of α-helix 4 of pTsr than binding of (S)-DHMA. Experiments demonstrated that (R)-DHMA and (S)-DHMA are both attractants, with (R)-DHMA being more potent than (S)-DHMA. This result validates our approach of making computational predictions to guide experimental design.
Introduction
It has recently become clear that chemotaxis in E. coli confers advantages other than the ability to migrate to higher concentrations of nutrients. Signaling molecules derived both from other microorganisms and from the host have been identified as potent attractants. Most notable in the first group is the general quorum-sensing signal AI-2, used to assess bacterial population density (1). It is sensed by a compound chemoreceptor (2) that involves the periplasmic AI-2-binding protein LsrB (3) and the membrane-associated methyl-accepting chemotaxis protein Tsr, which was originally annotated as the serine chemoreceptor (4). Chemotaxis to AI-2 has been shown to be important in the initiation of the cell aggregation that leads to biofilm development (5,6).
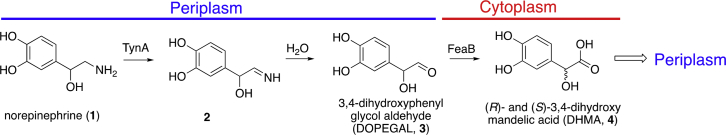
Included in the second group is the host neurotransmitter and adrenal hormone norepinephrine (NE), also sensed by Tsr (7). NE is converted to the actual attractant, 3,4-dihydroxymandelic acid (DHMA), by the action of two bacterial enzymes, the periplasmic monoamine oxidase TynA and the cytoplasmic aromatic aldehyde dehydrogenase FeaB (8). DHMA is sensed by binding to the periplasmic domain of Tsr (pTsr). Therefore, chemotaxis to NE requires that 3,4-dihydroxyphenyl-glycol-aldehyde (DOPEGAL) be formed by deamination of NE in the periplasm; that DOPEGAL be imported into the cytoplasm, where it is converted to DHMA; and that DHMA then be exported out to the periplasm (7,9) (Fig. 1). This scenario should mean that DHMA will be effective as a chemoattractant at much lower concentrations than NE, which is the case.
Figure 1.
Reaction schematic of the conversion of NE to DHMA. NE is converted by the periplasmic monoamine oxidase TynA into DOPEGAL, and DOPEGAL is converted by the cytoplasmic aromatic aldehyde dehydrogenase FeaB into (R)- or (S)-DHMA. To see this figure in color, go online.
DHMA also shares with NE the capability of inducing virulence genes contained in enterohemorrhagic E. coli (10). A number of other host-produced compounds also are active as either chemoattractants or repellents (11). NE is released in copious amounts in the gastrointestinal tract, with high concentrations found in gut-associated lymphoid tissue (12). The presence of detectable amounts of DHMA in the mouse gastrointestinal tract, however, requires the presence of the resident microbiota, as DHMA is not found in the feces or cecal contents of germ-free mice (10).
Our initial studies showed that DHMA is active as an attractant at very low concentrations but has the perplexing property of losing its activity at concentrations of 500 μM and above (7). DHMA interacts with the periplasmic serine-binding site of Tsr. A Tsr dimer having only one intact serine-binding site detects DHMA as an attractant as well as a fully intact dimer. However, the loss of attraction at high concentrations depends on having both serine-binding sites of the Tsr dimer intact. This result suggests that the attractant response is abrogated when both binding sites are occupied with DHMA.
DHMA is commercially available only as a racemic mixture of its two enantiomers (R)-DHMA and (S)-DHMA. The DHMA produced by E. coli is probably also a racemic mixture (Fig. 1). To determine whether one or both enantiomers of DHMA are active as attractants, we used computational docking and simulation-based approaches to study the binding of (R)- and (S)-DHMA to pTsr. Previous experimental (13) and computational (14) studies indicated that the binding of chemoattractants to Tsr or Tar results in a downward (toward-the-membrane) piston-like movement of α-helix 4 of the periplasmic domain. To study the effect of subtle physicochemical differences of the two chiral forms of DHMA on their interactions with pTsr, and to determine whether any binding differences between the two could result in potential structural and allosteric conformational differences linked to chemotaxis, we used highly accurate docking methods complemented with multiple, multi-nanosecond (multi-ns) simulations. First, we used an in-house computational protocol (15, 16, 17, 18, 19, 20) to investigate both enantiomers of DHMA binding to pTsr after an initial computational study focused on (R)-DHMA docking to pTsr (15). This method allows for nearly exhaustive docking of a compound in a receptor binding pocket, with the most energetically favorable binding mode (i.e., the one the lowest in binding energy) being revealed through molecular dynamics (MD) simulations and physics-based energy calculations (15, 16, 17, 18, 19, 20). Since its development, the method has been used to investigate several other ligand-protein complexes (16, 17, 18, 19, 20). Subsequently, we employed multiple multi-ns MD simulations to investigate the most energetically favorable binding modes of (R)- and (S)-DHMA in complex with pTsr. We focused our analysis on the interactions between the key residues of the protein with the two compounds, focusing on potential conformational changes induced by their binding.
Materials and Methods
Computational methods
We performed multi-ns, explicit-solvent MD simulations followed by structural, conformational, and energetic analyses to compare the binding properties of (R)- and (S)-DHMA complexed with E. coli pTsr. The starting structure for the MD simulations corresponded to the most energetically favorable binding mode of each enantiomer in complex with pTsr, determined independently using a docking-refinement protocol developed in house (15, 16, 17, 18, 19, 20). All analyses were performed using CHARMM (21), Wordom (22), and in-house FORTRAN programs.
Docking (S)-DHMA bound to pTsr
We used our in-house docking-refinement protocol to derive the most energetically favorable binding modes of (R)- and (S)-DHMA in complex with E. coli pTsr (15, 16, 17, 18, 19, 20). Specifically, the (R)-DHMA-pTsr complex structure was derived in our recent previous study (15), and the (S)-DHMA-pTsr complex structure was derived in the current study. The docking-refinement protocol (15, 16, 17, 18, 19, 20) can be divided into the following stages: 1) initial positioning of the ligand within the receptor binding pocket, based on available experimental data; 2) multiple short MD docking simulations of the ligand within the receptor binding pocket, with the ligand forced to rotate and sample different binding modes while being constrained in the pocket by harmonic and quartic spherical potentials and with the absence of any constraints on the binding-pocket residues; 3) interaction-energy calculations to select the most probable binding modes generated by the short docking simulations; and 4) multiple explicit-solvent MD simulations and physical-chemistry-based free energy calculations to identify the most favorable binding mode of the ligand-receptor complex (Fig. S1). The independent use of harmonic and quartic harmonic potential functions that constrain the ligand in the binding pocket with respect to its initial position encourage the exploration of binding modes both away from (quartic) and proximal to (harmonic) their initial positioning.
In this study, we docked (S)-DHMA in the serine-binding site of the experimentally resolved crystal structure of the serine-pTsr complex (Protein Data Bank, PDB: 3ATP (13)) using the same parameters as in our previous study of (R)-DHMA binding to pTsr (15). Details about the docking of (S)-DHMA are described in Supporting Material. From the short MD docking simulations, we collected 15 binding modes of the (S)-DHMA-pTsr complex. All 15 binding modes were investigated using 20-ns MD simulations, and the binding mode resulting in the most energetically favorable complex, as assessed by MM-GBSA association free energy calculations (23), was selected. We verified that all the other binding modes of the (S)-DHMA-pTsr complex were far less energetically favorable than the identified binding mode, and the identified binding mode was selected for further investigation. In the case of (R)-DHMA, the two most energetically favorable binding modes were similar in energy and also similar in structure (15); thus, we selected the binding mode resulting in the most favorable association free energy for further investigation. The selected binding modes of (S)-DHMA, derived in the current study, and (R)-DHMA, derived in our previous study (15), in complex with pTsr were used as initial structures in five replicate 100-ns MD simulations.
Investigating (R)-DHMA and (S)-DHMA in complex with pTsr using multiple MD simulations
Using the structures of the most energetically favorable binding modes as initial structures, we performed five independent 100-ns MD simulations of bound (R)- and (S)-DHMA. MD simulations refined intermolecular interactions and the complex structures for detailed examination of the key interactions between the enantiomers and pTsr. They also facilitated the investigation of the conformational properties of the pTsr protein, including allosteric effects, induced by the binding of the enantiomers. A short, 2-ns equilibration stage was introduced before each simulation. Subsequently, five 100-ns MD simulations were performed for each system, resulting in an aggregate duration of 0.5 μs per system. Multiple multi-ns MD simulations of the ligand-binding domain, as performed in (14), were preferred over longer coarse-grained simulations or shorter all-atom simulations of the entire Tsr protein bound to the enantiomers. The former lacks detail in the ligand-binding site, whereas the latter prevents reaching equilibrium in piston-like motions. Our simulated system provided a suitable balance between atomic details of the key interactions of pTsr with the two enantiomers and the observation of allosteric effects associated with attractant signaling (14). All MD simulations were performed using explicit water with periodic boundary conditions. Additional information on setup and parameters is detailed in (15) and in Supporting Material. Coordinates of (R)- and (S)-DHMA in complex with pTsr from the 100-ns MD simulations are provided (Data S1).
We performed root mean-squared deviation (RMSD) calculations of the ligand heavy atoms with respect to 1) the initial position of the ligand for each independent trajectory 2), the average ligand structure for each independent trajectory, and 3) the average ligand structure from the combined five replicate trajectories. The RMSD calculations were used to determine 1) whether the initial structure identified by our docking-refinement protocol was conserved in all multi-ns MD simulations, 2) whether the orientation of both ligands was stable within their respective simulations, and 3) whether the orientation of both ligands was reproduced in all five simulations. Before the RMSD calculations, the trajectories were aligned with the pTsr backbone atoms of both subunits.
We also identified key interactions associated with the binding of (R)- and (S)-DHMA through per-residue interaction free energy calculations, analogously to (16,17,19,20,24, 25, 26, 27, 28). This analysis complemented an examination of hydrogen bonds, ligand-protein contacts, and water-mediated interactions within the complex. This approach provided insights into the interplay of the interactions with the structure of the (R)- and (S)-DHMA-pTsr complexes. The analysis was performed to examine, and potentially differentiate, key interactions between the two enantiomers and pTsr. The determination of water-mediated interactions and per-residue interaction free energy calculations is described briefly in Supporting Material and described in detail in (16,17,19,20).
Investigating piston-like motions in pTsr caused by binding of (R)-DHMA and (S)-DHMA
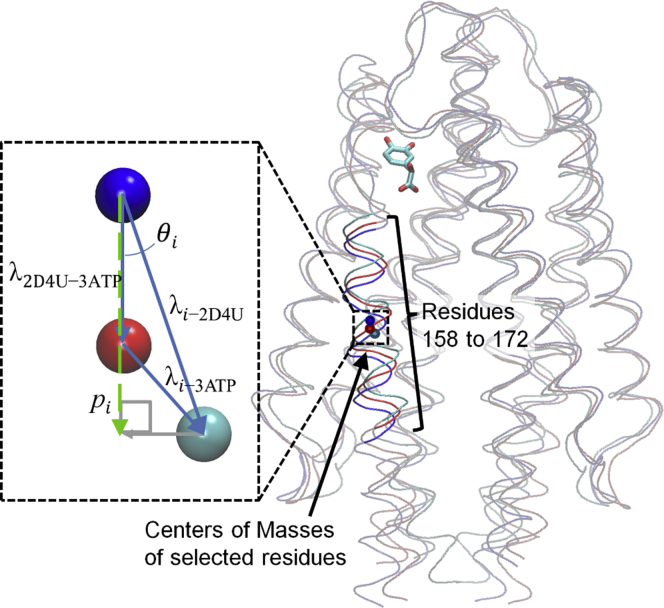
According to the x-ray studies of Tajima et al. (13), binding of L-serine to E. coli pTsr induces a conformational change in residues 158–172 of α-helix 4 (13). This helix is contiguous with the second transmembrane helix, which connects the periplasmic domain of Tsr to the cytoplasmic HAMP domain (29,30). To investigate the conformational dynamics within pTsr, we analyzed the dynamics of α-helix 4 residues 158–172 throughout the multiple multi-ns MD simulations using calculations quantifying the motions of these residues. Based on the quantified motions of α-helix 4 residues, we defined a metric to compare the potential signaling strength of (R)- and (S)-DHMA as described below.
We calculated the deviation between the position of α-helix 4 in each simulation snapshot and its position in the crystal structure of the unbound pTsr (extracted from PDB: 2D4U (13)) and compared it with the deviation in the crystal structure of serine-bound pTsr (PDB: 3ATP (13)). Before our analysis, the simulation snapshots of all simulated systems and the crystal structure of serine in complex with pTsr were aligned with the crystal structure of apo pTsr using all Cα atoms. To describe the position of the helix within a structure, we calculated the geometric center of the Cα atoms of residues 158–172 of the signaling subunit. As a point of reference, we calculated the vector of the Cα geometric center of the experimentally resolved apo pTsr to the Cα geometric center of the experimentally resolved serine-bound pTsr. This vector was used as a baseline for Eqs. 1 and 2. We defined a metric to evaluate the signaling strength of (R)- and (S)-DHMA based on the deviations projected onto the baseline vectors or the projected lengths. The vectors used to describe the position of the helix are shown in Fig. 2.
| (1) |
| (2) |
According to our definition, Eq. 1 isolates the direction of motion in α-helix 4 of the ligand-pTsr complex that is likely to contribute to pTsr signaling. Equation. 2 provides the magnitude of the motion of α-helix 4 in the direction of interest, defined by Eq. 1, normalized by the change in position observed in the crystal structures of the unbound and serine-bound pTsr. In Eq. 1, the angle between the vector of the helix in the crystal structure of serine-bound pTsr to the crystal structure in apo pTsr, and the vector of the helix in a simulation snapshot of pTsr bound to (R)- or (S)-DHMA and the crystal structure of apo pTsr, θi, is determined through the law of cosines. λ corresponds to the length between the helix in the crystal structure of the serine-bound pTsr and the crystal structure of the apo pTsr (3ATP-2D4U), the helix in a simulation snapshot of pTsr bound to (R)- or (S)-DHMA and the crystal structure of apo pTsr (i-2D4U), or the helix in a simulation snapshot of pTsr bound to (R)- or (S)-DHMA and the crystal structure of the serine-bound pTsr (i-3ATP). In Eq. 2, the length between the helix in a simulation snapshot of pTsr bound to (R)- or (S)-DHMA and the crystal structure of apo pTsr (λi-2D4U) is projected onto the baseline vector and normalized using the length between the helix in the crystal structure of serine-bound pTsr and the crystal structure of the apo pTsr (λ3ATP-2D4U) to give the reported projected length, pi. According to the defined metric, a pi value of 1 indicates a movement of α-helix 4 in response to binding of DHMA comparable to the movement induced by serine, a value > −1 indicates a movement of α-helix 4 greater than the movement induced by serine, and a value < −1 indicates a movement of α-helix 4 less than the movement induced by serine. The vectors used to describe the position of the helix are shown in Fig. 2.
Figure 2.
Schematic of the vectors used to calculate the projected lengths. Tsr is shown in a gray tube representation. Residues 158–172 of the crystal structure of apo and serine-bound pTsr are shown in blue and red tube representations. Residues 158–172 of the simulated snapshot of pTsr bound to (R)-DHMA or (S)-DHMA are shown in a cyan tube representation. The geometric centers of residues 158–172 are shown as blue, red, and cyan spheres in the crystal structure of apo pTsr, the crystal structure of serine-bound pTsr, and the simulated snapshot of pTsr bound to (R)-DHMA or (S)-DHMA, respectively. In the zoomed-in window, vectors between the centers of mass are indicated by blue arrows. The projection of the vector λi-2D4U onto the baseline vector, λ2D4U-3ATP, is indicated by a gray arrow. The resulting projected length, pi, which is shown as a dashed green arrow. To see this figure in color, go online.
Extracting collected motions and correlated motions of pTsr in complex with (R)-DHMA and (S)-DHMA using principal component analysis and dynamic cross correlation
Principal component analysis (PCA) was used to investigate collected motions within the multiple multi-ns MD simulations of (R)- and (S)-DHMA in complex with pTsr. PCA has been implemented in similarly modeled systems of the homologous pTar in complex with agonist compounds to extract piston-like motions (14), as well as applied to other proteins involved in chemotaxis (31), ATP generation (32), cancer (33), and HIV (34). Throughout the simulations of both (R)- and (S)-DHMA in complex with pTsr, we observed an oscillatory piston-like motion in α-helix 4 of pTsr and extracted the principal motions due to (R)- and (S)-DHMA binding. We performed PCA based on pTsr Cα atoms using Wordom (22). In preparation for PCA, the five simulation trajectories of (R)- and (S)-DHMA binding to pTsr were independently combined and aligned using Cα atoms. We calculated the covariance matrix of the deviations in Cα atom positions with respect to the average structure using the combined and aligned multiple multi-ns MD simulations. The covariance matrix was diagonalized to obtain an orthogonal set of eigenvectors. We subsequently analyzed the eigenvectors with the largest eigenvalues within the multiple multi-ns MD simulations.
We also performed dynamic cross correlation (DCC) analysis to examine the correlated motions between the residues within the ligand-binding site and the residues in α-helix 4 associated with the piston-like motions of pTsr. We created DCC maps of the dynamic cross-correlated displacements of Cα atoms throughout the multiple multi-ns MD simulations for both (R)- and (S)-DHMA, as in (34,35). The analysis does not provide information about the magnitude of the motions, which can be small local oscillations or large-scale collective motions. Complete correlation corresponds to motions with the same phase and period (36,37).
Experimental methods
Separation of (R)- and (S)-DHMA
Separation of (R)- and (S)-DHMA was achieved using high-performance liquid chromatography (HPLC) with a chiral stationary-phase column. We screened commercially available racemic DHMA (American Custom Chemicals, San Diego, CA) in several analytical columns with different chiral stationary phases and found that a CHIRALPAK ID-3 (150 × 4.6 mm I.D., 3 μm) column provided good resolution using methanol/water (10:90) with 0.1% formic acid as the mobile phase. On the analytical HPLC column, the enantiomers of DHMA had retention times of 4.0 and 5.9 min, and we assigned the (R)- and (S)-configuration, respectively, as described in Results. To isolate milligram quantities of each enantiomer, the separation was carried out on a semipreparative CHIRALPAK ID (250 × 20 mm I.D., 5 μm) column using methanol/water (20:80) with 0.1% formic acid as the mobile phase. We took advantage of the more-rapid reaction kinetics to prepare acetylated (S)-DHMA in high yield and to assign the absolute configuration to each enantiomer. Acetylated (S)-DHMA and nonacetylated (R)-DHMA were separated by reversed-phase semipreparative HPLC using a C18 column after workup of the reaction; DHMA was not stable to silica gel chromatography (see Supporting Material for details).
Tethered-cell assay
The tethered-cell assay was carried out as described by Berg et al. (38), with slight modifications. A ΔcheRB derivative of E. coli strain RP437 (39), called PL138 (this work), was employed in the assays. The strain carried a genomic variant of the native fliC allele that enabled tethering of the cells to glass surfaces (38). Cells were grown in tryptone broth at 33°C, harvested at an OD600 of 0.5, and washed and resuspended in motility buffer (10 mM potassium phosphate buffer, 67 mM NaCl, 0.1 mM EDTA, 10 mM sodium lactate, 0.001 mM L-methionine, pH 7.0). The flagellar filaments were sheared to stubs, and the cells were tethered to a glass coverslip, which was then placed onto the perfusion chamber (40). Cell rotation was imaged and recorded at 60 fps using an upright light microscope (Optihot-2; Nikon, Tokyo, Japan) with a 20× phase objective and a digital camera (UI-3240LE-M-GL; IDS Imaging Development Systems, Obersulm, Germany). The videos were analyzed with custom-written MATLAB codes to determine the rotational speed of the cells as a function of time, with negative and positive values indicating clockwise (CW) and counterclockwise (CCW) directions of rotation, respectively (41). The mean rotational biases of individual cells over time were determined from Gaussian fits to speed distributions.
Microfluidic chemotaxis assay
The motility migration coefficient (MMC) was determined as described by Pasupuleti et al. (7). Briefly, a strain of wild-type E. coli RP437 (39) expressing green fluorescent protein (GFP) from plasmid pCM18 (42) was grown at 30°C in tryptone broth containing 100 μg/mL erythromycin, harvested at an OD600 of 0.5, and washed and resuspended in motility buffer. Dead, RFP-expressing TG1 cells were mixed with the RP437 cells suspended in motility buffer, and the assay was performed within 20 min of preparation. The cell suspension was introduced into the microfluidic device in the middle of the flow channel, and DHMA solutions of equal concentration prepared in motility buffer were introduced on both sides of the introduced cells. The flow rate was maintained at 2.1 μL/min, and images of the cells 1.7 cm down the 2-cm channel were recorded every 2.5 s using a confocal fluorescence microscope (SP8; Leica, Wetzlar, Germany). 400 images were collected for each assay, and the location of the motile, GFP-expressing cells, determined by image analysis, was used to calculate the MMC. The dead, RFP-expressing TG1 cells remained in a thin stream in the exact middle of the channel, demonstrating that the flow was laminar.
Results
Computational results
Investigating (R)-DHMA and (S)-DHMA in complex with pTsr using multiple MD simulations
The initial structures of both (R)-DHMA and (S)-DHMA were conserved in all of their respective MD simulations (five independent 100-ns simulations per enantiomer). The low average RMSD of both enantiomers with respect to their initial docked position in each of the five simulations (ranging from 1.2 to 1.7 Å for (R)-DHMA and 1.3 to 1.7 Å for (S)-DHMA, Fig. S2, A and B) demonstrated that the two ligands do not deviate significantly from their docked positions throughout the individual simulations. The low average RMSD of both enantiomers with respect to their average structure within each of the individual simulations (ranging from 0.9 to 1.1 Å for (R)-DHMA and 0.7 to 1.2 Å for (S)-DHMA, Fig. S2, C and D) indicates that the ligands are stable throughout each of the individual simulations. The low average RMSD of both enantiomers with respect to their average structure within their combined five 100-ns MD simulations signifies that the orientation of the two ligands within their respective set of simulations (0.9 ± 0.4 Å for (R)-DHMA and 1.0 ± 0.5 Å for (S)-DHMA, Fig. S2, E and F) is reproduced across the multiple multi-ns simulations. However, the relatively high average RMSD of (R)-DHMA with respect to the average structure of (S)-DHMA and the high average RMSD of (S)-DHMA with respect to the average structure of (R)-DHMA (excluding the chiral hydroxyl group; 3.0 ± 0.2 and 3.0 ± 0.2 Å, respectively) demonstrates that the orientation of the two enantiomers within the binding site differs. Thus, specific interactions of key pTsr residues with (R)- and (S)-DHMA are expected to also differ.
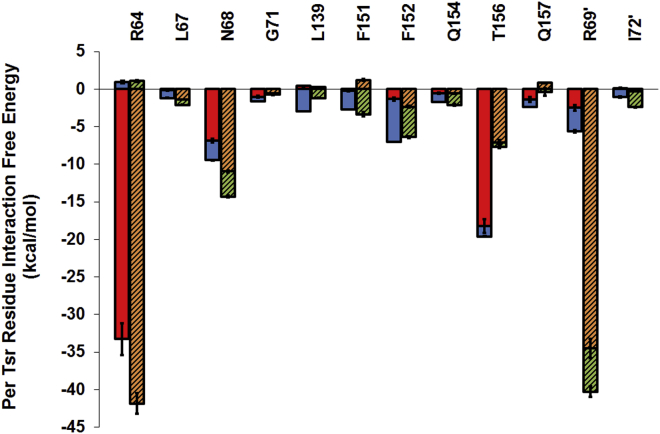
We used a combination of hydrogen bond, ligand-protein contact, and per-residue interaction free energy analyses to uncover the key interactions between pTsr residues and the two ligands and their energetic contribution per residue. These interactions persist throughout the duration of the five replicate simulations and are consistent across all five replicates; they are presented in Fig. 3. Within their simulations, both (R)-DHMA and (S)-DHMA form strong interactions with pTsr residues in the serine-binding site (Figs. 3 and 4). The negatively charged carboxyl group of both enantiomers forms salt bridges with the positively charged guanidino group of Arg64. Additionally, the carboxyl group of both enantiomers forms a hydrogen bond with the side-chain hydroxyl group of Thr156 and with Asn68. Hydrogen bonds are also formed by the three-hydroxyl group of the aromatic ring of both enantiomers with the backbone amide group of Phe151. In addition to their similar polar interactions, the aromatic rings of (R)- and (S)-DHMA form π-π interactions with the aromatic rings of Phe151 and Phe152. Both enantiomers are also involved in van der Waals interactions with Leu67, Leu139, and Ile72′.
Figure 3.
Per-pTsr residue interaction free energies for interactions with free energy values less than −1 kcal/mol. The first set of bars (solid red and blue) per pTsr residue corresponds to interaction free energies between the Tsr residue and (R)-DHMA. The second set of bars (striped orange and green) per pTsr residue corresponds to interaction free energies between the pTsr residue and (S)-DHMA. For each pTsr residue, the interaction free energy is decomposed into polar (gray red and orange) and nonpolar (blue and green bars) components. Error bars correspond to the standard deviation of the per-pTsr residue interaction free energy averaged over the number of simulation snapshots and the five replicate MD simulations. To see this figure in color, go online.
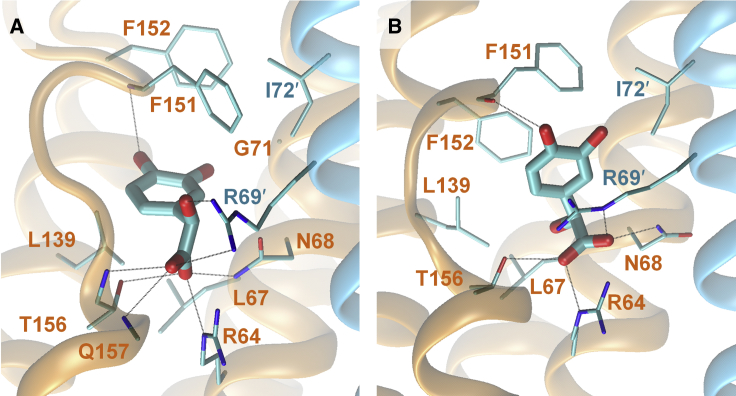
Figure 4.
Molecular graphic images of (A) (R)-DHMA and (B) (S)-DHMA in complex with E. coli pTsr. The majority binding site of pTsr is shown in an orange transparent cartoon representation; the minority binding site is shown in a cyan transparent cartoon representation. (R)-DHMA and (S)-DHMA are shown in cyan, with oxygen atoms in red. Key pTsr residues are shown in stick representations and are labeled in orange (majority side) or cyan (minority side). Black dotted lines indicate hydrogen bonds. To see this figure in color, go online.
Despite the structural similarity of (R)-DHMA and (S)-DHMA, they adopt different orientations within the pTsr binding pocket (Fig. 4), as indicated by the relatively large RMSD values of (R)-DHMA compared with (S)-DHMA, and vice versa. Compared with (R)-DHMA, (S)-DHMA forms a significantly stronger interaction with Arg69′ (Fig. 3). (R)-DHMA forms only a hydrogen bond with Arg69′, whereas (S)-DHMA forms both salt-bridge and cation-π interactions. Additionally, the hydrogen bond between (R)-DHMA and Arg69′ is mediated by water molecules in 19.6% of the simulation snapshots. On the other hand, the interaction free energy between Thr156 and (S)-DHMA is only half that of (R)-DHMA (Fig. 3). Whereas (R)-DHMA forms hydrogen bonds with both side-chain and backbone atoms of Thr156, (S)-DHMA forms only one hydrogen bond with the side-chain hydroxyl (Fig. 4). (R)-DHMA forms a hydrogen bond with the backbone amide group of Thr156 in 68.7% of the simulation snapshots, whereas (S)-DHMA forms a hydrogen bond with the backbone amide group of Thr156 in only 0.1% of the simulation snapshots. Additionally, although the carbonyl group of (R)-DHMA forms water-mediated interactions with either the side-chain or backbone amide group of Thr156 in only 2.4% of the simulation snapshots, the carbonyl group of (S)-DHMA forms water-mediated interactions with the side-chain and backbone amide groups of Thr156 in 34.7 and 18.2% of the simulation snapshots, respectively. Water molecules greatly weaken the interaction between two polar molecules (43). The interaction free energy calculations show that the interaction between Arg69′ and (R)-DHMA is weaker than the interaction between Arg69′ and (S)-DHMA and that the interaction between Thr156 and (S)-DHMA is weaker than the interaction between Thr156 and (R)-DHMA. These differences can partly be attributed to the presence of water mediating these interactions. These differences between the binding of (R)-DHMA and (S)-DHMA to pTsr likely lead to differences in the allosteric effects their binding elicits.
Investigating piston-like motions in pTsr caused by binding of (R)-DHMA and (S)-DHMA
The crystal structures of the unbound and serine-bound E. coli pTsr suggest that conformational changes in α-helix 4 of Tsr are important for signaling (13). We tracked the projected lengths of α-helix 4 throughout the simulations to understand the conformational dynamics of pTsr bound to (R)- and (S)-DHMA as well as to predict and compare the Tsr strength of the signal induced by the two ligands. The projected lengths with respect to time for each of the five replicate multi-ns simulations are plotted in Fig. 5. As controls, we also tracked projected lengths for five 100-ns MD simulations of serine in complex with pTsr and two 100-ns MD simulations of apo pTsr. The starting structure of the former corresponded to the experimentally resolved structure of the pTsr-serine complex (13), except that the second bound serine ligand with disordered electron density (13) was removed. The starting structure of the latter corresponded to the experimentally resolved structure of the pTsr-serine complex (13) with both bound serine ligands removed. For the simulations of serine in complex with pTsr, the average projected length for the downward motion of α-helix 4 was 0.92 ± 0.08 Å (Fig. S3 A), an average value that deviates from the crystal structure by only 0.08 Å. For the simulations of the apo pTsr, the projected length approached zero as the simulations progressed, and after 50 ns, the average projected length was only 0.11 ± 0.12 Å (Fig. S3 B). Thus, the simulated apo pTsr returned to the conformation of the apo pTsr crystal structure (13). These results suggest that the modeled systems and the simulation duration of (R)- and (S)-DHMA in complex with pTsr were sufficiently accurate to investigate signaling induced by the two ligands. They also indicate that the projected lengths can be used as a metric to predict the strength of the signal induced by the two ligands. Additional details on the MD simulations and subsequent analysis of serine-bound and apo pTsr are provided in Supporting Material.
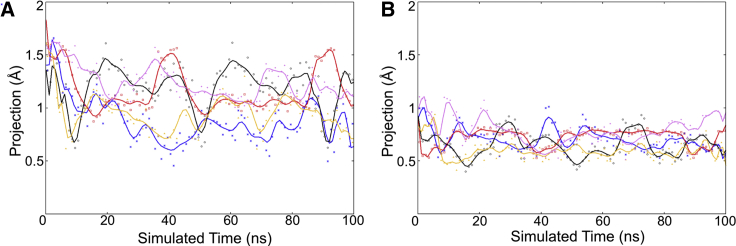
Figure 5.
Vector projections showing the normalized magnitude of piston-like motions (in Å) within α-helix 4 of pTsr bound to (A) (R)-DHMA and (B) (S)-DHMA with respect to the simulated time (in nanoseconds). The projection values plotted correspond to projected lengths, pi, calculated using Eq. 2 and extracted every 1 ns. Solid lines correspond to Bezier curves fitted to projected lengths extracted every 0.1 ns. Blue, red, black, yellow, and pink data points and lines correspond to data extracted from the first, second, third, fourth, and fifth simulations, respectively, for each of the complexes. The differences in initial points are attributed to the initial structure corresponding to the binding mode identified by our docking-refinement protocol as well as the use of a short equilibration stage before the 100-ns MD simulations. To see this figure in color, go online.
Our analysis shows that (R)-DHMA induces a larger motion of the α-helix 4 toward the membrane than (S)-DHMA. The average projected length for (R)-DHMA is 1.08 ± 0.15 Å, whereas that for (S)-DHMA is 0.69 ± 0.07 Å. By evaluating the projected lengths as a function of time, we observed that α-helix 4 oscillates throughout the MD simulations (Fig. 5). Although the simulations of both enantiomers in complex with pTsr show piston-like motions in the helix, the peak values with (R)-DHMA are higher than with (S)-DHMA; the maximal projected length after 50 ns with (R)-DHMA is 2.12 Å, whereas the maximal projected length with (S)-DHMA is 1.26 Å. The presence of larger piston-like motions with (R)-DHMA indicates that (R)-DHMA induces a stronger allosteric signal than (S)-DHMA.
Extracting collected motions of pTsr in complex with (R)-DHMA and (S)-DHMA using PCA
We performed PCA on the Cα atomic positions throughout the combined five replicate trajectories for both (R)- and (S)-DHMA to extract the principal motions occurring within the multiple multi-ns simulations. According to PCA, the first eigenvector of the simulations with (R)- and (S)-DHMA corresponds to a piston-like motion of α-helix 4 of pTsr (Fig. 6). The first eigenvector accounts for 35.35 and 24.76%, respectively, of the motions observed in the simulations of the (R)-DHMA-pTsr and (S)-DHMA-pTsr complexes. In line with our aforementioned analysis on the projected motions of the helix, the amplitude of the motion within the first eigenvector for residues 158–172 is larger with (R)-DHMA than with (S)-DHMA. These results complement the calculated projected lengths, providing further evidence that (R)-DHMA should be a stronger chemoattractant than (S)-DHMA.
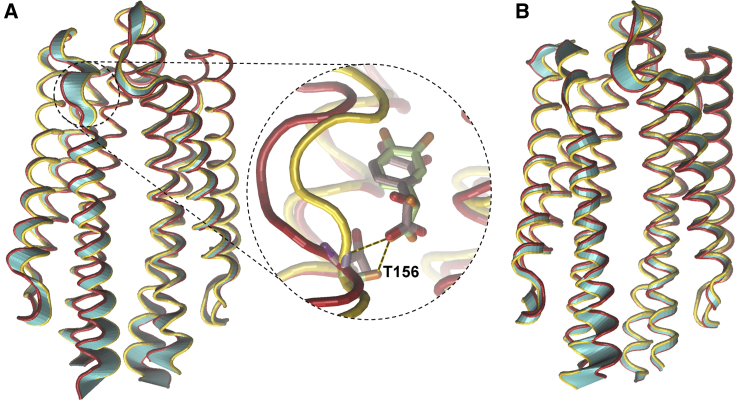
Figure 6.
Molecular graphics image of the principal motions of E. coli pTsr in complex with (A) (R)-DHMA and (B) (S)-DHMA. The beginning and ending structures of pTsr in the principal motion are shown in red and gold. When (R)-DHMA forms hydrogen bonds with both the side chain and backbone of Thr156 (indicated in black dotted lines within the zoomed-in window of the binding site), the pTsr loop containing residues 151–156 is pulled into the binding pocket, and α-helix 4 is pushed down (gold tube representation). When the hydrogen bond is absent, residues 151–156 move away from the binding pocket, and α-helix 4 relaxes upward relative to when the hydrogen bond is present (red tube representation). The motion between the relaxed and pushed-down states of α-helix 4 is represented by the cyan tube representation. To see this figure in color, go online.
Extracting correlated motions of pTsr in complex with (R)-DHMA and (S)-DHMA using DCC
To understand how, and to what extent, different interactions within the binding pocket correlate with the observed differences in the motion of α-helix 4, we performed DCC analysis. The DCC maps are shown in Fig. 7. The maps are quite similar for (R)- and (S)-DHMA. However, we observed a different correlation of motion for residues within the binding site (148–156) and residues in α-helix 4 (156–185). These are marked by the green box in Fig. 7. For both ligands, apart from the expected correlated motions among nearby residues in the pTsr sequence (the diagonal in Fig. 7), the motions of residues within the same α-helix and residues in proximal α-helices are correlated.
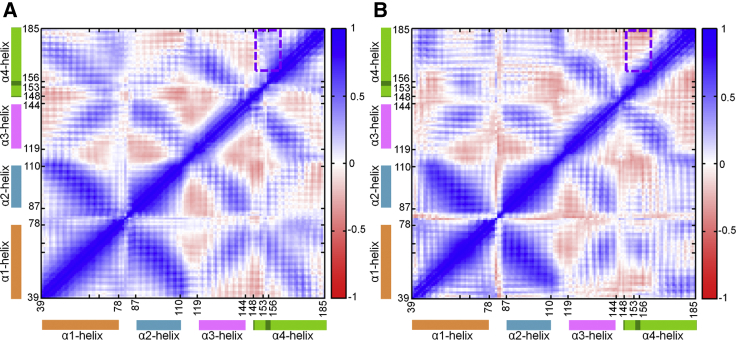
Figure 7.
Dynamic cross correlation maps for E. coli pTsr binding to (A) (R)-DHMA and (B) (S)-DHMA using Cα atom coordinates. The color range to the right of each map indicates the degree of correlation (or anticorrelation), with blue indicating fully correlated motions and red indicating fully anticorrelated motions. The orange, cyan, pink, and green blocks highlight residue motifs belonging to α-helix 1, α-helix 2, α-helix 3, and α-helix 4 of pTsr, respectively. The region encapsulated by green dotted lines denotes the key difference in correlated motions between pTsr bound to (R)-DHMA versus (S)-DHMA. To see this figure in color, go online.
The correlated motions of pTsr binding to (R)- and (S)-DHMA differ in the motions of the binding site residues and α-helix 4 residues (marked by the green box in Fig. 7). With (R)-DHMA, the motions of residues in α-helix 4 are highly correlated with the movement of residues in the binding site. In contrast, with (S)-DHMA, the motions of α-helix 4 residues are weakly correlated or anticorrelated with the movement of residues in the binding site. Specifically, with (R)-DHMA, the contraction of residues 148–156 into the binding site correlates with the downward motion of α-helix 4. Thus, differences in the piston-like motions observed in the simulations of (R)- and (S)-DHMA binding to pTsr are due to differences in their interactions with residues in the ligand-binding pocket.
To understand which interactions are most important for the contraction of the ligand-binding site residues, we extracted the simulation snapshots of pTsr binding to (R)-DHMA in which α-helix 4 was or was not pushed down. We observed that when the helix is pushed downward, (R)-DHMA forms a hydrogen bond with both the backbone amide group and the side-chain hydroxyl group of Thr156. When the helix is not pushed down, (R)-DHMA forms a hydrogen bond only with the side-chain hydroxyl group of Thr156. When (R)-DHMA forms hydrogen bonds with both the backbone and side chain of Thr156, the residues of the binding site contract into the binding site, and this contraction pushes α-helix 4 downward (Fig. 6). This event is not observed in the simulations of pTsr binding to (S)-DHMA. (R)-DHMA forms a hydrogen bond to the amide group of Thr156 in 68.7% of the simulation snapshots, whereas (S)-DHMA forms a hydrogen bond to the amide group of Thr156 in only 0.1% of the simulation snapshots. Thus, all of our computational analyses agree in suggesting that (R)-DHMA should be a stronger chemoattractant than (S)-DHMA. Thus, we conclude that (R)-DHMA was the key contributor to the previously reported chemotaxis to racemic DHMA (7,10). The final MD simulation snapshots are provided in PDB format as part of the Supporting Material.
Experimental results
Resolution and determination of the absolute configuration of (R)-DHMA and (S)-DHMA
The resolution of (R)- and (S)-DHMA was achieved using HPLC with a chiral stationary-phase column. We screened commercially available racemic DHMA in several analytical columns with different chiral stationary phases and found that a CHIRALPAK ID-3 (150 × 4.6 mm I.D., 3μm) column provided good resolution when methanol/water (10:90) containing 0.1% formic acid was used as the mobile phase. On the analytical HPLC column, the enantiomers of DHMA had retention times of 4.0 and 5.9 min, and we assigned them the (R) and (S) configurations, respectively. To isolate milligram quantities of each enantiomer, the separation was carried out by HPLC using a preparative CHIRALPAK ID (250 × 20 mm I.D., 5 μm) column with methanol/water (20:80) containing 0.1% formic acid as the mobile phase (see Supporting Material for details).
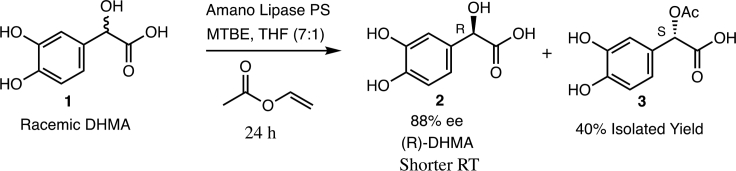
To assign the configuration of each isolated enantiomer, we used a lipase-catalyzed kinetic resolution technique described for secondary alcohols (44). We selected this method because of the structural similarity between DHMA and secondary alcohols whose configuration has been accurately assigned using this approach (45). Additionally, a significant difference in the size of the substituents attached to the secondary alcohol (phenyl versus carboxylic acid) greatly improves the reliability of the assignment. Thus, reaction of racemic DHMA with vinyl acetate and commercially available Amano Lipase PS for 24 h at room temperature provided acetate 3 in 41% isolated yield and enantiomerically enriched (R)-DHMA 2 in 45% yield (Fig. 8). Analysis of unreacted DHMA 2 by HPLC using the CHIRALPAK ID column indicated that it was highly enriched in the faster-eluting enantiomer (94:6 ratio, 88% enantiomeric excess (ee), RT = 6.9 min for the predominant isomer on the preparative column). Because the empirical rule of the lipase kinetic resolution method (44) predicts that the (S)-enantiomer of DHMA should react faster to form acetate 3, the unreacted enantiomer 2 was assigned the (R) configuration. This assignment agrees with the assignment reported for the kinetic resolution of structurally similar mandelic acid (45).
Figure 8.
Kinetic resolution of racemic DHMA. Reaction of racemic DHMA with vinyl acetate and commercially available Amano Lipase PS for 24 h at room temperature provided acetate 3 in 41% isolated yield and enantiomerically enriched (R)-DHMA 2 in 45% yield.
Tethered-cell assay
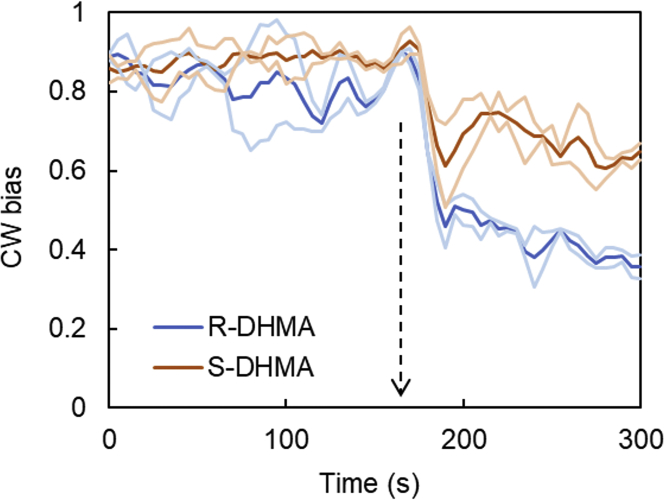
To determine the relative effectiveness of (R)- and (S)-DHMA as attractants, we employed the tethered-cell assay. The limited availability of the separated enantiomers required that we choose one optimum concentration, with 2 mM giving the clearest responses. The strain used in these experiments, PL138, was a derivative of strain RP437 (39) deleted for the cheRB genes (ΔcheRB) and containing a fliC allele that produces “sticky” flagella that facilitate spontaneous tethering to a glass surface. Tethered cells were first subjected to continuous flow of motility buffer in the microfluidic device, and the mean CW bias was analyzed over the initial 2 min. Cells used in the analysis were chosen to have a prestimulus CW bias of 0.8–0.9.
Cells were then exposed to a step stimulus by flowing 2 mM (R)- or (S)-DHMA in motility buffer into the chamber. The CW biases decreased immediately after introduction of either enantiomer and remained stable or continued to decrease slowly over the next 100 s. The response to (R)-DHMA was clearly stronger, with a stabilized CW bias of ∼0.4 (Fig. 9). The response to (S)-DHMA was also rapid, but the stabilized CW bias was ∼0.6 (Fig. 9), indicating a decreased attractant response compared with (R)-DHMA. The stronger response to (R)-DHMA is consistent with the predictions of the simulations.
Figure 9.
The tethered-cell response of an E. coli ΔcheRB strain to 2 mM (R)- and (S)-DHMA. The CW bias was calculated from the CW/CCW ratio of flagellar rotation. The response was averaged from two separate experiments, each including at least 12 motors. The faint lines represent the standard deviation. The arrow indicates the beginning of DHMA perfusion. To see this figure in color, go online.
The MMC assay
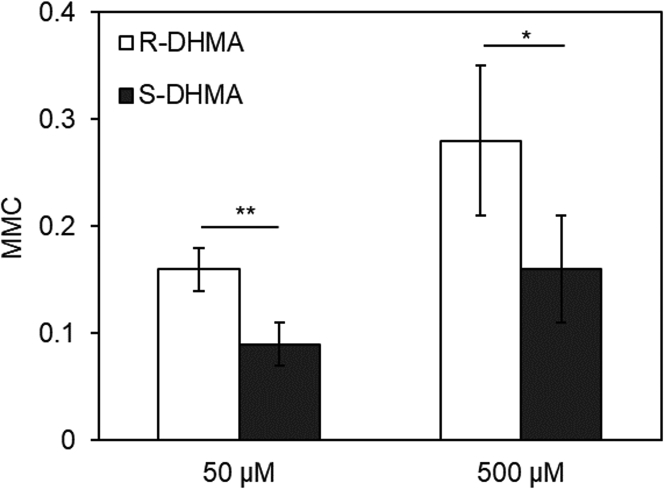
The responses to (R)- and (S)-DHMA were also measured in the MMC assay, which records the increased spreading of cells as a consequence of increased smooth swimming. Again, because of a limited supply of the two separated enantiomers, only data for the two concentrations that showed the clearest results are shown. In this assay, motile cells are introduced into the middle of the flow channel, and the distribution across the channel of the GFP-expressing RP437 cells 1.7 cm down the channel was determined by fluorescence microscopy. When the channel was perfused with the same motility buffer as that in which the cells were suspended, the MMC coefficient due to random motility was ∼0.08 (Fig. 10). When the channel was perfused with motility buffer containing (R)- or (S)-DHMA dissolved in motility buffer at 50 μM, the MMC value for (R)-DHMA doubled to ∼0.16, whereas the MMC value for S-DHMA remained at 0.08. When the experiment was repeated with 500 μM DHMA, the MMC value for (R)-DHMA rose to ∼0.27, and a smaller, but still significant, response of ∼0.16 was seen with (S)-DHMA. Thus, in agreement with the results from the tethered-cell assay, both (R)- and (S)-DHMA evoked a smooth-swimming (CCW-biased) response, but the response to (R)-DHMA appeared at a lower concentration and was of a greater magnitude.
Figure 10.
The chemotaxis response of wild-type E. coli to 50 and 500 μM of (R)-DHMA and (S)-DHMA. The response was averaged from three separate experiments. The error bars indicate the standard deviation. Statistical significance is indicated for the comparison of responses to (R)-DHMA and (S)-DHMA using the Student’s t-test at a significance level of p < 0.01 (∗∗) for 50 μM and of p < 0.04 (∗) for 500 μM DHMA.
Discussion
The study of protein-ligand interactions is key for prediction of protein function and drug design. With advances in computational power, molecular docking and MD simulation have become fundamental tools for investigating protein-ligand interactions at an atomic level (35,46, 47, 48, 49, 50). A number of docking methods have been released, which among others, include (51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66). Docking programs can predict docked poses with RMSDs averaging between 1.5 and 2 Å relative to the experimentally resolved structure (67). However, flexible receptor docking, particularly with flexibility in the receptor backbone, remains a major challenge for current docking algorithms (68). According to a recent review by Chaput and Mouawad on four popular docking programs (69), there is still a weakness in the ability of the scoring functions to detect the correct poses. Despite the associated additional computational cost, MD simulations combined with physics-based calculations of free energy can prove beneficial for detecting the lowest binding free energy mode of a protein-protein, peptide-protein, or compound-protein complex (15, 16, 17, 18, 19, 20,28,70, 71, 72, 73, 74, 75, 76). Furthermore, MD simulations are a useful tool for investigating allosteric changes in a receptor induced by ligand binding (14,31, 32, 33, 34,77, 78, 79, 80, 81, 82, 83, 84).
The catechol-containing compound that interacts with the E. coli Tsr chemoreceptor is DHMA (7). Initial studies used a racemic mixture of the (R) and (S) enantiomers, leaving open the question of which chiral form is active. Separation of the two enantiomers in amounts suitable for studies of chemotaxis is time-consuming and requires specialized chiral columns. To decide whether it was worthwhile to go to this effort, we first studied the binding of the two enantiomers to pTsr using a nearly exhaustive, highly accurate computational docking procedure combined with MD simulations and physics-based free energy calculations. This combination proved especially beneficial because short MD simulations introduced flexibility into pTsr and allowed refinement of the initial structures of the candidate binding modes before selecting the most energetically favorable ones for further investigation. The independent use of harmonic and spherical potentials constraining (R)- and (S)-DHMA within the binding pocket during the docking procedure enabled us to explore binding modes both away from (quartic) and in proximity to (harmonic) their initial position. Notably, the most energetically favorable binding mode of (R)-DHMA was derived using a quartic spherical potential (15), whereas the most energetically favorable binding mode of (S)-DHMA was derived using a harmonic spherical potential. A similar approach was used previously to determine how subtle differences between similar flavonoids binding to the aryl hydrocarbon receptor could explain their serving as agonists or antagonists (17,20). Multiple multi-ns MD simulations, using all-atom force fields (21) coupled with PCA and DCC analysis, allowed us to study in detail the complex formation and allosteric changes in pTsr caused by ligand binding.
In five replicate 100-ns simulations of serine in complex with pTsr, the average projected length deviated from the crystal structure by only 0.08 Å. Two supplementary 100-ns simulations of apo pTsr (with both resolved serine ligands removed), using the crystal structure of pTsr bound by serine (13) as the initial structure, show that the projected length approached zero as the simulations progressed. Thus, the simulated apo pTsr returned to the conformation of the apo pTsr crystal structure (13). These results demonstrate that our simulation system captures how differences in the binding site correlate with piston-like motions of α-helix 4. Thus, despite the fact that the entire Tsr protein was not used in the simulations, the modeled systems of (R)- and (S)-DHMA in complex with pTsr were sufficiently large to understand signaling induced by the two ligands.
The MD simulations and structural analyses strongly suggested that binding of (R)-DHMA causes a larger contraction of the binding pocket than binding of (S)-DHMA. This contraction is correlated with a larger downward piston-like motion of α-helix 4 than was seen with (S)-DHMA, leading to our prediction that (R)-DHMA is a stronger attractant than (S)-DHMA. (R)-DHMA forms weaker interactions to Arg69′ than (S)-DHMA. DHMA forms hydrogen bonds with both its chiral OH group and the negatively charged carboxyl group of pTsr. (S)-DHMA forms a single hydrogen bond to Arg69′ with its negatively charged carboxyl group and a cation-π interaction to Arg69′ with its aromatic ring. Because of the different orientations of (R)- and (S)-DHMA caused by their different chirality, (R)-DHMA forms hydrogen bonds to both the side chain and backbone of Thr156, whereas (S)-DHMA forms a hydrogen bond only with the side chain of Thr156. Hydrogen bonds between (R)-DHMA and Arg69′ as well as (S)-DHMA and Thr156 are occasionally mediated by water molecules, weakening their interaction.
We also observed correlated motions of residues within the binding site with residues in α-helix 4. Interactions of (R)-DHMA with both the side-chain and backbone atoms of Thr156 induce conformational changes within the binding pocket that displace α-helix 4 downward. The counterpart of Thr156 in the Tar chemoreceptor, Thr154, has been implicated in the piston-like movement of α-helix 4 in response to the binding of the attractant L-aspartate (14), and Thr156 of Tsr is essential for attractant responses to DHMA (7). Our computational prediction that (R)-DHMA should be a stronger chemoattractant than (S)-DHMA encouraged us to undertake the separation of the two enantiomers and to test their efficacy as chemoattractants.
Separation of (R)- and (S)-DHMA was achieved using HPLC with a chiral stationary-phase column (semipreparative CHIRALPAK ID). Analysis of the two isolated enantiomers demonstrated that the (R)-DHMA was >98% ee and the (S)-DHMA was in 94% ee. In this way, sufficient amounts of the two highly enriched enantiomers were generated for use in chemotaxis assays.
In the tethered-cell assay, addition of 2 mM (R)-DHMA to ΔcheRB cells caused a decrease in the CW bias from 80 to 40% that persisted for at least 100 s. Addition of 2 mM (S)-DHMA decreased the CW bias from 80 to 60%, again for a period of at least 100 s. Thus, although both enantiomers at an equivalent concentration increased CCW rotation, (R)-DHMA had a larger effect. In the MMC assay, exposure of wild-type (RP437) cells to (R)-DHMA produced an increase in smooth swimming at concentrations of 50 and 500 μM, with a larger increase seen with 500 μM. In comparison, (S)-DHMA did not increase smooth swimming at 50 μM, and at 500 μM, it was only as effective as (R)-DHMA at 50 μM. Thus, two different assays supported the prediction from the simulations that both enantiomers of DHMA might serve as chemoattractants but that (R)-DHMA would be more potent than (S)-DHMA.
DHMA is generated in the gut by the resident microbiota (10), presumably as a racemic mixture of the two enantiomers, although the chiral form(s) in the gut have not been determined experimentally. The racemic mixture in vitro loses its potency as an attractant at higher concentrations (7). This observation is consistent with the report that DHMA at high concentrations serves as a repellent (11). In a heterodimeric Tsr designed to bind DHMA at only one of its two ligand-binding sites, DHMA served as an attractant at all concentrations tested (7). In future studies, it will be interesting to determine how binding at the second site cancels out the attractant response and whether one enantiomer or both of them are responsible for this effect. It also remains to be determined how attractant and repellent responses to DHMA may serve to control the migration of E. coli, and potentially other motile gut bacteria, in the intestinal tract.
Our study demonstrates how refined computational docking and simulation studies can be combined with experiments to investigate biological phenomena in which subtle differences between ligands may lead to diverse binding-mediated outcomes. The computational results provided the impetus to carry out the separation of the two chiral forms of DHMA. After the two enantiomers were separated, they could be used to assay the function of each enantiomer as a chemoattractant. In this instance, the predictions from computational docking and simulations were supported by the experiments. Our study could serve as a model for additional collaborative efforts to address questions involving the different activities of closely related chemical compounds as signaling molecules or enzyme substrates.
Author Contributions
A.J., M.D.M., and P.T. conceived the project. A.J., M.D.M., and P.T. supervised the project. All authors contributed to the analysis and writing of the manuscript.
Acknowledgments
All MD simulations and free energy calculations were performed on the Ada supercomputing cluster at the Texas A&M High Performance Research Computing Facility and in-house facilities available to P.T. Support from Texas A&M University (Natural Products LINCHPIN Laboratory) and Baylor University (CPRIT Synthesis & Drug Lead Discovery Laboratory) where this work was conducted, is gratefully acknowledged.
This study is supported by the Texas A&M University Graduate Diversity Fellowship from the TAMU Office of Graduate and Professional Studies awarded to A.A.O. and by start-up funding from the Artie McFerrin Department of Chemical Engineering of Texas A&M awarded to P.T. P.P.L. acknowledges support from the National Institute of General Medical Sciences (R01-GM123085) and the DOD United States Army ACC-APG-RTP Division (W911NF1810353).
Editor: Julie Biteen.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.11.3382.
Contributor Information
Michael D. Manson, Email: mike@bio.tamu.edu.
Phanourios Tamamis, Email: tamamis@tamu.edu.
Supporting Citations
References (85, 86, 87, 88, 89, 90, 91, 92, 93, 94) appear in the Supporting Material.
Supporting Material
Coordinates of (R)- and (S)-DHMA in Complex with pTsr from the 100-ns MD Simulations
References
- 1.Surette M.G., Miller M.B., Bassler B.L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hegde M., Englert D.L., Jayaraman A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 2011;193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller S.T., Xavier K.B., Hughson F.M. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Springer M.S., Goy M.F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc. Natl. Acad. Sci. USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laganenka L., Colin R., Sourjik V. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat. Commun. 2016;7:12984. doi: 10.1038/ncomms12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jani S., Seely A.L., Manson M.D. Chemotaxis to self-generated AI-2 promotes biofilm formation in Escherichia coli. Microbiology. 2017;163:1778–1790. doi: 10.1099/mic.0.000567. [DOI] [PubMed] [Google Scholar]
- 7.Pasupuleti S., Sule N., Manson M.D. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4-dihydroxymandelic acid. J. Bacteriol. 2014;196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rankin L.D., Bodenmiller D.M., Spiro S. Escherichia coli NsrR regulates a pathway for the oxidation of 3-nitrotyramine to 4-hydroxy-3-nitrophenylacetate. J. Bacteriol. 2008;190:6170–6177. doi: 10.1128/JB.00508-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenhofer G., Kopin I.J., Goldstein D.S. Catecholamine metabolism: a contemporary view with implications for physiology and medicine. Pharmacol. Rev. 2004;56:331–349. doi: 10.1124/pr.56.3.1. [DOI] [PubMed] [Google Scholar]
- 10.Sule N., Pasupuleti S., Jayaraman A. The norepinephrine metabolite 3,4-dihydroxymandelic acid is produced by the commensal microbiota and promotes chemotaxis and virulence gene expression in enterohemorrhagic Escherichia coli. Infect. Immun. 2017;85 doi: 10.1128/IAI.00431-17. e00431-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopes J.G., Sourjik V. Chemotaxis of Escherichia coli to major hormones and polyamines present in human gut. ISME J. 2018;12:2736–2747. doi: 10.1038/s41396-018-0227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meirieu O., Pairet M., Ruckebusch M. Local release of monoamines in the gastrointestinal tract: an in vivo study in rabbits. Life Sci. 1986;38:827–834. doi: 10.1016/0024-3205(86)90599-0. [DOI] [PubMed] [Google Scholar]
- 13.Tajima H., Imada K., Kawagishi I. Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J. Biol. Chem. 2011;286:42200–42210. doi: 10.1074/jbc.M111.221887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu D., Ma X., Lai L. Both piston-like and rotational motions are present in bacterial chemoreceptor signaling. Sci. Rep. 2015;5:8640. doi: 10.1038/srep08640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orr A.A., Jayaraman A., Tamamis P. Molecular modeling of chemoreceptor:ligand interactions. Methods. Mol. Biol. 2018;1729:353–372. doi: 10.1007/978-1-4939-7577-8_28. [DOI] [PubMed] [Google Scholar]
- 16.Jin U.H., Cheng Y., Safe S. Short chain fatty acids enhance aryl hydro carbon (Ah) responsiveness in mouse colonocytes and Caco- 2 human colon cancer cells. Sci. Rep. 2017;7:10163. doi: 10.1038/s41598-017-10824-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin U.H., Park H., Safe S. Structure-dependent modulation of aryl hydrocarbon receptor-mediated activities by flavonoids. Toxicol. Sci. 2018;164:205–217. doi: 10.1093/toxsci/kfy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohan R.R., Wilson M., Morikis D. Virtual screening of chemical compounds for discovery of complement C3 ligands. ACS Omega. 2018;3:6427–6438. doi: 10.1021/acsomega.8b00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon K., Chen C.C., Safe S. Activation of COUP-TFI by a novel diindolylmethane derivative. Cells. 2019;8:220. doi: 10.3390/cells8030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park H., Jin U.H., Safe S. Isoflavones as Ah receptor agonists in colon-derived cell lines: structure-activity relationships. Chem. Res. Toxicol. 2019;32:2353–2364. doi: 10.1021/acs.chemrestox.9b00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks B.R., Brooks C.L., III, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seeber M., Felline A., Fanelli F. Wordom: a user-friendly program for the analysis of molecular structures, trajectories, and free energy surfaces. J. Comput. Chem. 2011;32:1183–1194. doi: 10.1002/jcc.21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes J.M., Archontis G. MM-GB(PB)SA calculations of protein-ligand binding free energies. In: Wang L., editor. Molecular Dynamics - Studies of Synthetic and Biological Macromolecules. InTech, Southern Illinois University Carbondale; 2012. pp. 171–190. [Google Scholar]
- 24.Tamamis P., Morikis D., Archontis G. Species specificity of the complement inhibitor compstatin investigated by all-atom molecular dynamics simulations. Proteins. 2010;78:2655–2667. doi: 10.1002/prot.22780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamamis P., Pierou P., Archontis G. Design of a modified mouse protein with ligand binding properties of its human analog by molecular dynamics simulations: the case of C3 inhibition by compstatin. Proteins. 2011;79:3166–3179. doi: 10.1002/prot.23149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamamis P., López de Victoria A., Archontis G. Molecular dynamics in drug design: new generations of compstatin analogs. Chem. Biol. Drug Des. 2012;79:703–718. doi: 10.1111/j.1747-0285.2012.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieslich C.A., Tamamis P., Morikis D. Exploring protein-protein and protein-ligand interactions in the immune system using molecular dynamics and continuum electrostatics. Curr. Phys. Chem. 2012;2:324–343. [Google Scholar]
- 28.Tamamis P., Floudas C.A. Elucidating a key component of cancer metastasis: CXCL12 (SDF-1α) binding to CXCR4. J. Chem. Inf. Model. 2014;54:1174–1188. doi: 10.1021/ci500069y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samanta D., Borbat P.P., Crane B.R. Bacterial chemoreceptor dynamics correlate with activity state and are coupled over long distances. Proc. Natl. Acad. Sci. USA. 2015;112:2455–2460. doi: 10.1073/pnas.1414155112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitanovic S., Ames P., Parkinson J.S. Mutational analysis of the control cable that mediates transmembrane signaling in the Escherichia coli serine chemoreceptor. J. Bacteriol. 2011;193:5062–5072. doi: 10.1128/JB.05683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassidy C.K., Himes B.A., Zhang P. CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling. eLife. 2015;4:e08419. doi: 10.7554/eLife.08419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papachristos K., Muench S.P., Paci E. Characterization of the flexibility of the peripheral stalk of prokaryotic rotary A-ATPases by atomistic simulations. Proteins. 2016;84:1203–1212. doi: 10.1002/prot.25066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng H., Dai Z., Lai L. Molecular mechanism of 15-lipoxygenase allosteric activation and inhibition. Phys. Chem. Chem. Phys. 2018;20:14785–14795. doi: 10.1039/c7cp08586a. [DOI] [PubMed] [Google Scholar]
- 34.López de Victoria A., Tamamis P., Morikis D. Insights into the structure, correlated motions, and electrostatic properties of two HIV-1 gp120 V3 loops. PLoS One. 2012;7:e49925. doi: 10.1371/journal.pone.0049925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B., Francis J., Feig M. Long-range signaling in MutS and MSH homologs via switching of dynamic communication pathways. PLoS Comput. Biol. 2016;12:e1005159. doi: 10.1371/journal.pcbi.1005159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCammon A.J., Harvey S.C. Cambridge University Press; Cambridge, UK: 1986. Dynamics of Proteins and Nucleic Acids. [Google Scholar]
- 37.Brooks C.L., III, Karplus M., Pettitt B.M. John Wiley and Sons; New York: 1988. Proteins: A Theoretical Perspective of Dynamics, Structure and Thermodynamics. [Google Scholar]
- 38.Berg H.C., Turner L. Torque generated by the flagellar motor of Escherichia coli. Biophys. J. 1993;65:2201–2216. doi: 10.1016/S0006-3495(93)81278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkinson J.S., Houts S.E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg H.C., Block S.M. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J. Gen. Microbiol. 1984;130:2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- 41.Chawla R., Ford K.M., Lele P.P. Torque, but not FliL, regulates mechanosensitive flagellar motor-function. Sci. Rep. 2017;7:5565. doi: 10.1038/s41598-017-05521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen M.C., Palmer R.J., Jr., Molin S. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology. 2001;147:1383–1391. doi: 10.1099/00221287-147-5-1383. [DOI] [PubMed] [Google Scholar]
- 43.Alberts B., Johnson A., Walter P. Fifth Edition. Garland Science; London, UK: 2008. Molecular Biology of the Cell. [Google Scholar]
- 44.Jing Q., Kazlauskas R.J. Determination of absolute configuration of secondary alcohols using lipase-catalyzed kinetic resolutions. Chirality. 2008;20:724–735. doi: 10.1002/chir.20543. [DOI] [PubMed] [Google Scholar]
- 45.Dabkowska D., Szewczyk K.W. Influence of temperature on the activity and enantioselectivity of Burkholderia cepacia lipase in the kinetic resolution of mandelic acid enantiomers. Biochem. Eng. J. 2009;46:147–153. [Google Scholar]
- 46.Naqvi A.A.T., Mohammad T., Hassan M.I. Advancements in docking and molecular dynamics simulations towards ligand-receptor interactions and structure-function relationships. Curr. Top. Med. Chem. 2018;18:1755–1768. doi: 10.2174/1568026618666181025114157. [DOI] [PubMed] [Google Scholar]
- 47.Burger V.M., Vandervelde A., Stultz C.M. Hidden states within disordered regions of the CcdA antitoxin protein. J. Am. Chem. Soc. 2017;139:2693–2701. doi: 10.1021/jacs.6b11450. [DOI] [PubMed] [Google Scholar]
- 48.Burger V.M., Arenas D.J., Stultz C.M. A structure-free method for quantifying conformational flexibility in proteins. Sci. Rep. 2016;6:29040. doi: 10.1038/srep29040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips C.M., Nerenberg P.S., Stultz C.M. Physical basis of metal-binding specificity in Escherichia coli NikR. J. Am. Chem. Soc. 2009;131:10220–10228. doi: 10.1021/ja9026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherjee S., Feig M. Conformational change in MSH2-MSH6 upon binding DNA coupled to ATPase activity. Biophys. J. 2009;96:L63–L65. doi: 10.1016/j.bpj.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grosdidier A., Zoete V., Michielin O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011;32:2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 53.Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pevzner Y., Frugier E., Woodcock H.L. Fragment-based docking: development of the CHARMMing Web user interface as a platform for computer-aided drug design. J. Chem. Inf. Model. 2014;54:2612–2620. doi: 10.1021/ci500322k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones G., Willett P., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 56.Österberg F., Morris G.M., Goodsell D.S. Automated docking to multiple target structures: incorporation of protein mobility and structural water heterogeneity in AutoDock. Proteins. 2002;46:34–40. doi: 10.1002/prot.10028. [DOI] [PubMed] [Google Scholar]
- 57.Repasky M.P., Shelley M., Friesner R.A. Flexible ligand docking with Glide. Curr. Protoc. Bioinformatics. 2007;Chapter 8:Unit 8.12. doi: 10.1002/0471250953.bi0812s18. [DOI] [PubMed] [Google Scholar]
- 58.Zhang N., Zhao H. Enriching screening libraries with bioactive fragment space. Bioorg. Med. Chem. Lett. 2016;26:3594–3597. doi: 10.1016/j.bmcl.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 59.Ruiz-Carmona S., Alvarez-Garcia D., Morley S.D. rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput. Biol. 2014;10:e1003571. doi: 10.1371/journal.pcbi.1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morris G.M., Huey R., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allen W.J., Balius T.E., Rizzo R.C. DOCK 6: impact of new features and current docking performance. J. Comput. Chem. 2015;36:1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jain A.N. Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput. Aided Mol. Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 63.Majeux N., Scarsi M., Caflisch A. Efficient electrostatic solvation model for protein-fragment docking. Proteins. 2001;42:256–268. doi: 10.1002/1097-0134(20010201)42:2<256::aid-prot130>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 64.Majeux N., Scarsi M., Caflisch A. Exhaustive docking of molecular fragments with electrostatic solvation. Proteins. 1999;37:88–105. [PubMed] [Google Scholar]
- 65.Lagarde N., Carbone A., Sacquin-Mora S. Hidden partners: using cross-docking calculations to predict binding sites for proteins with multiple interactions. Proteins. 2018;86:723–737. doi: 10.1002/prot.25506. [DOI] [PubMed] [Google Scholar]
- 66.Kynast P., Derreumaux P., Strodel B. Evaluation of the coarse-grained OPEP force field for protein-protein docking. BMC Biophys. 2016;9:4. doi: 10.1186/s13628-016-0029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z., Sun H., Hou T. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: the prediction accuracy of sampling power and scoring power. Phys. Chem. Chem. Phys. 2016;18:12964–12975. doi: 10.1039/c6cp01555g. [DOI] [PubMed] [Google Scholar]
- 68.Pagadala N.S., Syed K., Tuszynski J. Software for molecular docking: a review. Biophys. Rev. 2017;9:91–102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaput L., Mouawad L. Efficient conformational sampling and weak scoring in docking programs? Strategy of the wisdom of crowds. J. Cheminform. 2017;9:37. doi: 10.1186/s13321-017-0227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Athanasiou C., Vasilakaki S., Cournia Z. Using physics-based pose predictions and free energy perturbation calculations to predict binding poses and relative binding affinities for FXR ligands in the D3R Grand Challenge 2. J. Comput. Aided Mol. Des. 2018;32:21–44. doi: 10.1007/s10822-017-0075-9. [DOI] [PubMed] [Google Scholar]
- 71.Gkeka P., Eleftheratos S., Cournia Z. Free energy calculations reveal the origin of binding preference for aminoadamantane blockers of influenza A/M2TM pore. J. Chem. Theory Comput. 2013;9:1272–1281. doi: 10.1021/ct300899n. [DOI] [PubMed] [Google Scholar]
- 72.Radom F., Plückthun A., Paci E. Assessment of ab initio models of protein complexes by molecular dynamics. PLoS Comput. Biol. 2018;14:e1006182. doi: 10.1371/journal.pcbi.1006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamamis P., Floudas C.A. Elucidating a key anti-HIV-1 and cancer-associated axis: the structure of CCL5 (Rantes) in complex with CCR5. Sci. Rep. 2014;4:5447. doi: 10.1038/srep05447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tamamis P., Floudas C.A. Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. PLoS One. 2014;9:e95767. doi: 10.1371/journal.pone.0095767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamamis P., Kieslich C.A., Archontis G. Insights into the mechanism of C5aR inhibition by PMX53 via implicit solvent molecular dynamics simulations and docking. BMC Biophys. 2014;7:5. doi: 10.1186/2046-1682-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamamis P., Floudas C.A. Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop. Biophys. J. 2013;105:1502–1514. doi: 10.1016/j.bpj.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Renault P., Louet M., Floquet N. Molecular dynamics simulations of the allosteric modulation of the adenosine A2A receptor by a mini-G protein. Sci. Rep. 2019;9:5495. doi: 10.1038/s41598-019-41980-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dror R.O., Green H.F., Shaw D.E. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;503:295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- 79.Lückmann M., Trauelsen M., Frimurer T.M. Molecular dynamics-guided discovery of an ago-allosteric modulator for GPR40/FFAR1. Proc. Natl. Acad. Sci. USA. 2019;116:7123–7128. doi: 10.1073/pnas.1811066116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Denisov I.G., Grinkova Y.V., Sligar S.G. Allosteric interactions in human cytochrome P450 CYP3A4: the role of phenylalanine 213. Biochemistry. 2019;58:1411–1422. doi: 10.1021/acs.biochem.8b01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thayer K.M., Lakhani B., Beveridge D.L. Molecular dynamics-markov state model of protein ligand binding and allostery in CRIB-PDZ: conformational selection and induced fit. J. Phys. Chem. B. 2017;121:5509–5514. doi: 10.1021/acs.jpcb.7b02083. [DOI] [PubMed] [Google Scholar]
- 82.Stetz G., Verkhivker G.M. Dancing through life: molecular dynamics simulations and network-centric modeling of allosteric mechanisms in Hsp70 and Hsp110 chaperone proteins. PLoS One. 2015;10:e0143752. doi: 10.1371/journal.pone.0143752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kasahara K., Fukuda I., Nakamura H. A novel approach of dynamic cross correlation analysis on molecular dynamics simulations and its application to Ets1 dimer-DNA complex. PLoS One. 2014;9:e112419. doi: 10.1371/journal.pone.0112419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gkeka P., Papafotika A., Cournia Z. Exploring a non-ATP pocket for potential allosteric modulation of PI3Kα. J. Phys. Chem. B. 2015;119:1002–1016. doi: 10.1021/jp506423e. [DOI] [PubMed] [Google Scholar]
- 85.Vainio M.J., Puranen J.S., Johnson M.S. ShaEP: molecular overlay based on shape and electrostatic potential. J. Chem. Inf. Model. 2009;49:492–502. doi: 10.1021/ci800315d. [DOI] [PubMed] [Google Scholar]
- 86.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vanommeslaeghe K., Hatcher E., Mackerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huggins D.J., Tidor B. Systematic placement of structural water molecules for improved scoring of protein-ligand interactions. Protein Eng. Des. Sel. 2011;24:777–789. doi: 10.1093/protein/gzr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Orr A.A., Wördehoff M.M., Tamamis P. Uncovering the binding and specificity of β-Wrapins for Amyloid-β and α-Synuclein. J. Phys. Chem. B. 2016;120:12781–12794. doi: 10.1021/acs.jpcb.6b08485. [DOI] [PubMed] [Google Scholar]
- 90.Orr A.A., Shaykhalishahi H., Tamamis P. Elucidating the multi-targeted anti-amyloid activity and enhanced islet amyloid polypeptide binding of β-wrapins. Comput. Chem. Eng. 2018;116:322–332. doi: 10.1016/j.compchemeng.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Im W., Lee M.S., Brooks C.L., III Generalized born model with a simple smoothing function. J. Comput. Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 92.Knight J.L., Brooks C.L., III Surveying implicit solvent models for estimating small molecule absolute hydration free energies. J. Comput. Chem. 2011;32:2909–2923. doi: 10.1002/jcc.21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin L.N., Li J., Weis R.M. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry. 1994;33:6564–6570. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- 94.Iwama T., Kawagishi I., Imae Y. In vivo sulfhydryl modification of the ligand-binding site of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 1995;177:2218–2221. doi: 10.1128/jb.177.8.2218-2221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coordinates of (R)- and (S)-DHMA in Complex with pTsr from the 100-ns MD Simulations