Abstract
Background:
Previous studies have evaluated the impact of MTHFR 677C>T polymorphism on susceptibility to ovarian and cervical cancers in women, but the conclusions are still controversial. To get a more precise evaluation of the association between MTHFR 677C>T polymorphism and risk of ovarian and cervical cancers, we performed a meta-analysis of the association of all eligible studies.
Methods:
A comprehensive search performed in PubMed, Google Scholar, CNKI, and Web of Science databases to identify the relevant studies up to October 15, 2018. The strength of the association was estimated by odds ratios (OR) with 95% confidence interval (CI).
Results:
A total of 27 case-control studies including eleven studies with 4990 cases 7730 controls on ovarian cancer and 16 studies with 4990 cases and 7730 controls on cervical cancer were selected. Pooled data revealed that the MTHFR 677C>T polymorphism not significantly associated with an increased risk of ovarian and cervical cancers under all five genetic models. However, stratified analysis by ethnicity showed that the MTHFR 677C>T polymorphism was significantly associated with risk of ovarian cancer in Asians. No publication bias was found in the current meta-analysis.
Conclusions:
The results of this meta-analysis proposes that the MTHFR 677C>T polymorphism may not play a role in development of ovarian and cervical cancers in overall population. Further well-designed studies are necessary to clarify the precise role of the MTHFR 677C>T polymorphism on ovarian and cervical cancers risk.
Key Words: Ovarian cancer, cervical cancer, MTHFR gene, polymorphism, meta-analysis
Introduction
Gynecological cancers are among the most common cancers in women and hence a major health problem worldwide (Maheshwari et al., 2016). Ovarian and cervical cancers are the most common gynecological cancers affecting women worldwide (Torre et al., 2017). Ovarian cancer is the second most common cancer and main cause of death with gynecological tumors worldwide (Qin et al., 2013; Zhu and Sun, 2017). Moreover, cervical cancer is the third most frequent neoplasm among women worldwide (Rocha et al., 2017). In 2017, a study showed that ovarian cancer (47%) followed by cervical cancer (29%) are the most common gynecological malignancy among Pakistan women (Manzoor et al., 2017). Despite continuous advances in cancer biology research, the etiology of ovarian and cervical cancer are not known exactly, partly because of the inconsistency of findings among epidemiological studies (Yu et al., 2013). A reappraisal of Genome-wide association studies (GWAS) and genetic association studies suggested a strong genetic component to susceptibility to ovarian and cervical cancers (Fearon et al., 2013).
Epidemiological studies had identified that Methylene tetrahydrofolate reductase (MTHFR) was a potential genetic marker of different malignancies (Yi et al., 2016; He and Shen, 2017). The human MTHFR gene is located on chromosome 1p36.3, consist of 11 exons and spans 2.2 kb of genomic DNA (Abedinzadeh et al., 2015; Azarpira et al., 2018). It is encodes the vital enzyme which plays a key role in the folate/homocysteine metabolic pathway and regulates the intracellular folate level for the synthesis and methylation of DNA (Azarpira et al., 2018; Kamali et al., 2018). In humans, the MTHFR 677C>T (in exon 4) polymorphism has been heavily studied in different disease. The MTHFR 677C>T is associated with reduced enzyme activity and arise an elevated plasma homocysteine level. Moreover, the MTHFR 677C>T polymorphism leads to increased heat liability and reduced enzymatic capability for methylation of Homocysteine (Rozycka et al., 2014). The mutant homozygous genotype (TT) of MTHFR 677C>T polymorphism was particularly common in northern China (20%), southern Italy (26%), and Mexico (32%) (Wilcken, 2003).
Over the past decades, a large number of epidemiological studies and meta-analyses have evaluated the association between the MTHFR 677C>T polymorphism and susceptibility to ovarian and cervical cancer (Yi et al., 2016; He and Shen, 2017). However, the results were conflicting and inconclusive, presumably due to small sample size in each published study, various genetic backgrounds and possible selection bias (He and Shen, 2017). Subsequently, a few novel studies have recently been performed to estimate the associations of MTHFR 677C>T polymorphism with risk of ovarian and cervical cancer and provide new evidences that were not included in the previous meta-analyses. Thus, this meta-analysis covering all potentially eligible studies was performed to get a more precise evaluation of the association between MTHFR 677C>T polymorphism and risk of ovarian and cervical cancers.
Materials and Methods
Literature Search Strategy
A comprehensive searched in PubMed, Google scholar, Web of Science, EMBASE, Chinese Biomedical database, and China National Knowledge Infrastructure (CNKI) databases was performed to obtain the all relevant studies investigated association of MTHFR 677C>T polymorphism with risk of ovarian and cervical cancers up to October 15, 2018. The following keywords and terms were used: (‘’Gynecological Cancer’’ OR ‘’Ovarian Cancer’’ OR ‘’Epithelial Ovarian Cancer’’ OR ‘’Cervical Cancer’’) AND (‘’Methylene tetrahydrofolate reductase’’ OR ‘’MTHFR’’ OR ‘’677C>T’’ OR ‘’rs1801133’’) AND (“Polymorphism” OR ‘’SNP’’ OR ‘’Mutation’’ OR “Variant” OR “Variation”). Moreover, the references of the retrieved articles manually checked for other potential studies that possibly have been missed in the initial search.
Inclusion Criteria and Data Extraction
Studies were included in the current meta-analysis only if they met all of the following criteria: a) studies with case-control or cohort design; b) only published studies; c) evaluated the association of MTHFR 677C>T polymorphism with ovarian cancer and cervical cancer; and d) the number of MTHFR 677C>T polymorphism genotypes in the cases and healthy control was reported to estimates odds ratio (OR) and 95% confidence interval (CI). The exclusion criteria were: a) abstracts, case reports, reviews, previous meta-analyses, posters, letters to editor and commentaries; b) animal studies; c) case only studies; d) linkage or sibling studies; e) studies did not calculated MTHFR polymorphisms genotype frequencies or which the number of genotypes and alleles could not be ascertained; f) studies on other polymorphisms of MTHFR gene; and g) overlapping studies and studies duplicate or containing previously published data. Moreover, if studies had overlapping data, only the study with the largest population or more recently published data was finally selected.
Data Extraction
Two authors (H.A and A.H) independently assessed the articles for their eligibility for inclusion and the needed data were carefully extracted based on the inclusion criteria above using a standard form. Any disagreements were solved by discussion with a third author (E.S). The following data were extracted for each study: first author’s name, publication year, country, ethnicity, source of control (hospital-based or population-based), genotyping methods, and the number of alleles and genotypes in the cases and controls, minor allele frequency (MAF) among controls, and P-value for Hardy-Weinberg equilibrium (HWE).
Statistical Analysis
The strength of association between MTHFR 677C>T polymorphism and ovarian and cervical cancers was assessed by using odds ratios (ORs) and 95% confidence intervals (CIs). The P-value of the pooled ORs was considered significant if less than 0.05, which was examined by Z-test. The pooled ORs were calculated under all five genetic models, i.e., allele (T vs. C), homozygote (TT vs. CC), heterozygote (TC vs. CC), dominant (TT+TC vs. CC), and recessive (TT vs. TC+CC), in which the ‘’C’’ represents the major allele and the ‘’T’’ represents the minor allele. The Cochran’s Q-test was used to access the between-study heterogeneity. Moreover, the effects of heterogeneity was we quantified using I2 statistic (ranges from 0 to 100%), in which detected variations among studies due to heterogeneity rather than chance (I2= 0-25%, no heterogeneity; I2=25-50%, moderate heterogeneity; I2=50–75%, large heterogeneity; I2=75-100%, extreme heterogeneity) (Higgins 2003, Hippel 2015). A chi-square test was used to determine Hardy-Weinberg equilibrium (HWE) in controls, which p-value less than 0.05 was representative of statistical significance. Subgroup analyses were performed to explore possible sources of heterogeneity by ethnicity, source of controls, genotyping method and HWE status. Sensitivity analyses were performed by sequential removal of each study and by excluding those studies deviation from HWE to test the stability and reliability of the results. Visual inspection of asymmetry in funnel plots and Begg’s rank correlation statistically were used to test whether publication bias existed or not, in which P<0.05 was considered to be represented of statistically significant. All statistical analyses were performed using the Comprehensive Meta-Analysis (CMA) software version 2.0 (Biostat, USA). Two-sided P<0.05 was considered statistically significant.
Results
Characteristics of Studies
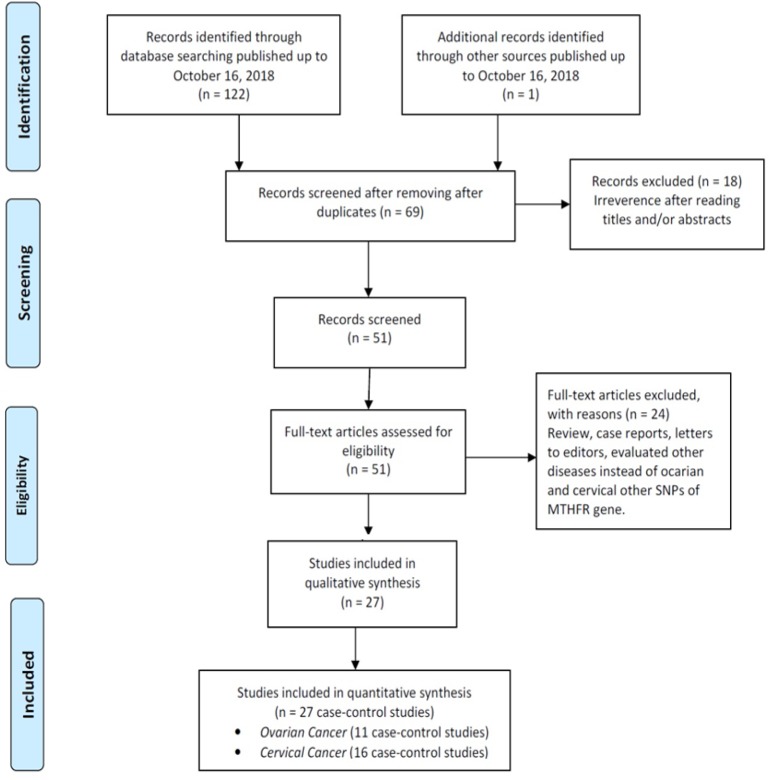
The flow diagram of study selection process was presented in Figure 1. According to the initial searches, 123 studies were identified, which after removing duplicates and irrelevant studies, there were 69 studies left. Then, the titles and abstracts of the remaining articles were reviewed, 51 full-text articles were considered eligible. After carefully reviewing the remaining studies, 24 of them were excluded because did not reported sufficient data, were not case-control studies, overlapped by other studies, and not relevant to the MTHFR 677C>T polymorphism (Figure 1). Finally, 27 case-control studies with 7856 ovarian and cervical cancer cases and 11,263 controls were included. Among these studies, eleven case-control studies with 4990 cases 7730 controls were on ovarian cancer (Jakubowska et al., 2007; Wu et al., 2007; Terry et al., 2010; Prasad and Wilkhoo, 2011; Webb et al., 2011; Gao et al., 2012; Pawlik et al., 2012; Zhang et al., 2012; Özkılıç et al., 2016) and 16 case-control studies with 2,866 cases 3,533 controls were on cervical cancer (Lambropoulos et al., 2003; Sull et al., 2004; Zoodsma et al., 2005; Kang et al., 2005; Delgado-Enciso et al., 2006; Ma et al., 2006; Shekari et al., 2008; Nandan et al., 2008; Kohaar et al., 2010; Tong et al., 2011; von Keyserling et al., 2011; Yang et al., 2011; Mostowska et al., 2011; Prasad and Wilkhoo, 2011; Hajiesmaeil et al., 2016; Gong et al., 2018). The main characteristics of included studies were listed in Table 1. The studies were published from 2004 to 2018, and the sample sizes in cases groups ranged from 22 to 1,638. For ovarian cancer, seven studies were conducted in Caucasians and four in Asians. For cervical cancer, eleven studies were conducted in Asians, four in Caucasian, and one study in mixed population. All the 27 included studies were case-control studies, 24 of them were in a Hospital-Based (HB) and the remaining was Population-Based (PB) design. Five different genotyping methods were used including: PCR-PFLP, TaqMan, MassARRAY, SNapShot and LDR-PCR. The genotype distribution of the healthy subjects in all included studies was in agreement with the Hardy-Weinberg equilibrium (HWE), except one study for cervical cancer (Table 1).
Figure 1.
The Flow Diagram of the Included and Excluded Studies
Table 1.
Characteristics of Studies Included in the Meta-analysis
| First Author | Country (Ethnicity) | SOC | Genotyping Technique |
Case/Control | Cases |
Controls |
MAFs | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes | Allele | Genotypes | Allele | |||||||||||||
| Ovarian Cancer | CC | CT | TT | C | T | CC | CT | TT | C | T | ||||||
| Wu 2007 | China (Asian) | HB | NA | 81/8 | 17 | 40 | 24 | 74 | 88 | 32 | 35 | 13 | 99 | 61 | 0.381 | 0.515 |
| Terry 2010 | USA (Caucasian) | HB | TaqMan | 1059/1125 | 427 | 492 | 140 | 1346 | 772 | 499 | 488 | 138 | 1486 | 764 | 0.340 | 0.270 |
| Terry 2010 | USA (Caucasian) | HB | TaqMan | 158/496 | 71 | 72 | 10 | 184 | 92 | 210 | 217 | 55 | 637 | 327 | 0.339 | 0.925 |
| Terry 2010 | USA (Caucasian) | HB | TaqMan | 364/412 | 164 | 167 | 33 | 495 | 233 | 193 | 168 | 51 | 554 | 270 | 0.328 | 0.130 |
| Webb 2011 | Australia (Caucasian) | PB | MassARRAY | 1638/1278 | 744 | 709 | 185 | 2197 | 1079 | 571 | 568 | 139 | 1710 | 846 | 0.331 | 0.898 |
| Prasad 2011 | India (Asian) | HB | PCR-PFLP | 80/125 | 72 | 3 | 5 | 147 | 13 | 116 | 8 | 1 | 240 | 10 | 0.040 | 0.062 |
| Pawlik 2011 | Poland (Caucasian) | PB | PCR-PFLP | 136/16 | 67 | 55 | 13 | 189 | 81 | 63 | 79 | 18 | 205 | 115 | 0.359 | 0.360 |
| Jakubowska 2012 | Poland (Caucasian) | HB | PCR-PFLP | 985/335 | 423 | 446 | 116 | 1292 | 678 | 1447 | 1481 | 422 | 4375 | 2325 | 0.347 | 0.156 |
| Zhang 2012 | China (Asian) | HB | PCR-PFLP | 215/218 | 102 | 94 | 19 | 298 | 132 | 115 | 92 | 11 | 322 | 114 | 0.261 | 0.170 |
| Gao 2012 | China (Asian) | HB | PCR-PFLP | 224/432 | 97 | 100 | 27 | 294 | 154 | 232 | 178 | 22 | 642 | 222 | 0.257 | 0.100 |
| Özkılıç 2016 | Turkey (Caucasian) | HB | PCR-PFLP | 50/54 | 18 | 28 | 4 | 258 | 36 | 19 | 30 | 5 | 68 | 40 | 0.370 | 0.160 |
| Total | 4990/773 | 2202 | 2206 | 450 | 6610 | 3106 | 3497 | 3344 | 875 | 10338 | 5094 | 0.330 | 0.077 | |||
| Cervical Cancer | CC | CT | TT | C | T | CC | CT | TT | C | T | ||||||
| Lambropoulos 2003 | Greece (Caucasian) | HB | PCR-PFLP | 21/91 | 11 | 8 | 2 | 30 | 12 | 42 | 37 | 12 | 121 | 61 | 0.335 | 0.403 |
| Sull 2004 | Korea (Asian) | HB | SNapShot | 246/454 | 73 | 115 | 58 | 261 | 231 | 153 | 221 | 80 | 527 | 381 | 0.420 | 0.989 |
| Kang 2005 | Korea (Asian) | HB | PCR-PFLP | 79/74 | 27 | 32 | 20 | 86 | 72 | 30 | 32 | 12 | 92 | 56 | 0.378 | 0.487 |
| Zoodsma 2005 | Netherlands (Caucasian) | HB | TaqMan | 636/592 | 357 | 230 | 49 | 944 | 328 | 273 | 262 | 57 | 808 | 376 | 0.318 | 0.608 |
| Ma 2006 | China (Asian) | HB | PCR-PFLP | 111/111 | 20 | 53 | 38 | 93 | 129 | 33 | 60 | 18 | 126 | 96 | 0.432 | 0.286 |
| Delgado 2006 | Mexico (Mixed) | HB | PCR-PFLP | 70/89 | 18 | 34 | 14 | 70 | 62 | 20 | 49 | 20 | 89 | 69 | 0.500 | 0.340 |
| Nandan 2008 | India (Asian) | HB | PCR-PFLP | 62/77 | 36 | 0 | 26 | 72 | 52 | 53 | 0 | 24 | 106 | 48 | 0.312 | ≤0.001 |
| Shekari 2008 | India (Asian) | HB | PCR-PFLP | 200/2 | 125 | 68 | 7 | 318 | 82 | 170 | 28 | 2 | 368 | 32 | 0.080 | 0.489 |
| Kohaar 2010 | India (Asian) | HB | SNapShot | 164/231 | 113 | 47 | 4 | 273 | 55 | 161 | 65 | 5 | 387 | 75 | 0.162 | 0.598 |
| Yang 2010 | China (Asian) | HB | PCR-PFLP | 391/382 | 229 | 85 | 77 | 530 | 234 | 182 | 166 | 34 | 536 | 234 | 0.306 | 0.658 |
| Mostowska 2011 | Poland (Caucasian) | HB | PCR-PFLP | 124/168 | 56 | 59 | 9 | 171 | 77 | 69 | 81 | 18 | 219 | 117 | 0.348 | 0.420 |
| Prasad 2011 | India (Asian) | PB | PCR-PFLP | 62/125 | 57 | 5 | 0 | 119 | 5 | 116 | 8 | 1 | 240 | 10 | 0.040 | 0.062 |
| Tong 2011 | Korea (Asian) | HB | TaqMan | 146/427 | 53 | 65 | 28 | 171 | 121 | 152 | 198 | 77 | 502 | 342 | 0.412 | 0.373 |
| Keyserling 2011 | Germany (Caucasian) | HB | LDR-PCR | 386/328 | 164 | 188 | 34 | 516 | 256 | 165 | 136 | 27 | 466 | 190 | 0.290 | 0.890 |
| Hajiesmaeil 2016 | Iran (Asian) | HB | PCR-PFLP | 22/74 | 13 | 7 | 2 | 21 | 11 | 34 | 36 | 4 | 104 | 44 | 0.297 | 0.157 |
| Gong 2018 | China (Asian) | HB | TaqMan | 146/11 | 34 | 70 | 42 | 138 | 154 | 20 | 52 | 38 | 92 | 128 | 0.582 | 0.764 |
| Total | 2866/3533 | 1386 | 1066 | 410 | 3813 | 1881 | 1673 | 1431 | 429 | 4783 | 2259 | 0.324 | ≤0.001 | |||
Abbreviation, SOC, Source Of Control; HB, Hospital Based; PB, Population Based; NA, Not applicable; PCR-RFLP, Polymerase Chain Reaction-Restriction Fragment Length Polymorphism; AS-PCR, Allele-Specific PCR; MAF, Minor Allele Frequency; HWE, Hardy-Weinberg Equilibrium.
Quantitative Synthesis
Ovarian Cancer
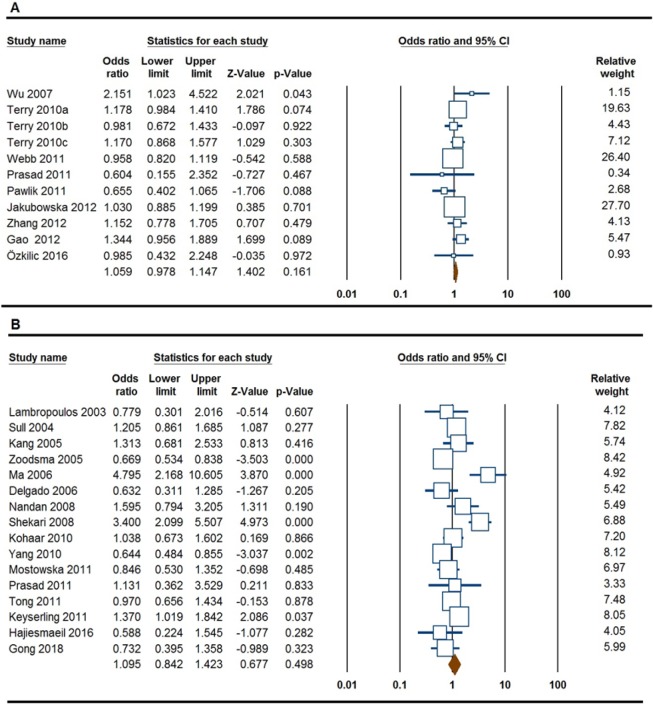
The results of meta-analysis for association between the MTHFR 677C>T polymorphism and ovarian cancer were listed in Table 2. In overall, pooled data showed that the MTHFR 677C>T polymorphism did no significantly associated with an increased risk of ovarian cancer under all five genetic models, i.e., allele (T vs. C: OR = 0.894, 95% CI 0.421-1.896, p = 0.770), homozygote (TT vs. CC: OR = 1.178, 95% CI 0.894-1.551, p = 0.244); heterozygote (TC vs. CC: OR = 1.059, 95% CI 0.978-1.147, p = 0.161, Figure 2A), dominant (TT+TC vs. CC: OR = 1.087, 95% CI 0.956-1.236, p = 0.202), and recessive (TT vs. TC+CC: OR = 1.110, 95% CI 0.876-1.406, p = 0.389). When stratified by ethnicity, there was a significant association between the MTHFR 677C>T polymorphism and an increased risk of ovarian cancer in Asians under all five genetic models, i.e., allele (T vs. C: OR = 1.132, 95% CI 1.259-1.772, p ≤0.001), homozygote (TT vs. CC: OR = 1.212, 95% CI 1.857-4.277, p ≤0.001); heterozygote (TC vs. CC: OR = 0.985, 95% CI 1.023-1.651, p = 0.032), dominant (TT+TC vs. CC: OR = 1.095, 95% CI 1.183-1.859, p = 0.001), and recessive (TT vs. TC+CC: OR = 1.410, 95% CI 1.575-3.445, p ≤0.001), but not in Caucasians (Table 2).
Table 2.
Pooled Results for Association of MTHFR 677C>T Polymorphism with Risk of Ovarian and Cervical Cancer
| Subgroup | Genetic Model | Type of Model | Heterogeneity |
Odds Ratio |
Publication Bias |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| Ovarian Cancer | ||||||||||
| Overall | T vs. C | Random | 99.38 | ≤0.001 | 0.894 | 0.421-1.896 | -0.293 | 0.770 | 0.212 | 0.388 |
| TT vs. CC | Random | 68.02 | 0.001 | 1.178 | 0.894-1.551 | 1.165 | 0.244 | 0.275 | 0.290 | |
| TC vs. CC | Fixed | 26.73 | 0.190 | 1.059 | 0.978-1.147 | 1.402 | 0.161 | 1.000 | 0.798 | |
| TT+TC vs. CC | Random | 50.52 | 0.027 | 1.087 | 0.956-1.236 | 1.276 | 0.202 | 0.640 | 0.414 | |
| TT vs. TC+CC | Random | 61.22 | 0.004 | 1.110 | 0.876-1.406 | 0.862 | 0.389 | 0.350 | 0.295 | |
| Ethnicity | ||||||||||
| Caucasian | T vs. C | Random | 99.59 | ≤0.001 | 0.634 | 0.240-1.676 | -0.919 | 0.358 | 0.071 | 0.737 |
| TT vs. CC | Fixed | 10.34 | 0.350 | 0.975 | 0.854-1.113 | -0.377 | 0.706 | 0.763 | 0.106 | |
| TC vs. CC | Fixed | 15.31 | 0.313 | 1.032 | 0.948-1.123 | 0.728 | 0.467 | 0.548 | 0.543 | |
| TT+TC vs. CC | Fixed | 21.59 | 0.265 | 1.018 | 0.939-1.103 | 0.429 | 0.668 | 0.367 | 0.339 | |
| TT vs. TC+CC | Fixed | 0.00 | 0.423 | 0.958 | 0.845-1.085 | -0.675 | 0.500 | 1.000 | 0.127 | |
| Asian | T vs. C | Fixed | 9.75 | 0.344 | 1.493 | 1.259-1.772 | 4.595 | ≤0.001 | 0.734 | 0.351 |
| TT vs. CC | Fixed | 0.00 | 0.578 | 2.818 | 1.857-4.277 | 4.869 | ≤0.001 | 0.308 | 0.377 | |
| TC vs. CC | Fixed | 11.47 | 0.335 | 1.300 | 1.023-1.651 | 2.147 | 0.032 | 0.734 | 0.880 | |
| TT+TC vs. CC | Fixed | 3.27 | 0.376 | 1.483 | 1.183-1.859 | 3.415 | 0.001 | 1.000 | 0.547 | |
| TT vs. TC+CC | Fixed | 0.00 | 0.608 | 2.329 | 1.575-3.445 | 4.233 | ≤0.001 | 1.000 | 0.324 | |
| Cervical Cancer | ||||||||||
| Overall | T vs. C | Random | 73.78 | ≤0.001 | 1.132 | 0.956-1.341 | 1.434 | 0.151 | 0.620 | 0.232 |
| TT vs. CC | Random | 49.57 | 0.013 | 1.212 | 0.924-1.590 | 1.388 | 0.165 | 0.964 | 0.802 | |
| TC vs. CC | Random | 77.11 | ≤0.001 | 0.985 | 0.755-1.284 | -0.113 | 0.910 | 0.843 | 0.438 | |
| TT+TC vs. CC | Random | 79.60 | ≤0.001 | 1.095 | 0.842-1.423 | 0.677 | 0.498 | 0.752 | 0.215 | |
| TT vs. TC+CC | Random | 83.79 | ≤0.001 | 1.410 | 0.913-2.176 | 1.551 | 0.121 | 0.620 | 0.867 | |
| Ethnicity | ||||||||||
| Caucasian | T vs. C | Fixed | 45.38 | 0.160 | 1.071 | 0.891-1.287 | 0.733 | 0.464 | 1.000 | 0.431 |
| TT vs. CC | Fixed | 8.40 | 0.336 | 0.996 | 0.637-1.559 | -0.016 | 0.987 | 1.000 | 0.439 | |
| TC vs. CC | Fixed | 27.78 | 0.250 | 1.197 | 0.930-1.540 | 1.393 | 0.164 | 1.000 | 0.413 | |
| TT+TC vs. CC | Fixed | 44.89 | 0.163 | 1.162 | 0.912-1.480 | 1.214 | 0.225 | 1.000 | 0.406 | |
| TT vs. TC+CC | Fixed | 0.00 | 0.573 | 0.913 | 0.594-1.403 | -0.415 | 0.678 | 1.000 | 0.470 | |
| Asian | T vs. C | Random | 77.58 | ≤0.001 | 1.173 | 0.958-1.438 | 1.542 | 0.123 | 0.427 | 0.119 |
| TT vs. CC | Random | 55.31 | 0.008 | 1.295 | 0.944-1.776 | 1.602 | 0.109 | 0.854 | 0.530 | |
| TC vs. CC | Random | 79.26 | ≤0.001 | 0.967 | 0.705-1.325 | -0.211 | 0.833 | 0.945 | 0.264 | |
| TT+TC vs. CC | Random | 82.20 | ≤0.001 | 1.119 | 0.816-1.532 | 0.697 | 0.486 | 0.427 | 0.116 | |
| TT vs. TC+CC | Random | 86.09 | ≤0.001 | 1.594 | 0.961-2.642 | 1.806 | 0.071 | 0.582 | 0.924 | |
Figure 2.
Forest Plot for the Association of MTHFR 677C>T Polymorphism with Risk of Ovarian and Cervical Cancer. A, ovarian cancer (heterozygote model, TC vs. CC); B, cervical cancer (dominant model, TT+TC vs. CC)
Cervical Cancer
Table 2 also summarizes the results of association between the MTHFR 677C>T polymorphism and cervical cancer. Pooled data failed to show a significant association between MTHFR 677C>T polymorphism and risk of cervical cancer under all five genetic models, i.e., allele (T vs. C: OR = 1.132, 95% CI 0.956-1.341, p = 0.151), homozygote (TT vs. CC: OR = 1.212, 95% CI 0.924-1.590, p = 0.165); heterozygote (TC vs. CC: OR = 0.985, 95% CI 0.755-1.284, p = 0.910), dominant (TT+TC vs. CC: OR = 1.095, 95% CI 0.842-1.423, p = 0.498, Figure 2B), and recessive (TT vs. TC+CC: OR = 1.410, 95% CI 0.913-2.176, p = 0.121). Similarly, stratified analysis by ethnicity did not show a significant association between the MTHFR 677C>T polymorphism and cervical cancer under all five genetic models in Asians and Caucasians (Table 2).
Between-Study Heterogeneity Test
In the current meta-analysis, there was obvious between-study heterogeneity under four genetic models for ovarian cancer and all five genetic models for cervical cancer in overall population (Table 2). Therefore, we performed subgroup analyses by ethnicity to assess the potential source of between-study heterogeneity. In subgroup analysis, between-study heterogeneity was disappeared for ovarian cancer in the Caucasian and Asian population, as well as in the Caucasian subgroup for cervical cancer. The subgroup analysis results showed that ethnicity might be the major source of between-study heterogeneity for both ovarian and cervical cancer in the current meta-analysis.
Sensitivity Analysis
Moreover, sensitivity analysis was performed to assess the influence of each independent study on the pooled ORs by the sequential removal of each individual study form the analysis. However, the results of the sensitivity analysis for both ovarian cancer and cervical cancer did not materially changed by removing any of each individual study. Moreover, sensitivity analysis was carried out by excluding the HWE-violating study (Nandan et al., 2008) for cervical cancer. However, excluding the study did not significantly affect the pooled ORs for cervical cancer, indicating the robustness and reliability of this meta-analysis.
Publication Bias
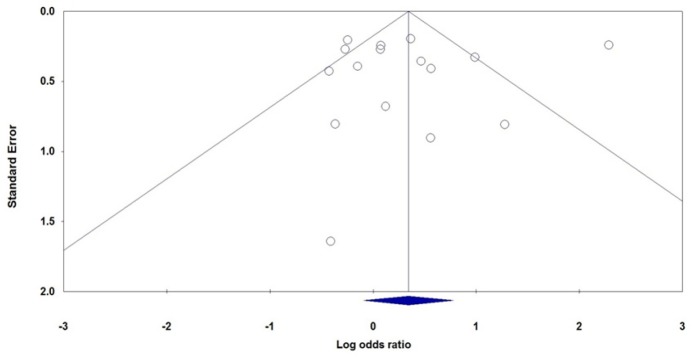
Begg’s and Egger’s tests were used to examine the potential publication bias in assessment of the association of MTHFR 677C>T polymorphism with ovarian and cervical cancer risk in all genetic models. No asymmetry was observed in the Begg’s rank correlation among the studies on ovarian and cervical cancer. Figure 3 showed the shape of the Begg’s funnel plots for association between MTHFR 677C>T polymorphism and risk of cervical cancer in recessive model (TT vs. TC+CC). Moreover, the Egger’s linear regression test did not show any statistical evidence of publication bias among the studies on ovarian and cervical cancer (Table 2).
Figure 3.
Begg’s Funnel Plot for Association between MTHFR 677C>T Polymorphism and Cervical Cancer Risk under the Recessive Genetic Model (TT vs. TC+CC).
Discussion
MTHFR is an important enzyme which has an important role in the regulation of methionine and homocysteine levels in folate metabolism. The MTHFR 677C>T polymorphism is one of the most studied functional polymorphism in cancer development, which could reduce the production of MTHFR and affect enzyme activity (Kamali et al., 2018). Thus, the current meta-analysis was performed to evaluate the association of MTHFR 677C>T polymorphism with susceptibility to ovarian and cervical cancers in women. Finally, 27 case-control studies including eleven studies on ovarian cancer and 16 studies on cervical cancer were selected. Therefore, the current meta-analysis was the largest scale study so far on MTHFR 677C>T polymorphism association with ovarian and cervical cancers. This meta-analysis pooled data revealed that the MTHFR 677C>T polymorphism was not associated with an increased risk of ovarian and cervical cancers in overall population. The negative results in this pooled analysis agrees with the previous meta-analysis on cervical cancer (Yi et al., 2016). However, He and Shen (2017) in meta-analysis of eight studies found that the MTHFR 677C>T polymorphism was associated with ovarian cancer risk. The main strength of this meta-analysis in comparison with the previous meta-analyses was the total number of cases and healthy controls selected. Therefore, this meta-analysis more power to detect the small effects of the polymorphism than previous studies. However, we recommend increasing the sample size in future studies in order to increase the power to detect small effects of the MTHFR 677C>T polymorphism on risk of ovarian and cervical cancers.
When stratified analysis by ethnicity was performed the results showed that the MTHFR 677C>T polymorphism was significantly associated with ovarian cancer in Asians, but not in Caucasians. Similarly, He and Shen (2017) showed that the MTHFR 677C>T polymorphism is a risk factor for ovarian cancer and also breast cancer in Asians. It seems that due to the some genetic and environmental differences between Asian and Caucasian populations, the MTHFR 677C>T polymorphism might play a different role in the development of ovarian cancer in the two populations. However, the subgroup analysis did not show a significant association between MTHFR 677C>T polymorphism and cervical cancer by ethnicity. However, the previous meta-analysis revealed that MTHFR 677C>T polymorphism was significantly associated with cervical cancer in Asians (Yi et al., 2016).
Between-study heterogeneity refers to the variation in study results between different studies, which could affects pooled results of a meta-analysis (Kamali et al., 2017) and a significant problem when interpreting of a meta-analysis (Forat-Yazdi et al., 2017; Jafari-Nedooshan et al., 2017). Several factors such as sample size, ethnicity, source of controls, genotyping methods, participants demographic and lifestyle might lead to the heterogeneity among studies (Mehdinejad et al., 2017; Yazdi et al., 2017). Thus, to explore the potential sources of heterogeneity among studies, we conducted subgroup analyses by ethnicity, cancer, source of control, HWE and genotyping methods. However, the subgroup analysis results showed that only ethnicity was the main source of heterogeneity in this meta-analysis.
To the best knowledge, this meta-analysis was the most comprehensive and convicting on the association of the MTHFR 677C>T polymorphism with susceptibility to ovarian and cervical cancer. However, there were some limitations in our meta-analysis which must be described. First, we have only focused those published studies in English and Chinese in the current meta-analysis. Second, in the current meta-analysis the number of studies and the sample size in the studies by other ethnicities such as Africans, Latinos and mixed populations were small. Therefore, the lack of power due to the small number of studies leaves it an open field by ethnicity. Third, our meta-analysis was not adjusted by the potential confounders, such as age, gender and lifestyle, because not all of the studies reported adjusted ORs potential confounders. Finally, the potential effect of MTHFR 677C>T polymorphism might be affected by gene-gene and gene-environment interactions. However, due to the lack of original data limited further evaluation of potential gene-gene and gene-environment interactions and also interactions of other polymorphisms of MTHFR gene.
In summary, this meta-analysis demonstrated that the MTHFR 677C>T polymorphism was not associated with an increased risk to ovarian and cervical cancer in overall population. However, MTHFR 677C>T polymorphism was significantly associated with ovarian cancer in Asians, but not in Caucasians. Moreover, considering the limitations of the study, large well-designed studies from different ethnicities should be conducted to provide a better understanding of the association of MTHFR 677C>T polymorphism with risk of ovarian and cervical cancer.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
References
- Abedinzadeh M, Zare-Shehneh M, Neamatzadeh H, et al. Association between MTHFR C677T polymorphism and risk of prostate cancer: Evidence from 22 studies with 10,832 cases and 11,993 controls. Asian Pac J Cancer Prev. 2015;16:4525–30. doi: 10.7314/apjcp.2015.16.11.4525. [DOI] [PubMed] [Google Scholar]
- Azarpira MR, Ghilian MM, Sobhan MR, et al. Association of MTHFR and TNF-α genes polymorphisms with susceptibility to Legg-Calve-Perthes disease in Iranian children: A case-control study. J Orthop. 2018;15:984–7. doi: 10.1016/j.jor.2018.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Enciso I, Martínez-Garza SG, Rojas-Martínez A, et al. The effect of MTHFR polymorphisms, pregnancy and first intercourse on cervical cancer in a population from the Northeastern Mexico. Rev Invest Clin. 2006;58:462–9. [PubMed] [Google Scholar]
- Fearon AE, Gould CR, Grose RP. FGFR signalling in women’s cancers. Int J Biochem Cell Biol. 2013;45:2832–42. doi: 10.1016/j.biocel.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Forat-Yazdi M, Jafari M, Kargar S, et al. Association between SULT1A1 Arg213His (Rs9282861) polymorphism and risk of breast cancer: A systematic review and meta-analysis. J Res Health Sci. 2017;17:e00396. [PubMed] [Google Scholar]
- Gao S, Liu N, Ma Y, et al. Methylenetetrahydrofolate reductase gene polymorphisms as predictive and prognostic biomarkers in ovarian cancer risk. Asian Pac J Cancer Prev. 2012;13:569–73. doi: 10.7314/apjcp.2012.13.2.569. [DOI] [PubMed] [Google Scholar]
- Gong J-M, Shen Y, Shan W-W, et al. The association between MTHFR polymorphism and cervical cancer. Sci Rep. 2018;8:7244. doi: 10.1038/s41598-018-25726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajiesmaeil M, Tafvizi F, Sarmadi S. The effect of methylenetetrahydrofolate reductase polymorphisms on susceptibility to human papilloma virus infection and cervical cancer. Infect Genet Evol. 2016;46:1–6. doi: 10.1016/j.meegid.2016.10.012. [DOI] [PubMed] [Google Scholar]
- He L, Shen Y. MTHFR C677T polymorphism and breast, ovarian cancer risk: a meta-analysis of 19,260 patients and 26,364 controls. Onco Targets Ther. 2017;10:227–38. doi: 10.2147/OTT.S121472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JPT. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippel PT von. The heterogeneity statistic I2 can be biased in small meta-analyses. BMC Med Res Methodol. 2015;15:35. doi: 10.1186/s12874-015-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari-Nedooshan J, Kargar S, Neamatzadeh H, et al. Lack of association of the fat mass and obesity associated (FTO) gene rs9939609 polymorphism with breast cancer risk: a systematic review and meta-analysis based on case - control studies. Asian Pac J Cancer Prev. 2017;18:1031–7. doi: 10.22034/APJCP.2017.18.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska A, Gronwald J, Menkiszak J, et al. Methylenetetrahydrofolate reductase polymorphisms modify BRCA1-associated breast and ovarian cancer risks. Breast Cancer Res Treat. 2007;104:299–308. doi: 10.1007/s10549-006-9417-3. [DOI] [PubMed] [Google Scholar]
- Kamali M, Hamadani S, Neamatzadeh H, et al. Association of XRCC2 rs3218536 polymorphism with susceptibility of breast and ovarian cancer: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:1743–9. doi: 10.22034/APJCP.2017.18.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamali M, Hantoushzadeh S, Borna S, et al. Association between thrombophilic genes polymorphisms and recurrent pregnancy loss susceptibility in the Iranian population: a systematic review and meta-analysis. Iran Biomed J. 2018;22:78–89. doi: 10.22034/ibj.22.2.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Kim JW, Kang GH, et al. Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecol Oncol. 2005;96:173–80. doi: 10.1016/j.ygyno.2004.09.031. [DOI] [PubMed] [Google Scholar]
- von Keyserling H, Bergmann T, Schuetz M, et al. Analysis of 4 single-nucleotide polymorphisms in relation to cervical dysplasia and cancer development using a high-throughput ligation-detection reaction procedure. Int J Gynecol Cancer. 2011;21:1664–71. doi: 10.1097/IGC.0b013e31822b6299. [DOI] [PubMed] [Google Scholar]
- Kohaar I, Kumar J, Thakur N, et al. Homocysteine levels are associated with cervical cancer independent of methylene tetrahydrofolate reductase gene (MTHFR) polymorphisms in Indian population. Biomarkers. 2010;15:61–8. doi: 10.3109/13547500903295881. [DOI] [PubMed] [Google Scholar]
- Lambropoulos AF, Agorastos T, Foka ZJ, et al. Methylenetetrahydrofolate reductase polymorphism C677T is not associated to the risk of cervical dysplasia. Cancer Lett. 2003;191:187–91. doi: 10.1016/s0304-3835(02)00675-4. [DOI] [PubMed] [Google Scholar]
- Ma H, Jin G, Hu Z, et al. Variant genotypes of CDKN1A and CDKN1B are associated with an increased risk of breast cancer in Chinese women. Int J Cancer. 2006;119:2173–8. doi: 10.1002/ijc.22094. [DOI] [PubMed] [Google Scholar]
- Maheshwari A, Kumar N, Mahantshetty U. Gynecological cancers: A summary of published Indian data. South Asian J Cancer. 2016;5:112–20. doi: 10.4103/2278-330X.187575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor H, Naheed H, Ahmad K, et al. Pattern of gynaecological malignancies in south western region of Pakistan: An overview of 12 years. Biomed Rep. 2017;7:487–91. doi: 10.3892/br.2017.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdinejad M, Sobhan MR, Mazaheri M, et al. Genetic association between ERCC2, NBN, RAD51 gene variants and osteosarcoma risk: A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;2017:18. doi: 10.22034/APJCP.2017.18.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowska A, Myka M, Lianeri M, et al. Folate and choline metabolism gene variants and development of uterine cervical carcinoma. Clin Biochem. 2011;44:596–600. doi: 10.1016/j.clinbiochem.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Nandan NK, Wajid S, Biswas S, et al. Allelic variations in 5, 10-methylenetetrahydrofolate reductase gene and susceptibility to cervical cancer in Indian women. Drug Metab Lett. 2008;22:18–22. doi: 10.2174/187231208783478498. [DOI] [PubMed] [Google Scholar]
- Özkılıç AÇ, Çetin A, Bayoğlu B, et al. Association between MTHFR C677T polymorphism and folate, vitamin B12, homocysteine, and DNA fragmentation in patients with ovarian cancer. Turk Biyokim Derg. 2016;41:459–65. [Google Scholar]
- Pawlik P, Mostowska A, Lianeri M, et al. Folate and choline metabolism gene variants in relation to ovarian cancer risk in the Polish population. Mol Biol Rep. 2012;39:5553–60. doi: 10.1007/s11033-011-1359-0. [DOI] [PubMed] [Google Scholar]
- Prasad VVTS, Wilkhoo H. Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Onkologie. 2011;34:422–6. doi: 10.1159/000331131. [DOI] [PubMed] [Google Scholar]
- Qin X, Lu Y, Qin A, et al. Vitamin D receptor BsmІ polymorphism and ovarian cancer risk. Int J Gynecol Cancer. 2013;23:1178–83. doi: 10.1097/IGC.0b013e31829db839. [DOI] [PubMed] [Google Scholar]
- Rocha TAH, Silva NC da, Thomaz EBAF, et al. Primary health care and cervical cancer mortality rates in Brazil: A longitudinal ecological study. J Ambul Care Manage. 2017;40:24–34. doi: 10.1097/JAC.0000000000000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozycka A, Jagodzinski P, Kozubski W, et al. Homocysteine level and mechanisms of injury in Parkinson’s disease as related to MTHFR, MTR, and MTHFD1 Genes polymorphisms and LDopa Treatment. Curr Genomics. 2014;14:534–42. doi: 10.2174/1389202914666131210210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekari M, Sobti RC, Kordi Tamandani DM, et al. Impact of methylenetetrahydrofolate reductase (MTHFR) codon (677) and methionine synthase (MS) codon (2756) on risk of cervical carcinogenesis in North Indian population. Arch Gynecol Obstet. 2008;278:517–24. doi: 10.1007/s00404-008-0623-6. [DOI] [PubMed] [Google Scholar]
- Sull JW, Jee SH, Yi S, et al. The effect of methylenetetrahydrofolate reductase polymorphism C677T on cervical cancer in Korean women. Gynecol Oncol. 2004;95:557–63. doi: 10.1016/j.ygyno.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Terry KL, Tworoger SS, Goode EL, et al. MTHFR polymorphisms in relation to ovarian cancer risk. Gynecol Oncol. 2010;95:557–63. doi: 10.1016/j.ygyno.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Kim MK, Lee JK, et al. Common polymorphisms in methylenetetrahydrofolate reductase gene are associated with risks of cervical intraepithelial neoplasia and cervical cancer in women with low serum folate and vitamin B12. Cancer Causes Control. 2011;22:63–72. doi: 10.1007/s10552-010-9675-6. [DOI] [PubMed] [Google Scholar]
- Torre LA, Islami F, Siegel RL, et al. Global cancer in women: Burden and trends. Cancer Epidemiol Biomarkers Prev. 2017;26:444–457. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- Webb PM, Ibiebele TI, Hughes MC, et al. Australian cancer study (Ovarian Cancer), Australian ovarian cancer study group Folate and related micronutrients folate-metabolising genes and risk of ovarian cancer. Eur J Clin Nutr. 2011;65:1133–40. doi: 10.1038/ejcn.2011.99. [DOI] [PubMed] [Google Scholar]
- Wilcken B. Geographical and ethnic variation of the 677C>T allele of 5,10 methylenetetrahydrofolate reductase (MTHFR): findings from over 7000 newborns from 16 areas world wide. J Med Genet. 2003;40:619–25. doi: 10.1136/jmg.40.8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang J, Zuo W. Association between genetic polymorphisms of methylenetetrahydrofolate reductase C677T and susceptibility to ovarian cancer. Prog Obstet Gynecol. 2007;16:811–3. [Google Scholar]
- Yang F, Zhou Y, Jiang Y. Study on the correlation between polymorphism of MTHFR gene and the pathogenesis of cervical cancer. Maternal Child Health Care China. 2011;26:4087–9 (article in Chinese). [Google Scholar]
- Yazdi MM, Jamalaldini MH, Sobhan MR, et al. Association of ESRα Gene Pvu II T>C, XbaI A>G and BtgI G>A polymorphisms with Knee osteoarthritis susceptibility: A systematic review and meta-analysis based on 22 case-control studies. Arch Bone Jt Surg. 2017;5:351–62. [PMC free article] [PubMed] [Google Scholar]
- Yi K, Yang L, Lan Z, et al. The association between MTHFR polymorphisms and cervical cancer risk: a system review and meta analysis. Arch Gynecol Obstet. 2016;294:579–88. doi: 10.1007/s00404-016-4037-6. [DOI] [PubMed] [Google Scholar]
- Yu Z, Liu Q, Huang C, et al. The interleukin 10 −819C/T polymorphism and cancer risk: A HuGE review and meta-analysis of 73 studies including 15,942 cases and 22,336 controls. OMICS. 2013;17:200–14. doi: 10.1089/omi.2012.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu W, Hao Q, et al. Folate intake and methylenetetrahydrofolate reductase gene polymorphisms as predictive and prognostic biomarkers for ovarian cancer risk. Int J Mol Sci. 2012;13:4009–20. doi: 10.3390/ijms13044009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X-M, Sun W-F. Association between matrix metalloproteinases polymorphisms and ovarian cancer risk: A meta-analysis and systematic review. PLoS One. 2017;12: e0185456. doi: 10.1371/journal.pone.0185456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoodsma M, Nolte IM, Schipper M, et al. Methylenetetrahydrofolate reductase (MTHFR) and susceptibility for (pre)neoplastic cervical disease. Hum Genet. 2005;116:247–54. doi: 10.1007/s00439-004-1233-4. [DOI] [PubMed] [Google Scholar]