Abstract
Background:
MicroRNAs are mentioned as a small non-coding RNAs groups and aberrant miRNA expression was found in hepatocellular carcinoma (HCC) patients.
Aim:
To evaluate role of plasma MicroRNA-215 as a diagnostic tool in HCC patients.
Methods:
A prospective study included 195 subjects: healthy controls (group I), cirrhotic patients (group II), and patients with HCC (group III). Clinical examination, radiological and laboratory investigations which included quantification of miR-215 by Real-time qPCR were done for all cases.
Results:
Spearman’s rank correlation revealed that in HCC group, there was a negative correlation between MiRNA-215 and serum AFP levels and focal size lesion (cm) (rs = -0.72, - 0.94 respectively, p<0.001). Receiver operating characteristics analysis for discrimination between cirrhosis and HCC groups regarding microRNA-215 displayed 78.3% sensitivity, 88.0% specificity at cutoff value of ≤ 1.90. Area under the curve (AUC) was 0.87 (p< 0.001). As regards AFP, it had a sensitivity of 81.7%, a specificity of 66.7 at cutoff value of ≥ 11.50 (ng/mL).
Conclusions:
Plasma level of miR-215 may be a promising biomarker in HCC diagnosis. Moreover, if miR-215 combined with AFP, it can be used as a diagnostic biomarker, for early detection of HCC.
Key Words: hepatocellular carcinoma (HCC), chronic hepatitis C virus (chronic HCV), MicroRNA-215 (miR-215)
Introduction
Hepatocellular carcinoma (HCC) is one of the common cancers worldwide as it represents the 5th one in men and the 7th one in women (Bosetti et al., 2014) and the 3rd one of mortality related tumors. Most patients with hepatic cirrhosis may develop HCC and finally death may occur (Salgia and Singal, 2014). The prognosis of HCC still being unsatisfactory in spite of development in surgical and non-surgical treatments compared to other cancer tumors (W Pang and TP Poon, 2012).
Number of HCV patients worldwide is in the range of 170 to 200 million. Genotype 4 is the most commonly HCV genotype in Egypt and it represents about 90% of cases, research is not enough, because its localization is restricted to Africa and Middle East (Shelbaya et al., 2015). Significant numbers of infected patients are at high risk of developing liver cirrhosis and subsequently hepatocellular carcinoma. About 10% to 15% of HCC cases are resecTablebecause of the late diagnosis.
MicroRNAs (miRNAs) are one of evolutionarily conserved small non-coding RNAs which make a contribution in the gene expression regulation and protein translation, and incriminated in differentiation, cell evolution, and occurrence of diverse types of cancers. miRNAs are released to extracellular areas, and they are extremely constant in body fluids, which includes plasma or serum, in which they are packaged into several microparticles or related with RNA-binding proteins because of passive leakage that caused by the deceased cell, and also they are actively secreted through exosomes from cells (Aravalli et al., 2013; Li et al., 2015). Different researches have confirmed that circulating miRNAs could be considered as a biomarker for HCC prognosis and diagnosis (Zhang et al., 2015).
Sun et al., (2013) reported that, deregulation of various miRNAs in exclusive types of cancer such as HCC.
It is common to found down regulation of miRNAs subsets as some types of them may act as assumed tumor suppressor genes in HCC. Tumor suppressive miRNAs restorations blocks cell cycle, increased apoptosis, decrease tumor angiogenesis and metastasis by stopping migration and invasion. Of these miRNAs, miR-122 miR-199, and miR-215 appear to be particularly important in HCC (Hou et al., 2011; Ashmawy et al., 2017). MiRNAs control expression of many target genes in HCC and their profiling reveals molecular mechanisms of pathogenesis and hidden visions into detection and treatment of HCC (Song He, 2015). Our aim was to evaluate role of plasma miR-215 as a diagnostic tool in Egyptian patients with HCC.
Materials and Methods
Our research was done in hepatology and gastroenterology department at the National Liver Institute, Menoufia University in the period from November 2016 till December 2017. It included 135 patients and 60 healthy subjects serving as control group. We have 3 groups:
Group I (Control)
This group included 60 apparently healthy individuals. They were 32 males and 28 females, whose ages ranged from 39 to 67 years old (mean ±SD= 51.67 ± 6.40 years).
Group II (Cirrhosis)
This group included 75 patients with liver cirrhosis due to chronic HCV infection. They were 41 males and 34 females whose ages ranged from 41 to 68 years old (mean ±SD= 54±6.73 years).
Group III (HCC)
Sixty patients with HCC were included. They were 35 males and 25 females, whose ages ranged from 41 to 70 years old (mean ±SD= 53.97±6.15 years).
All studied subjects provided informed written consent and the study approval by local ethics committee of National Liver Institute, Menoufia University was obtained.
Exclusion criteria
All patients with the following: hepatitis B virus (HBV) or HIV, immunosuppression, organ transplantation, autoimmune disease, Schistosomiasis, all other malignancies and lastly patients who were under chemotherapy and antiviral treatment were excluded.
All individuals were subjected to complete clinical history and examination, laboratory investigations and abdominal ultrasonography.
Radiological study
Abdominal ultrasound was done to assess liver size, coarseness of parenchyma, liver surface nodularity, lymph nodes enlargement and size, spleen size (which if enlarged can suggest portal hypertension), patency and flow of veins and arteries and focal lesions (which if present can suggest hepatocellular carcinoma).
All HCC patients were diagnosed by characteristic vascular enhancement pattern detected by multislice triphasic spiral CT scan or MRI according to established diagnostic criteria (Galle et al., 2018).
Laboratory Investigations
- Complete blood picture was determined by the Sysmex® Automated Hematology Analyzer KX-21N (Sysmex Corporation, Kobe 651-0073, Japan). Liver tests: aspartate transaminase (AST) and alanine transaminase (ALT), serum albumin, total and direct bilirubin, alkaline phosphatase and INR were measured. Creatinine level in serum was measured in (mg/dl). The analysis of serum alpha fetoprotein (AFP) (ng/ml) was done by IMMULITE 1000 system supplied by Siemens kit (SIEMENS Medical Solutions Diagnostics, USA). Chronic HCV infection was diagnosed by HCV antibodies detection and also HCV RNA level detection by real time PCR. Anti-HCV antibodies were founded by 3rd generation enzyme immunoassay (Ortho HCV version 3.0 ELISA; Ortho- Clinical Diagnostics INC., Raritan. NJ, USA). Quantification of HCV RNA level was performed by COBAS Taqman 84 (Roche) real time HCV RNA assay with lower detection limit 15 IU/ml.
- Model of End-Stage Liver Disease (MELD): It was measured according to next formula: MELD =3.78×ln [serum bilirubin (mg/dL)] + 11.2×ln [INR] + 9.57×ln [serum creatinine (mg/dL)] + 6.43 (Malinchoc et al., 2000).
MiRNA Quantification by Real-Time qPCR (RT-qPCR): Blood samples were collected in EDTA tubes, centrifuged at 4,000 rpm for 10min at 4oC, plasma separated, and stored at −70oC.
RNA Extraction and cDNA Synthesis
Total RNA was taken from 200 µl plasma by the miRNeasy extraction kit (Qiagen, Germany). RNA purity was evaluated by the RNA concentration and quantified by Nano Drop Spectrophotometer (NanoDrop ND-1000, United States). Reverse transcription was done for cDNA synthesis using miRNeasy Plasma Reverse Transcription Kit (Qiagen, Germany) regarding to manufacturer’s instructions.
Amplification and Quantification
The expressions miR-215 was evaluated by RT-qPCR analysis regarding to manufacturer’s protocol. The housekeeping miRNAU6 (RNU6) was used as the endogenous control. For qRT-PCR, 2 μL of diluted reverse transcription products was mixed with 12.5 μL SYBR® Green Real-time PCR Master Mix (Qiagen, Germany), 2.5 μL of forward primer, 2.5 μL reverse primers and 5 μL RNase-free water (in syper green box) in a final volume of 25 μL in each strip tube according to the manufacturer’s instructions. The reaction was performed on AB Applied Biosystem Real-Time System (Model: 7500 system, USA), with the following reaction conditions: 95°C for 15 min, followed by 40 cycles at 94°C for 15 sec, 55 °C for 30 sec, and 70°C for 34 sec.
Array plate for PCR was full of reverse and forward miRNA-specific primers.
The primer sequences were used for miR-215: forward primer:

Statistical analysis
Results were analyzed by using statistical package of social sciences (SPSS 22.0, IBM/SPSS Inc., Chicago, IL). The mean ± standard deviation (SD) was used for normally distributed data whereas interquartile range (IQR) and median was for skewed data. Categorical data was demonstrated as frequency with percentage. For continuous variables, ANOVA test was used to compare between several groups when homogeneity and normality assumptions were met, instead, its non-parametric equivalent Kruskal-Wallis test was applied upon violation. For multiple pairwise comparisons, an appropriate Post Hoc test was used as Tukey-HSD test with significant ANOVA or Bonferroni test with significant Kruskal-Wallis test. The Chi-square (X2) test was used to compare categorical variables. Binary logistic regression analysis was used for deriving the best fitting models that describe relation between presence of HCC and combined biomarkers measurement. We used Receiver operating characteristic analysis for assessment of diagnostic performance of plasma miR-215 in HCC detection. The p-values < 0.05 were considered as significant ones.
Results
This study included 135 patients who were selected from outpatient clinics and inpatient unit of hepatology and gastroenterology department of the National Liver Institute, Menoufia University and were classified into 2 groups, 75 cirrhotic patients (cirrhosis group) and 60 patients with HCC (HCC group) and control group which included 60 healthy volunteers with matched age and gender.
Regarding age and gender distribution among studied groups as shown in Table 1, there was no statistically significant difference among those group (p= 0.071, 0.848 respectively).
Table 1.
Age and Gender Distribution among Studied Groups
| Age (years) | GI | GII | GIII | significant test | p-value |
|---|---|---|---|---|---|
| Control (n = 60) | Cirrhosis (n = 75) | HCC (n = 60) | |||
| Median (IQR) | 51.0 (8.0) | 53.0 (9.0) | 54.0 (8.0) | F = 2.68a | 0.071 NS |
| Mean ± SD | 51.67 ± 6.40 | 54.00 ± 6.73 | 53.97 ± 6.15 | ||
| Range (min-max) | 39.00 - 67.00 | 41.00 - 68.00 | 41.00 - 70.00 | ||
| Gender [n (%)] | |||||
| Male | 32 (53.3) | 41 (54.7) | 35 (58.3) | x 2 = 0.33b | 0.071 NS |
| Female | 28 (46.7) | 34 (45.3) | 25 (41.7) |
NS, Non significant at p-value ≥ 0.05; IQR, Interquartile range; n, Number; SD, Standard deviation; a, ANOVA test; b, Chi-Square test
Descriptive and analytical statistics of liver tests and serum creatinine in all groups were represented in Table 2. There was significant difference between cirrhosis and HCC groups as regards ALP (p=0.004), ALT, AST and GGT (p< 0.001).
Table 2.
Laboratory Investigations among the Studied Groups
| Parameters | GI | GII | GIII | Test of significance | Post hoc test |
|---|---|---|---|---|---|
| Control (n = 60) | Cirrhosis (n = 75) | HCC (n = 60) | (Kruskal- Wallis) | ||
| ALT (U/L) | |||||
| Median (IQR) | 16.00 (7.75) | 30.00 (39.0) | 62.00 (29.75) | x 2 = 93.81 | p1< 0.001HS |
| Mean ± SD | 17.08 ± 5.55 | 41.72 ± 35.02 | 63.45 ± 22.33 | p-value | p2< 0.001HS |
| Range (min-max) | 8.00 - 33.00 | 8.00 - 200.0 | 22.00 - 119.00 | <0.001HS | p3< 0.001HS |
| AST (U/L) | |||||
| Median (IQR) | 19.00 (4.75) | 37.00 (47.00) | 63.00 (26.00) | x 2 = 86.65 | p1< 0.001HS |
| Mean ± SD | 18.58 ± 3.77 | 49.24 ± 36.75 | 65.05 ± 26.72 | p-value | p2< 0.001HS |
| Range (min-max) | 10.00 - 30.00 | 9.00 - 180.00 | 20.00 - 170.00 | <0.001HS | p3< 0.001HS |
| ALP (U/L) | |||||
| Median (IQR) | 63.50 (18.75) | 98.00 (33.00) | 121.50 (52.75) | x 2= 96.29 | p1< 0.001HS |
| Mean ± SD | 61.47 ± 10.51 | 98.65 ± 26.85 | 122.53 ± 35.03 | p-value | p2< 0.001HS |
| Range (min-max) | 41.00 - 80.00 | 46.00 - 148.00 | 55.00 - 197.00 | <0.001HS | p3= 0.004 HS |
| g GT (U/L) | |||||
| Median (IQR) | 20.00 (7.00) | 42.00 (39.00) | 80.00 (66.00) | x 2 = 65.39 | p1< 0.001HS |
| Mean ± SD | 19.50 ± 5.97 | 43.80 ± 21.71 | 76.35 ± 40.06 | p-value | p2< 0.001HS |
| Range (min-max) | 9.00 - 39.00 | 15.00 - 105.00 | 18.00 - 169.00 | <0.001HS | p3< 0.001HS |
| Total bilirubin (mg/dL) | |||||
| Median (IQR) | 0.54 (0.32) | 1.30 (2.10) | 1.28 (1.19) | x 2 = 71.08 | p1< 0.001HS |
| Mean ± SD | 0.57 ± 0.19 | 1.87 ± 1.55 | 1.91 ± 1.65 | p-value | p2< 0.001HS |
| Range (min-max) | 0.20 - 0.90 | 0.20 - 7.27 | 0.60 - 7.50 | <0.001HS | p3= 0.360 NS |
| Direct bilirubin (mg/dL) | |||||
| Median (IQR) | 0.10 (0.06) | 0.70 (1.50) | 0.40 (0.62) | x 2 = 94.83 | p1< 0.001HS |
| Mean ± SD | 0.12 ± 0.06 | 1.07 ± 1.03 | 0.80 ± 0.97 | p-value | p2< 0.001HS |
| Range (min-max) | 0.01 - 0.25 | 0.10 - 4.60 | 0.10 - 4.40 | <0.001HS | p3= 1.000 NS |
| Albumin (g/dL) | |||||
| Median (IQR) | 4.30 (0.75) | 3.00 (1.80) | 3.45 (0.80) | x 2 = 64.28 | p1< 0.001HS |
| Mean ± SD | 4.33 ± 0.43 | 3.23 ± 0.97 | 3.40 ± 0.56 | p-value | p2< 0.001HS |
| Range (min-max) | 3.60 - 5.10 | 1.60 - 4.90 | 2.10 - 4.40 | <0.001HS | p3= 1.000 NS |
| Platelets (103 /mL) | |||||
| Median (IQR) | 266.00 (91.75) | 151.00 (95.00) | 108.00 (61.75) | x 2 = 12.47 | p1= 0.001HS |
| Mean ± SD | 272.83 ± 49.63 | 172.77 ± 82.05 | 118.92 ± 65.57 | p-value | p2< 0.407 NS |
| Range (min-max) | 184.00 - 360.00 | 39.00 - 333.00 | 22.00 - 340.00 | =0.002 HS | p3= 0.160 NS |
| Creatinine (mg/dL) | |||||
| Median (IQR) | 0.81 (0.29) | 0.90 (0.35) | 1.00 (0.30) | x 2 = 12.19 | p1< 0.030S |
| Mean ± SD | 0.84 ± 0.17 | 0.96 ± 0.27 | 0.97 ± 0.20 | p-value | p2< 0.002 HS |
| Range (min-max) | 0.50 - 1.24 | 0.60 – 1.90 | 0.60 - 1.60 | <0.002HS | p3 = 0.997NS |
a, Multiple pairwise comparisons by Post hoc test; -p1:p-value for the difference between control and cirrhosis group (GI vs. GII); p2, p-value for the difference between control and HCC group (GI vs. GIII); p3, p-value for the difference between cirrhosis and HCC group (GII vs. GIII); IQR, Interquartile range; NS, Non significant at p-value ≥ 0.05; S, Significant at p-value < 0.05; HS, Highly significant at p-value < 0.01
In control group, miRNA-215 ranged from 1.60 to 21.30 with a median value of 6.89. In cirrhotic patients group, it ranged from 0.70 to 14.65 with a median value of 2.85. In patients with HCC group, it ranged from 0.03 to 10.95 with a median value of 0.52. MiRNA-215 and AFA mean levels showed a statistically significant difference (p <0.001) between the three studied groups (Table 3).
Table 3.
The Median of microRNA -215 and AFP among the Studied Groups
| Parameters | GI | GII | GIII | Test of significance Kruskal-Wallis | Post hoc test |
|---|---|---|---|---|---|
| Control | Cirrhosis | HCC | |||
| (n = 60) | (n = 75) | (n = 60) | |||
| MiRNA-215 | |||||
| Median (IQR) | 6.89 (7.33) | 2.85 (2.96) | 0.52 (1.63) | x 2 = 93.42 | p1< 0.001HS |
| p-value | p2< 0.001HS | ||||
| Range (min-max) | 1.60 - 21.30 | 0.70 - 14.65 | 0.03 - 10.95 | <0.001HS | p3< 0.001HS |
| AFP (ng/mL) | |||||
| Median (IQR) | 3.95 (2.53) | 5.50 (21.20) | 38.00 (184.76) | x 2 = 69.63 | p1= 0.001HS |
| p-value | p2< 0.001HS | ||||
| Range (min-max) | 0.80 - 10.50 | 1.01 - 110.00 | 1.80 - 3130.00 | <0.001HS | p3< 0.001HS |
a, Multiple pairwise comparisons by Post hoc test; -p1, p-value for the difference between control and cirrhosis group (GI vs. GII); -p2, p-value for the difference between control and HCC group (GI vs. GIII); p3, p-value for the difference between cirrhosis and HCC group (GII vs. GIII); IQR, Interquartile range; NS, Non significant at p-value ≥ 0.05; S, Significant at p-value < 0.05; HS, Highly significant at p-value < 0.01.
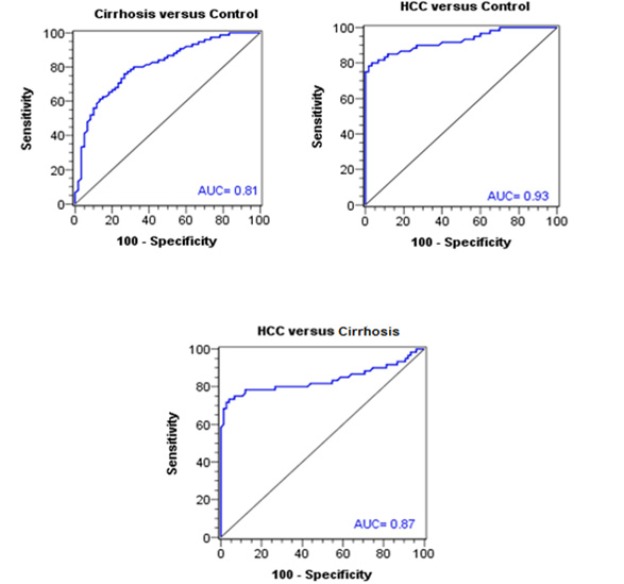
Figure 4.
ROC Curves of Circulating miRNA-215 for Pairwise Dicrimination between the Three Studied Groups
Table 5.
Logistic Regression Models of Combined Circulating microRNA-215 and AFP Measurements
Spearman’s rank correlation revealed that in cirrhosis group, there was a negative correlation between miRNA-215 level and serum AFP level (rs= -0.27; p<0.020) as illustrated in Figure 1 and in HCC group, there was a negative correlation between MiRNA-215 level and serum AFP level, (rs= -0.72; p<0.001) as illustrated in Figure 2. Also, there was a negative correlation between miRNA-215 level and focal size lesion (cm) in HCC group (rs = - 0.94, p< 0.001) as shown in Figure 3.
Figure 1.
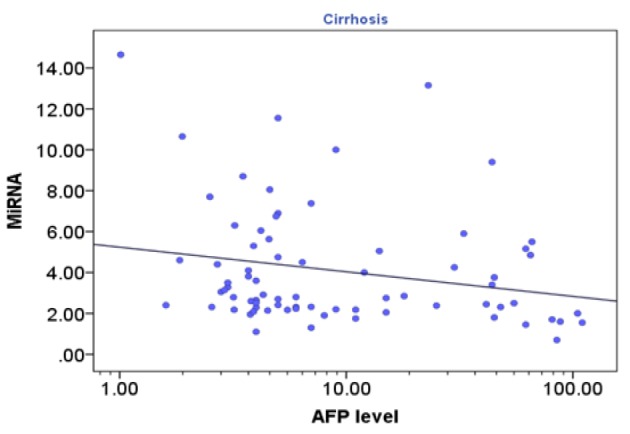
Correlation between Circulating microRNA-215 and AFP Serum Level (log10 scaled) in Cirrhosis Group (rs= -0.27; p<0.020)
Figure 2.

Correlation between Circulating Microrna-215 and AFP Serum Level (Log10 Scaled) in HCC Group
Regarding MELD score, there was no correlation between miRNA-215 level and MELD score in both cirrhosis (rs = -0.13, p=0.261) and HCC (rs = -0.01, p=0.921) groups.
Receiver operating characteristics (ROC) analysis was used to assess the diagnostic performance of circulating microRNA-215 and AFP in HCC, individually and combined. For discrimination between cirrhosis and HCC groups regarding microRNA-215, a discrimination was displayed with 78.3% sensitivity, 88.0% specificity, 83.9% PPV, 83.5% NPV, and 83.2% accuracy at the best cutoff value of ≤ 1.90. The area under the curve (AUC) was 0.87 with SE of 0.04 (p-value < 0.001) (Table 4 and Figure 4). As regards AFP, it had a sensitivity of 81.7%, a specificity of 66.7%, a PPV of 66.2%, a NPV of 82.0% and an accuracy of 74.17% at the best cutoff value of ≥ 11.50 (ng/mL). The AUC was 0.77 with SE of 0.04 (p-value < 0.001) (Table 4).
Table 4.
ROC Curves of Circulating miRNA-215 for Pairwise Dicrimination between the Three Studied Groups
| Outcome Disease | Model Variables | Coefficient | SE | Wald Square | p-value a | OR (95% CI) |
|---|---|---|---|---|---|---|
| Cirrhosis vs. control | AFP (log10) | 2.18 | 0.72 | 9.18 | 0.002 HS | 8.82 (2.16-36.03) |
| miRNA (log10) | -3.98 | 0.85 | 21.74 | < 0.001 HS | 0.02 (0.003-0.10) | |
| Constant | 1.41 | 0.79 | 3.18 | 0.074 NS | - | |
| Equation | Logit (p)*= 2.18 × (log10 AFP) - 3.98 × (log10 miRNA) +1.41 | |||||
| x 2- value | 54.92 | |||||
| p-value b | < 0.001 HS | |||||
| HCC vs. control | AFP (log10) | 3.21 | 1.1 | 8.59 | 0.003 HS | 24.83 (2.9-212.72) |
| miRNA (log10) | -3.01 | 0.91 | 10.86 | 0.001 HS | 0.05 (0.01-0.29) | |
| Constant | -1.44 | 1.17 | 1.51 | 0.219 NS | - | |
| Equation | Logit(p)*= 3.21 × (log10 AFP) - 3.01 × (log10 miRNA) - 1.44 | |||||
| x 2- value | 105.11 | |||||
| p-value b | < 0.001 HS | |||||
| HCC vs. cirrhosis | AFP (log10) | 0.55 | 0.43 | 1.61 | 0.204 NS | 1.74 (0.74-4.06) |
| miRNA (log10) | -2.89 | 0.64 | 20.38 | < 0.001 HS | 0.06 (0.02-0.20) | |
| Constant | -0.29 | 0.64 | 0.21 | 0.646 NS | - | |
| Equation | Logit(p)*= 0.55 × (log10 AFP) - 2.89 × (log10 miRNA) -0.29 | |||||
| x 2- value | 67.87 | |||||
| p-value b | < 0.001 HS | |||||
Predicted probability: p=1/(1+e^(-logit(p)) ); * logit(p)=ln(p/(1-p)); x2 value, model chi square; a, p-value of Wald test squared; b, p-value corresponds to overall chi square of the model; NS, Non significant at p-value ≥ 0.05; S, Significant at p-value < 0.05; HS, Highly significant at p-value < 0.01; OR (95% CI), Odds ratio with 95 % confidence interval
Figure 3.

Correlation between Circulating microRNA-215 and Focal Size of Focal Lesion in HCC Group (R= - 0.94, p< 0.001)
Binary logistic regression analysis was conducted to evaluate the contribution of combined measurement of miRNA-215 and AFP to assign individuals to the outcome disease whether cirrhosis or HCC.
Table 5 showed variables included in the models, the estimated equation derived from each model, and models Chi square values with the associated p-values indicating the overall significant (p <0.001) fit of the models.
Table 5.
Logistic Regression Models of Combined Circulating microRNA-215 and AFP Measurements
| Outcome Disease | Model Variables | Coefficient | SE | Wald Square | p-value a | OR (95% CI) |
|---|---|---|---|---|---|---|
| Cirrhosis vs. control | AFP (log10) | 2.18 | 0.72 | 9.18 | 0.002 HS | 8.82 (2.16-36.03) |
| miRNA (log10) | -3.98 | 0.85 | 21.74 | < 0.001 HS | 0.02 (0.003-0.10) | |
| Constant | 1.41 | 0.79 | 3.18 | 0.074 NS | - | |
| Equation | Logit (p)*= 2.18 × (log10 AFP) - 3.98 × (log10 miRNA) +1.41 | |||||
| x 2- value | 54.92 | |||||
| p-value b | < 0.001 HS | |||||
| HCC vs. control | AFP (log10) | 3.21 | 1.1 | 8.59 | 0.003 HS | 24.83 (2.9-212.72) |
| miRNA (log10) | -3.01 | 0.91 | 10.86 | 0.001 HS | 0.05 (0.01-0.29) | |
| Constant | -1.44 | 1.17 | 1.51 | 0.219 NS | - | |
| Equation | Logit(p)*= 3.21 × (log10 AFP) - 3.01 × (log10 miRNA) - 1.44 | |||||
| x 2- value | 105.11 | |||||
| p-value b | < 0.001 HS | |||||
| HCC vs. cirrhosis | AFP (log10) | 0.55 | 0.43 | 1.61 | 0.204 NS | 1.74 (0.74-4.06) |
| miRNA (log10) | -2.89 | 0.64 | 20.38 | < 0.001 HS | 0.06 (0.02-0.20) | |
| Constant | -0.29 | 0.64 | 0.21 | 0.646 NS | - | |
| Equation | Logit(p)*= 0.55 × (log10 AFP) - 2.89 × (log10 miRNA) -0.29 | |||||
| x 2- value | 67.87 | |||||
| p-value b | < 0.001 HS | |||||
Predicted probability: p=1/(1+e^(-logit(p)) ); * logit(p)=ln(p/(1-p)); x2 value, model chi square; a, p-value of Wald test squared; b, p-value corresponds to overall chi square of the model; NS, Non significant at p-value ≥ 0.05; S, Significant at p-value < 0.05; HS, Highly significant at p-value < 0.01; OR (95% CI), Odds ratio with 95 % confidence interval
In the first model, the outcome disease is cirrhosis against control, the p-values for the regression coefficients relevant to log10 AFP (p= 0.002, OR = 8.82, 95% CI: 2.16-36.03) and log10 miRNA-215 (p < 0.001, OR = 0.02, 95% CI: 0.003-0.10) revealed that both biomarkers contributed significantly to assign patients to cirrhosis group. For the second model, the outcome disease is HCC against control. The p-values for the regression coefficients of log10 AFP (p= 0.003, OR = 24.83, 95% CI: 2.90-212.72) and log10 miRNA-215 (p =0.001, OR = 0.05, 95% CI: 0.01-0.29) indicated that both biomarkers contributed significantly to assign patients to HCC group.
In the third model, the outcome disease is HCC against cirrhosis, the p-value associated with the regression coefficients of log10 miRNA-215 (p <0.001, OR = 0.06, 95% CI: 0.02-0.20) indicated a highly significant contribution of miRNA-215 measurement. However, log10 AFP (p= 0.204, OR = 1.74, 95% CI: 0.74 - 4.06) revealed non-significant contribution of AFP measurement indicating superiority of miRNA-215 over AFP regarding HCC prediction versus cirrhosis.
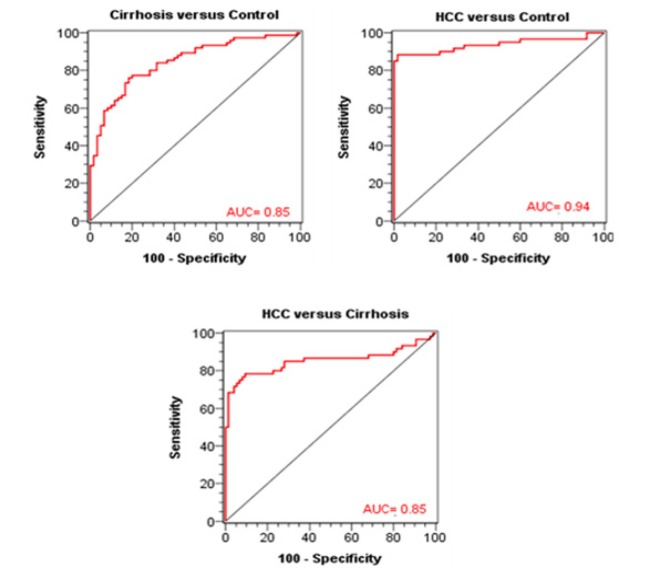
The characteristics of the combined MiRNA-215 and AFP (both were log10 transformed) were illustrated in Table6 and graphically by ROC curves in figure 5. For discrimination between control and HCC, the estimates were 88.3% sensitivity, 98.3% specificity, 98.1% PPV, 89.4% NPV, and 93.3% accuracy at the best cutoff value of ≥ 0.52(ng/mL). The AUC was 0.94 with SE of 0.03 (p-value < 0.001).
Table 6.
Test Characteristics of Combined Tests of miRNA-215 and AFP
| Test characteristics | Combined Tests |
||
|---|---|---|---|
| Cirrhosis vs. Control | HCC vs. Control | HCC vs. Cirrhosis |
|
| Best cutoff value | ≥ 0.56 | ≥ 0.52 | ≥ 0.47 |
| AUC | 0.85 | 0.94 | 0.85 |
| SE | 0.03 | 0.03 | 0.04 |
| p-value | < 0.001HS | < 0.001HS | < 0.001HS |
| Sensitivity % | 76 | 88.3 | 78.3 |
| Specificity % | 81.7 | 98.3 | 90.7 |
| PPV % | 83.8 | 98.1 | 87 |
| NPV % | 73.1 | 89.4 | 84 |
| Accuracy % | 78.8 | 93.3 | 84.5 |
SE, Standard error; PPV, Positive predictive value; NPV, Negative predictive value; NS, Non significant at p-value ≥ 0.05; S33, Significant at p-value < 0.05; HS, Highly significant at p-value < 0.01
Between cirrhosis and HCC groups, the ROC analysis estimated a sensitivity of 78.3%, a specificity of 90.7%, a PPV of 87.0%, a NPV of 84.0% and an accuracy of 84.5% at the best cutoff value of ≥ 0.47 (ng/mL). The AUC was 0.85 with SE of 0.04 (p-value < 0.001).
The mean ± SD of serum MiRNA-215 was significantly lower in patients with hepatocellular carcinoma who had multiple focal lesions than in those with single focal lesion (p< 0.001) as shown in Table 7.
Table 7.
Statistical Analysis of miRNA-215 Regarding Focal Lesion Number in HCC Group
| MiRNA | Single | Multiple | Mann-Whitney test | p-value |
|---|---|---|---|---|
| Median (IQR) | 3.75 (5.43) | 0.17 (0.46) | Z = 6.10 | < 0.001HS |
| Mean ± SD | 4.67 ± 3.11 | 0.33 ± 0.31 | ||
| Range (min-max) | 1.27 - 10.95 | 0.03 - 1.11 |
Discussion
The HCC incidence is rising all through the world because of a rising incidence of HBV and HCV infection, due to an increase in prevalence of non-alcoholic fatty liver disorder (Njei et al., 2015). In Egypt, HCC is the fourth common cancer and is the second reason of most cancers mortality in both sexes (Demerdash et al., 2017). The burden of HCC increased with a doubling within the occurrence rate in the past 10 years and this was attributed to excessive occurrence of HCV. Egypt has incidence of HCC approximately 21% in cirrhotic sufferers.
Serum AFP measurement remains combined with ultrasound to reduce the hazard of missing small lesions within the cirrhotic liver that were not detected via ultrasound. Alternative or additional biomarkers could be beneficial tools for surveillance or as a decisional device in medical practice to discover patients that will benefit from advanced imaging techniques in a surveillance setting to augment the percentage of patients presented with HCC diagnosed in an early tumor stage (Schütte et al., 2015).
As for the relationship among miRNAs and HCC, several research have proven that the atypical miRNAs expression can be found in plasma and serum of HCC patients or HCC cells and tissues, and miRNAs have proven great promise as prognostic and diagnostic markers for HCC (Morishita and Masaki, 2015; Qi et al., 2013; Demerdash et al., 2017).
We found that the mean age was over 50 years old in both cirrhosis and HCC groups. Estes et al., (2015) reported that anti-HCV prevalence in Egypt was totally 14.7%, and prevalence of viremia was 9.8%. Prevalence in patients aged >59 years was similar to prevalence in those with age between 55 and 59 years.
Regarding the liver profile ALT, AST, GGT, total and direct bilirubin and ALP were higher in HCC and cirrhotic patients than in control group. In addition, Albumin values were lower to a significant level in HCC and cirrhotic patients than in control group. Our results agreed with Awadallah et al., (2011) who reported a significant deterioration in the liver function in cirrhosis and HCC compared to control group. However, there was no significant difference between HCC and cirrhotic groups regarding total and direct bilirubin and albumin results. Serum ALT, AST, GGT and ALP was higher in HCC patients compared to cirrhotic patients. Our findings were in accordance to Sleisenger et al., (2002) and Thapa and Walia ( 2007) who declared that aminotransferases are the most frequently utilized indicators of hepatocellular necrosis and expected to be elevated in liver cell injury whatever the cause and the conventional tests of hepatic function don’t distinguish HCC from cirrhosis and so they contribute little to the diagnosis of the tumor with the exception of serum ALP, which was significantly elevated in HCC group when compared with cirrhotic group due to the more prominent cholestatic effect of the tumor.
We found a significantly higher levels of AFP in HCC patients (median= 294.9 ng/mL) compared to the cirrhosis patients (median= 19.5 ng/mL) and control group (median= 4.3ng/mL). This was in agreement with Spadaro et al.; El-Assaly et al. and Awadallah et al. who reported a significant elevation in serum AFP in HCC group compared to cirrhosis patients and control group (Spadaro et al., 2005; El-Assaly et al., 2008; Awadallah et al., 2011).
Our results revealed a significantly lower levels of MicroRNA-215 in HCC patients (median= 1.63) compared to the cirrhosis patients (median= 4.0) and control group (median= 8.6). This was in agreement with Ashmawy et al., (2017) who reported a significant lower expression of MicroRNA-215 level in HCC group when compared to cirrhotic group and control group (Ashmawy et al., 2017).
Significantly high correlation found between tumor grades and miR-215 level was consistent with numerous studies indicating its role in tumor progression. Similarly, we found that mean ± SD of serum MiRNA-215 was significantly lower in HCC patients who had multiple focal lesions than in those with single focal lesion (p< 0.001) and also, there was a negative correlation between miRNA-215 level and focal lesion size (cm) in HCC group.
Our results was not in agreement with Zhang et al., (2014) who reported a significant higher level of MicroRNA-215 in HCC group compared to cirrhosis patients and control group and the expression of miR-215 in plasma was significantly up-regulated in patients with chronic hepatitis and HCC. This is may be happened by genotypes. Demerdash et al., (2017) reported that about 90% of Egyptian patients suffering from HCV belong to genotype-4 and this differs from other countries at which HCC patients suffering from other genotypes.
The cut-off value of AFP for discriminating HCC patients versus cirrhosis and control groups was 8.9 ng/mL. This had a diagnostic sensitivity of 83.3%, specificity 95%. The cut-off value of AFP for HCC diagnosis has been a matter of debate. AFP had a specificity of 76%-94% and a sensitivity of 39%-65% for the presence of HCC in previously published research (Trevisani et al., 2001). The variation in specificity and sensitivity of AFP in the research may be because of the variety of patient populations who were examined, various study designs and different normal cut-off levels (Gomaa et al., 2009) .
Serum AFP is related to different problems; First, the rise in AFP level which occurred transiently in patients with chronic liver disease especially during exacerbation of hepatitis (serum level >100ng/mL) (low specificity) (Kobeisy et al., 2012), mild rise in acute hepatitis, chronic hepatitis and cirrhosis and overlaps can cause diagnostic difficulties. The second one is that AFP levels may be normal in up to 40% of patients with HCC, especially for the duration of the early stages (low sensitivity) (El-Assaly et al., 2008). However, El-Assaly et al. suggested a different cutoff values for AFP. At a cutoff value >100ng/mL the diagnostic sensitivity was 66.6%, specificity was 93.6% and at a cutoff 200ng/mL low sensitivity (32.2%) and high specificity (100%) were recorded. When the cutoff increase >400 ng/mL, the sensitivity decreased to 13.5%. When increasing the cutoff of serum AFP, the sensitivity decrease and the specificity increase but at the cost of a progressive increase in the false-negative results (El-Assaly et al., 2008).
From (ROC) curve analysis, we found that the best cut-off level of MicroRNA-215 for discriminating HCC patients versus control groups was 2.30. This had a diagnostic sensitivity of 80%, specificity 96.7%. For discrimination between cirrhosis and HCC groups regarding microRNA-215, a discrimination was displayed with 78.3% sensitivity, 88.0% specificity at the best cutoff value of ≤ 1.90. The AUC was 0.87 (p-value < 0.001).
When we combined both microRNA-215 and AFP to discriminate between control and cirrhosis groups, the ROC analysis showed 76.0% sensitivity, 81.7% specificity at the best cutoff value of ≥ 0.56. The AUC was 0.85. Between cirrhosis and HCC groups, the ROC analysis showed a sensitivity of 78.3%, a specificity of 90.7% at the best cutoff level of ≥ 0.47. The AUC was 0.85 (p-value < 0.001).
In conclusion, MicroRNA-215 was proved to be significantly lower in patients with HCC compared to cirrhotic patients and control group. This marker might be used as a potential marker for HCC diagnosis. An observation worth further investigation is that the combined use of (microRNA-215 and AFP), increase the diagnostic performance for detection of HCC which suggested that the inclusion of MicroRNA-215 to the current standard tests for early detection of HCC may improve the ability to identify patients who might be missed by current diagnostic methods and thus might provide a better therapeutic outcome. Moreover, this miRNA have an elevated advantage over traditional biomarkers since plasma miRNAs are very stable and remaining for long time and their determination techniques are easy to perform. Additionally, their detection in plasma can clearly define disease progression.
Acknowledgments
Compliance with Ethical Standards
Funding
No funding was received for this study.
Conflict of Interest
Author Hussein Ahmed El Mahdy declares that he has no conflict of interest. Ismail Abdelshafy Abdelhamid declares that he has no conflict of interest. Author Ahmed Ibrahim Amen declares that he has no conflict of interest. Author Eman Abdelsameea declares that she has no conflict of interest and Author Mona M. Hassouna declares that she has no conflict of interest.
Research involving human participants and/or animals
- Ethical approval: All procedures performed in our study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
- Informed consent: an informed consent was obtained from all individual participants included in the study.
- It is a part of an approved thesis.
References
- Aravalli RN, Cressman EN, Steer CJ. Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch Toxicol. 2013;87:227–47. doi: 10.1007/s00204-012-0931-2. [DOI] [PubMed] [Google Scholar]
- Ashmawy AM, Elgeshy KM, Salam E-STA, et al. Crosstalk between liver-related microRNAs and Wnt/β-catenin pathway in hepatocellular carcinoma patients. Arab J Gastroenterol. 2017;18:144–50. doi: 10.1016/j.ajg.2017.09.001. [DOI] [PubMed] [Google Scholar]
- Awadallah AM, Issa HA, Soliman MS. Evaluation of serum chromogranin a as a useful tumor marker for diagnosis of hepatocellular carcinoma. J Am Sci. 2011;7:999–1007. [Google Scholar]
- Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753–70. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Demerdash HM, Hussien HM, Hassouna E, Arida EA. Detection of MicroRNA in hepatic cirrhosis and hepatocellular carcinoma in hepatitis C genotype-4 in Egyptian patients. BioMed Rese Int. 2017;2017 doi: 10.1155/2017/1806069. doi: 10.1155/2017/1806069. Epub 2017 May 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assaly N, El Ashry I, Mostafa I, El Ghannam M, Attia M. Serum chromogranin-A and serum PIVKA-II as useful complementary and diagnostic markers for HCC. Res J Med Med Sci. 2008;4:391–401. [Google Scholar]
- Estes C, Abdel-Kareem M, Abdel-Razek W, et al. Economic burden of hepatitis C in Egypt: the future impact of highly effective therapies. Aliment Pharmacol Ther. 2015;42:696–706. doi: 10.1111/apt.13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle PR, Forner A, Llovet JM, et al. EASL clinical practice guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- Gomaa AI, Khan SA, Leen EL, Waked I, Taylor-Robinson SD. Diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2009;15:1301. doi: 10.3748/wjg.15.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–43. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Kobeisy MA, Morsy KH, Galal M, et al. Clinical significance of elevated alpha-foetoprotein (AFP) in patients with chronic hepatitis C without hepatocellular carcinoma in upper EGYPT. Arab J Gastroenterol. 2012;13:49–53. doi: 10.1016/j.ajg.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Li G, Shen Q, Li C, et al. Identification of circulating MicroRNAs as novel potential biomarkers for hepatocellular carcinoma detection: a systematic review and meta-analysis. Clin Translational Oncol. 2015;17:684–93. doi: 10.1007/s12094-015-1294-y. [DOI] [PubMed] [Google Scholar]
- Malinchoc M, Kamath PS, Gordon FD, et al. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology. 2000;31:864–71. doi: 10.1053/he.2000.5852. [DOI] [PubMed] [Google Scholar]
- Morishita A, Masaki T. mi RNA in hepatocellular carcinoma. Hepatol Res. 2015;45:128–41. doi: 10.1111/hepr.12386. [DOI] [PubMed] [Google Scholar]
- Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61:191–9. doi: 10.1002/hep.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang J, Katayama H, Sen S, Liu S-m. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135. doi: 10.4149/neo_2013_018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgia R, Singal AG. Hepatocellular carcinoma and other liver lesions. Med Clin. 2014;98:103–18. doi: 10.1016/j.mcna.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Schütte K, Schulz C, Link A, Malfertheiner P. Current biomarkers for hepatocellular carcinoma: Surveillance, diagnosis and prediction of prognosis. World J Hepatol. 2015;7:139. doi: 10.4254/wjh.v7.i2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelbaya A, Kuznik A, Salem M, Mankola W, Sadik K. P1265: Estimating the epidemiologic and economic impact of different treatment rates for hepatitis C virus (HCV) in Egypt. J Hepatol. 2015;62:832–3. [Google Scholar]
- Song He D-CZ, Cheng Wei. MicroRNAs as biomarkers for hepatocellular carcinoma diagnosis and prognosis. Clin Res Hepatol Gastroenterol. 2015;39:426–34. doi: 10.1016/j.clinre.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Spadaro A, Ajello A, Morace C, et al. Serum chromogranin-A in hepatocellular carcinoma: diagnostic utility and limits. World J Gastroenterol. 2005;11:1987. doi: 10.3748/wjg.v11.i13.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa B, Walia A. Liver function tests and their interpretation. Indian J Pediat. 2007;74:663–71. doi: 10.1007/s12098-007-0118-7. [DOI] [PubMed] [Google Scholar]
- Trevisani F, D’Intino PE, Morselli-Labate AM, et al. Serum α-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol. 2001;34:570–5. doi: 10.1016/s0168-8278(00)00053-2. [DOI] [PubMed] [Google Scholar]
- WC Pang R, TP Poon R. Cancer stem cell as a potential therapeutic target in hepatocellular carcinoma. Curr Cancer Drug Tar. 2012;12:1081–94. doi: 10.2174/156800912803987995. [DOI] [PubMed] [Google Scholar]
- Zhang Y-C, Xu Z, Zhang T-F, Wang Y-L. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. 2015;21:9853. doi: 10.3748/wjg.v21.i34.9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-q, Meng H, Wang N, et al. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Curr Diagn Pathol. 2014;9:135. doi: 10.1186/1746-1596-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]