Abstract
Enterovirus D68 (EV-D68) was detected in 93 patients from five European countries between 1 January 2019 and 15 January 2020, a season with expected low circulation. Patients were primarily children (n = 67, median age: 4 years), 59 patients required hospitalisation and five had severe neurologic manifestations. Phylogenetic analysis revealed two clusters in the B3 subclade and subclade A2/D. This circulation of EV-D68 associated with neurological manifestations stresses the importance of surveillance and diagnostics beyond expected peak years.
Keywords: enterovirus D68, acute flaccid myelitis, severe respiratory infection, novel strains, surveillance
Enterovirus D68 (EV-D68) is primarily a respiratory virus. Previously published data have suggested circulation with a biennial epidemic cycle in Europe and North America [1-3], but surveillance is not consistent. The Danish enterovirus surveillance detected two cases of EV-D68 infection in August 2019 and a further case in early September. Colleagues in other European countries were contacted, and France, Germany, the Netherlands and Sweden responded that they had also seen cases. Here we report on the start of seasonal circulation of EV-D68 in five European countries, a circulation which is still ongoing with further cases detected since the initial submission of this report.
Epidemiological trend and description of cases
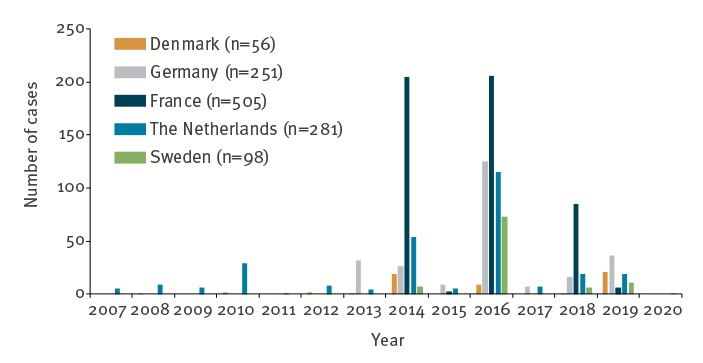
Denmark, France, Germany, the Netherlands and Sweden have seen continuous circulation of EV-D68 with variable upsurges since 2007 (Figure 1). However, in 2018, only one case was detected in Denmark, 16 in Germany, 19 in the Netherlands and six in Sweden, whereas other European countries experienced large outbreaks with between 21 and at least 114 cases (as reported by November 2018 by Public Health Wales), overall including at least 11 cases with AFM [4-8].
Figure 1.
Yearly variation in detection of enterovirus D68 in five European countries, 1 January 2007–15 January 2020 (n = 1,191)
For Sweden, cases in 2014 include only those confirmed by sequencing, and only a limited number of samples were screened (n = 30). Data from Germany represent five sites for the period 2013 to 2018 (Bonn, Düsseldorf, Freiburg, Leipzig and Würzburg), but only three for 2019 and 2020 (Bonn, Freiburg and Leipzig).
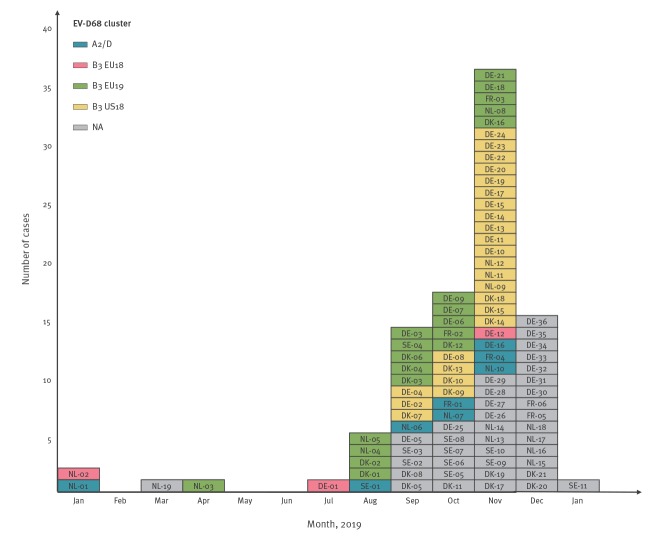
A total of 93 EV-D68 infections were reported between 1 January 2019 and 15 January 2020 (Denmark n = 21, France n = 6, Germany n = 36, the Netherlands n = 19, Sweden n = 11). Epidemiological and clinical information was collected for cases where possible (Table 1). As surveillance samples and data are collected retrospectively, data for December 2019 and January 2020 are not complete.
Table 1. Cases of enterovirus D68 in five European countries, 1 January 2019–15 January 2020 (n = 93).
| Case number | Sample date (month-year) | Age (years) | Sex | Hospitalised | Pre-existing disease | Travel abroad < 2 weeks before sampling | Fever | Enteric symptoms | Respiratory symptoms | Neurological symptoms | Dermatological symptoms | Co-infections |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DK-01 | Aug-19 | 15 | F | Y | N | N | Y | None | Bronchiolitis | N | N | N |
| DK-02 | Aug-19 | 2 | F | N | N | N | Y | None | Common cold | N | N | N |
| DK-03 | Sep-19 | 3 | F | Y | Y | N | Y | None | Acute bronchitis | N | N | N |
| DK-04 | Sep-19 | 1 | M | Y | N | N | Y | None | ILI | AFM | NA | Adenovirus, parechovirus |
| DK-05 | Sep-19 | 2 | M | Y | Y | N | Y | None | Obstructive bronchiolitis | N | Rash | Adenovirus |
| DK-06 | Sep-19 | 29 | F | Y | Y | N | Y | None | Common cold | Cranial nerve palsies, dysphagia, dysatria | N | N |
| DK-07 | Sep-19 | 0 | F | N | N | NA | Y | None | ILI | N | Rash | N |
| DK-08 | Sep-19 | 0 | F | Y | N | NA | Y | Diarrhoea, rumbling stomach | Common cold | N | N | Bordetella pertussis |
| DK-09 | Oct-19 | 0 | F | Y | Y | N | Y | None | Pneumonia | N | N | Moraxella catarrhalis |
| DK-10 | Oct-19 | 27 | M | Y | Y | N | Y | Diarrhoea | Pneumonia | Reduced strength in right leg, attenuated patellar and plantar reflexes | N | Suspected bacterial infection |
| DK-11 | Oct-19 | 50 | F | N | NA | NA | N | None | Common cold | N | N | N |
| DK-12 | Oct-19 | 30 | M | N | N | N | Y | None | ILI | N | N | N |
| DK-13 | Oct-19 | 0 | F | Y | N | N | N | None | Pneumonia | N | N | Haemophilus influenzae, Moraxella catarrhalis |
| DK-14 | Nov-19 | 2 | M | Y | Y | N | Y | None | Pneumonia | N | N | N |
| DK-15 | Nov-19 | 2 | F | Y | Y | N | Y | N | Obstructive bronchiolitis | N | N | Parechovirus, adenovirus, Haemophilus influenzae |
| DK-16 | Nov-19 | 1 | M | Y | N | NA | N | Diarrhoea | N | N | N | N |
| DK-17 | Nov-19 | 16 | F | Y | N | N | N | Nausea, vomiting | Y | Ataxic cerebral palsy, diplopia, hypoaesthesia | N | N |
| DK-18 | Nov-19 | 2 | M | Y | Y | NA | Y | None | Common cold | N | N | Haemophilus influenzae, Pseudomonas aeruginosa |
| DK-19 | Nov-19 | 2 | M | Y | Y | NA | N | N | Respiratory distress | N | N | N |
| DK-20 | Dec-19 | 2 | F | Y | Y | NA | NA | NA | Bilateral pneumonia | N | N | Pneumococcus |
| DK-21 | Dec-19 | 1 | M | N | Y | NA | NA | NA | Obstructive bronchiolitis | N | NA | Rhinovirus C22 Haemophilus influenzae |
| NL-01 | Jan-19 | 61 | M | NA | NA | Y | NA | NA | NA | NA | NA | NA |
| NL-02 | Jan-19 | 65 | F | N | Y | N | NA | Yes | N | N | N | NA |
| NL-03 | Apr-19 | 1 | F | Y | NA | Y | NA | NA | Respiratory insufficiency | NA | NA | NA |
| NL-04 | Aug-19 | 5 | F | Y | NA | Y | NA | NA | Respiratory insufficiency | NA | NA | NA |
| NL-05 | Aug-19 | 75 | F | NA | NA | NA | NA | NA | Pneumonia | NA | NA | NA |
| NL-06 | Sep-19 | 46 | M | N | N | N | Y | Diarrhoea | ILI | NA | NA | NA |
| NL-07 | Oct-19 | 61 | F | N | N | N | N | N | ARI, dyspnoea | NA | NA | NA |
| NL-08 | Nov-19 | 1 | F | NA | NA | NA | NA | N | Common cold, dyspnoea | NA | NA | NA |
| NL-09 | Nov-19 | 1 | M | NA | NA | N | Y | N | ARI, dyspnoea | NA | NA | Respiratory syncytial virus type A |
| NL-10 | Nov-19 | 89 | M | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NL-11 | Nov-19 | 4 | F | NA | NA | NA | NA | NA | Dyspnoea | NA | NA | NA |
| NL-12 | Nov-19 | 2 | F | NA | NA | N | Y | N | ARI, dyspnoea | NA | NA | NA |
| NL-13 | Nov-19 | 0 | M | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NL-14 | Nov-19 | 2 | F | NA | NA | NA | NA | NA | Pneumonia and bronchial hyperreactivity | NA | NA | NA |
| NL-15 | Dec-19 | 4 | M | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NL-16 | Dec-19 | 53 | M | NA | N | N | Y | N | ILI, dyspnoea | NA | NA | NA |
| NL-17 | Dec-19 | 7 | F | NA | N | N | N | N | ARI | NA | NA | NA |
| NL-18 | Dec-19 | 22 | M | NA | N | N | N | N | ILI | NA | NA | NA |
| NL-19 | Mar-19 | 60 | M | N | N | N | Y | N | ILI, dyspnoea | N | NA | Influenza virus A(H1N1)pdm09 |
| SE-01 | Aug-19 | 70 | F | NA | NA | NA | NA | NA | NA | NA | NA | Haemophilus influenzae |
| SE-02 | Sep-19 | 2 | M | NA | NA | NA | NA | NA | NA | NA | NA | Rhinovirus |
| SE-03 | Sep-19 | 0 | F | N | N | N | Y | N | Y | N | NA | N |
| SE-04 | Sep-19 | 5 | F | NA | NA | NA | NA | NA | NA | NA | NA | N |
| SE-05 | Oct-19 | 61 | M | N | N | Y | Y | N | Y | N | NA | Streptococcus pneumoniae |
| SE-06 | Oct-19 | 2 | M | NA | NA | NA | NA | NA | NA | NA | NA | N |
| SE-07 | Oct-19 | 4 | F | Y | N | N | N | N | Y | N | NA | N |
| SE-08 | Oct-19 | 2 | M | Y | N | N | Y | N | Y | N | NA | Adenovirus Rhinovirus Haemophilus influenzae |
| SE-09 | Nov-19 | 2 | M | NA | NA | NA | NA | NA | NA | NA | NA | N |
| SE-10 | Nov-19 | 5 | M | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| SE-11 | Jan-20 | 69 | M | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| FR-01 | Oct-19 | 0 | F | Y | N | N | Y | N | Bronchiolitis | N | N | N |
| FR-02 | Oct-19 | 33 | M | Y | Y | N | Y | N | Pneumonia with acute respiratory distress | N | N | Streptococcus pneumoniae |
| FR-03 | Nov-19 | 1 | M | Y | N | N | Y | Diarrhoea | N | AFM | N | N |
| FR-04 | Nov-19 | 0 | F | Y | N | N | Y | Diarrhoea | Bronchiolitis | N | N | N |
| FR-05 | Dec-19 | 51 | F | Y | N | Y | Y | N | Bronchitis | N | N | N |
| FR-06 | Dec-19 | 66 | M | Y | Y | N | Y | N | Respiratory distress | N | N | N |
| DE-01 | Jul-19 | 1 | M | Y | N | N | Y | N | Obstructive bronchitis | N | N | N |
| DE-02 | Sep-19 | 1 | M | Y | N | N | Y | N | Obstructive bronchitis | N | N | Rhinovirus C17 |
| DE-03 | Sep-19 | 20 | F | Y | Y | N | Y | Y | Y | N | N | Multiple bacterial and viral infections |
| DE-04 | Sep-19 | 9 | M | Y | Y | N | Y | N | Asthma exacerbation | N | N | N |
| DE-05 | Sep-19 | 1 | M | Y | N | N | Y | N | Obstructive bronchitis | N | N | Rhinovirus A49 |
| DE-06 | Oct-19 | 0 | M | Y | N | N | N | N | Obstructive bronchitis | N | N | N |
| DE-07 | Oct-19 | 0 | F | Y | N | N | Y | N | Y | N | N | N |
| DE-08 | Oct-19 | 6 | F | Y | N | N | Y | N | Y | Bilateral lower limb paralysis | N | N |
| DE-09 | Oct-19 | 12 | M | Y | N | NA | Y | NA | NA | NA | NA | N |
| DE-10 | Nov-19 | 2 | M | Y | N | N | Y | N | Obstructive bronchitis | N | N | N |
| DE-11 | Nov-19 | 7 | F | Y | Y | N | Y | N | Asthma exacerbation | N | N | N |
| DE-12 | Nov-19 | 2 | M | Y | N | N | Y | N | Obstructive bronchitis | N | N | N |
| DE-13 | Nov-19 | 5 | F | Y | Y | N | Y | N | Acute bronchitis | Seizures | N | Clostridioides difficile |
| DE-14 | Nov-19 | 22 | F | Y | N | N | N | N | Obstructive bronchitis | N | N | N |
| DE-15 | Nov-19 | 1 | F | Y | N | N | Y | N | Acute bronchitis | N | N | N |
| DE-16 | Nov-19 | 8 | F | Y | Y | NA | NA | N | Y | NA | NA | Adenovirus |
| DE-17 | Nov-19 | 78 | F | N | Y | N | N | N | Y | N | N | N |
| DE-18 | Nov-19 | 8 | M | Y | N | NA | NA | N | Y | NA | N | N |
| DE-19 | Nov-19 | 0 | F | Y | Y | N | Y | N | Y | N | N | N |
| DE-20 | Nov-19 | 5 | F | Y | N | N | N | N | Obstructive bronchitis | N | N | N |
| DE-21 | Nov-19 | 4 | F | Y | Y | N | Y | N | Y | N | N | N |
| DE-22 | Nov-19 | 2 | M | Y | NA | NA | Y | NA | Y | NA | NA | N |
| DE-23 | Nov-19 | 29 | M | Y | Y | N | N | N | Y | N | N | N |
| DE-24 | Nov-19 | 7 | M | Y | NA | NA | NA | NA | Y | NA | NA | N |
| DE-25 | Oct-19 | 74 | F | Y | Y | N | N | N | Y | N | NA | N |
| DE-26 | Nov-19 | 71 | M | N | Y | Y | N | N | N | N | N | N |
| DE-27 | Nov-19 | 1 | M | Y | N | N | N | N | Obstructive bronchitis | N | N | N |
| DE-28 | Nov-19 | 3 | M | Y | N | NA | Y | N | Y | N | N | N |
| DE-29 | Nov-19 | 40 | F | N | N | NA | Y | N | Y | NA | NA | N |
| DE-30 | Dec-19 | 2 | F | Y | N | N | N | N | Obstructive bronchitis | N | N | N |
| DE-31 | Dec-19 | 1 | F | Y | N | N | Y | N | Obstructive bronchitis | N | N | N |
| DE-32 | Dec-19 | 6 | M | Y | N | N | Y | N | Pneumonia | N | N | N |
| DE-33 | Dec-19 | 2 | M | Y | N | N | Y | N | Bronchitis and pneumonia | N | N | Rhinovirus C55 |
| DE-34 | Dec-19 | 1 | M | Y | N | N | N | Y | Pneumonia | N | N | Bocavirus 1 Respiratory syncytial virus |
| DE-35 | Dec-19 | 5 | F | Y | Y | N | N | N | Obstructive bronchitis | Seizures | N | N |
| DE-36 | Dec-19 | 2 | M | Y | N | Y | N | N | URTI | N | N | N |
AFM: acute flaccid myelitis; ARI: acute respiratory infection; DK: Denmark; FR: France; DE: Germany; ILI: influenza-like illness; N: no; NA: not available; NL: the Netherlands; SE: Sweden; URTI: upper respiratory infection; Y: yes.
Cases were identified through enterovirus surveillance (n = 31), rhinovirus surveillance (n = 3), influenza and respiratory infection community surveillance (n = 12) or hospital-based diagnostics of respiratory infections (n = 47). Following diagnostic testing of respiratory samples for enterovirus and/or rhinovirus using commercial or in-house assays, EV-D68 cases were identified by either EV-D68-specific real-time PCR or partial sequencing of the VP1 and/or VP4/VP2 region of the genome [9-11]. Most of the cases were identified during the usual enterovirus season starting in late summer (August n = 5, September n = 14, October n = 17, November n = 36 and December n=15; Figure 2).
Figure 2.
Epicurve of enterovirus D68 cases by phylogenetic cluster, five European countries, 1 January 2019–15 January 2020 (n = 93)
DK: Denmark; EU18: 2019 sequences with most recent common ancestor in large B3 cluster of European sequences from 2018; EU19: novel B3 cluster identified in this study; FR: France; DE: Germany; NA: sequence data for clade allocation not available or phylogenetic analysis not done; NL: the Netherlands; SE: Sweden; US18: 2019 sequences with most recent common ancestor in large B3 cluster predominantly containing sequences from the United States in 2018.
For clade allocation of cases see section Phylogenetic analysis of enterovirus D68 strains.
Of the Dutch cases, two became ill in Turkey and were diagnosed with EV-D68 after being hospitalised for respiratory support upon their return to the Netherlands. One Swedish patient had a recent travel history to East Asia and one French patient had travelled to Portugal. Most of the cases were children (n = 67), with a median age of 4 years (range: 16 days–89 years). Forty-six patients were female.
Clinical manifestations of cases
Five patients, from Denmark, France and Germany, presented with severe neurological symptoms. Of these, one was a 1 year-old, previously healthy boy who presented with acute flaccid myelitis (AFM) following a febrile respiratory infection. The paralysis was asymmetric, included all four limbs and the torso, with severe paraesthesia in affected limbs. The second patient was a 15-month-old boy who presented with AFM following a digestive prodromal illness. The third patient was a 29-year-old woman who presented with cranial nerve palsy. She underwent a caesarean section in week 40 of pregnancy because of the acute neurological symptoms. The fourth patient was a 6-year-old girl who presented with paralysis of both legs and the bladder. The fifth patient was a 16-year-old girl who presented with loss of balance and coordination, double vision and loss of sensation. In addition to the severe neurological cases, one further patient, a 27-year-old man, presented with a discrete unilateral paresis of the right leg. Two patients suffered seizures, most likely related to underlying conditions.
The clinical manifestations for the remaining patients ranged from mild cold-like symptoms of the upper respiratory tract to severe pneumonia requiring continuous positive airway pressure and respirator support (Table 1). Fifty-nine patients (among the 73 for whom this information was available) required hospitalisation, either because of severity of symptoms or underlying medical conditions. Twenty-one patients had underlying medical conditions, including asthma, cancer, non-HIV-related immunodeficiency, epilepsy and trisomy 14 mosaicism with growth retardation, cognitive impairment and multiple malformations. For one Dutch case, EV-D68 was identified in bronchoalveolar lavage and this patient also had a pneumococci-positive antigen test in urine. The patient died after 11 days of hospitalisation from bilateral pneumonia.
Phylogenetic analysis of enterovirus D68 strains
Full- or nearly full-length genome sequencing was successfully carried out for seven strains (Denmark and the Netherlands: in-house protocols, Sweden [12]). Sequences which were available up to and including 6 December 2019 are available on GenBank (Table 2).
Table 2. GenBank accession numbers and available sequence for phylogenetic analysis, enterovirus D68 strains from five European countries, 1 January–6 December 2019 (n = 67).
| Case number | GenBank accession number | Sequence |
|---|---|---|
| DK-01 | MN896974 | Partial genome |
| DK-02 | MN896975 | Partial genome |
| DK-03 | NA | NA |
| DK-04 | MN896976 | Partial VP1 |
| DK-05 | NA | NA |
| DK-06 | MN896977 | Partial VP1 |
| DK-07 | MN896978 | Full genome |
| DK-08 | NA | NA |
| DK-09 | MN896979 | Partial VP4/VP2 |
| DK-10 | MN896980 | Partial VP1 |
| DK-11 | NA | NA |
| DK-12 | MN896981 | Partial VP1 |
| DK-13 | MN896982 | Partial VP1 |
| DK-14 | MN896983 | Partial VP1 |
| DK-15 | MN896985 | Partial VP1 |
| DK-16 | MN896984 | Partial VP1 |
| DK-17 | NA | NA |
| DK-18 | MN896986 | Partial VP1 |
| NL-01 | MN764886 | Partial VP1 |
| NL-02 | MN764887 | Partial VP1 |
| NL-03 | MN764888 | Partial VP1 |
| NL-04 | MN764889 | Partial VP1 |
| NL-05 | MN726800 | Full genome |
| NL-06 | MN726801 | Full genome |
| NL-07 | MN726798 | Full genome |
| NL-08 | MN726799 | Partial VP1 |
| NL-09 | MN809623 | Complete VP1 |
| NL-10 | MN809624 | Partial VP1 |
| NL-11 | MN809625 | Partial VP1 |
| NL-12 | MN809626 | Partial VP1 |
| SE-01 | MN935869 | Full genome |
| SE-02 | NA | NA |
| SE-03 | NA | NA |
| SE-04 | MN935870 | Full genome |
| SE-05 | NA | NA |
| SE-06 | NA | NA |
| SE-07 | NA | NA |
| SE-08 | NA | NA |
| SE-09 | NA | NA |
| FR-01 | LR743438 | Complete VP1 |
| FR-02 | LR743439 | Complete VP1 |
| FR-03 | LR743440 | Complete VP1 |
| FR-04 | LR743441 | Complete VP1 |
| DE-01 | MN814240 | Partial VP1 |
| DE-02 | MN814241 | Partial VP1 |
| DE-03 | MN814242 | Partial VP1 |
| DE-04 | MN814243 | Partial VP1 |
| DE-05 | NA | NA |
| DE-06 | MN814244 | Partial VP1 |
| DE-07 | MN832475 | Partial VP4/VP2 |
| DE-08 | MN812202 | Partial VP1 |
| DE-09 | MN832476 | Partial VP4/VP2 |
| DE-10 | MN814245 | Partial VP1 |
| DE-11 | MN814246 | Partial VP1 |
| DE-12 | MN814247 | Partial VP1 |
| DE-13 | MN814248 | Partial VP1 |
| DE-14 | MN814249 | Partial VP1 |
| DE-15 | MN814250 | Partial VP1 |
| DE-16 | MN832477 | Partial VP4/VP2 |
| DE-17 | MN832478 | Partial VP4/VP2 |
| DE-18 | MN832479 | Partial VP4/VP2 |
| DE-19 | MN832480 | Partial VP4/VP2 |
| DE-20 | MN814251 | Partial VP1 |
| DE-21 | MN832481 | Partial VP4/VP2 |
| DE-22 | MN832482 | Partial VP4/VP2 |
| DE-23 | MN832483 | Partial VP4/VP2 |
| DE-24 | MN832484 | Partial VP4/VP2 |
NA: not available.
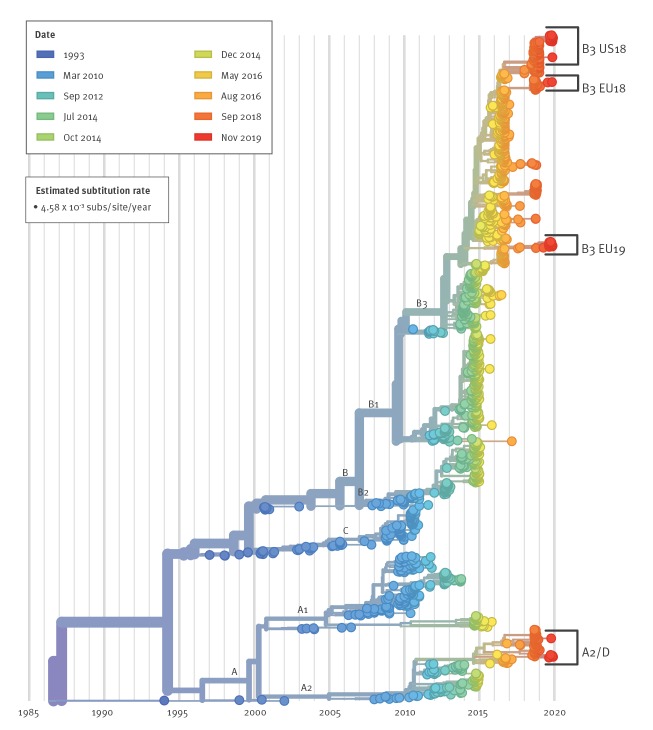
Phylogenetic analysis of VP1 sequence data was carried out using the Nextstrain augur pipeline [13]. We included samples from this study with ≥ 300 bp in VP1 (Table 2), alongside all available VP1 sequences in GenBank of ≥ 700 bp, randomly down-sampled to 20 samples per country per month to avoid sampling bias and overrepresentation of some countries (particularly during the 2014 and 2016 epidemics; no 2019 samples were down-sampled). The code is available at https://github.com/enterovirus-phylo/evd68-2019; the analysis can be viewed at https://nextstrain.org/community/enterovirus-phylo/evd68-2019/vp1-300. This analysis will be updated with new sequence data as this becomes available. EV-D68 has been characterised into the major clades A, B, and C, with A and B divided into the subclades A1–A2, and B1–B3. Some studies designate A2 as D and subdivide it into D1 and D2 (Figure 3).
Figure 3.
Phylogenetic analysis of enterovirus D68 with Nextstrain using partial VP1 sequences (n = 1,693)
EU18: 2019 sequences with most recent common ancestor in large B3 cluster of European sequences from 2018; EU19: novel B3 cluster identified in this study; US18: 2019 sequences with most recent common ancestor in large B3 cluster predominantly containing sequences from the United States in 2018.
For sequences with a sample date up to and including 2018, VP1 fragments of ≥ 700 bp in length were included, for sequences from 2019, fragments of ≥ 300 bp were included.
Divergence between major subclade divisions is shown, sequences are colour-coded according to date of sampling. Brackets identify where sequences reported in this study cluster with previous samples.
The dominant subclades have varied between years of upsurge: the 2014 outbreak was dominated by B1 strains, whereas the 2016 and 2018 outbreaks consisted only of the B3 and A2/D subclades. Some clades and subclades may no longer be circulating, as evidenced by the lack of clade C strains since 2010 and the absence of A1, B1 and B2 strains since 2014/15. In the current study, there was a close genetic relationship between 15 strains from all five countries (Figure 4A). These formed a distinct cluster within clade B3 (EU19 in Figures 2 and 3), which did not include previously detected B3 strains from these countries and was not well represented in the 2016 or 2018 epidemic (see Nextstrain analysis, https://nextstrain.org/community/enterovirus-phylo/evd68-2019/vp1-300). Fourteen of the sequences clustered closely, with an estimated most recent common ancestor (MRCA) in September 2018, suggesting that this cluster may have circulated in Europe during the 2018 season. Four German strains clustered within this group in the VP4/VP2 region (https://nextstrain.org/community/enterovirus-phylo/evd68-2019/vp4vp2). NL-03 (infection presumably acquired in Turkey) was more distant, with an estimated MRCA with the larger 2019 European B3 subclade of January 2016.
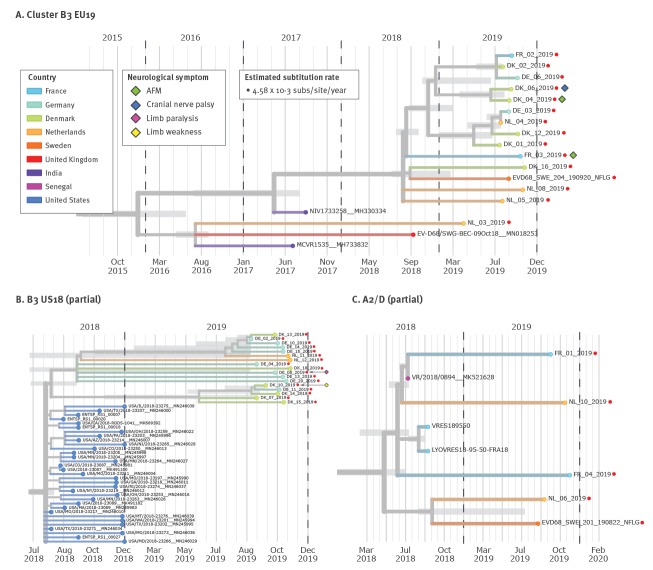
Figure 4.
Phylogenetic analysis of three clusters of partial VP1 sequences of enterovirus D68 strains using Nextstrain, five European countries, 1 January–6 December 2019 (n = 36)
AFM: acute flaccid myelitis.
Zoomed in view of three phylogenetic clusters containing 2019 EV-D68 sequences. Nodes are coloured by country; the colour legend is the same for all three panels. Date estimate confidence intervals are shown as light grey bars for nodes leading to 2019 strains. Years are marked with dashed vertical lines. Red dots: sequences from this study; diamond shapes: neurological symptoms for 2019 cases.
Panel A shows a novel 2019 B3 cluster which is poorly represented in previous years. Panel B shows two 2019 clusters which descend from a large, United States-dominated, 2018 B3 cluster. The partial VP1 sequences of the upper seven strains with nodes having wide date estimate confidence intervals are identical. Panel C shows 2019 A2/D strains clustering with strains that circulated in France and Senegal in 2018.
Seventeen other strains (US18 in Figures 2 and 3) formed two further clusters within clade B3, one cluster with five strains and an MRCA of May 2019, the other with 12 strains and an MRCA of September 2018 (Figure 4B). These two clusters had a common MRCA in August 2018. This common ancestor fell within a large American cluster circulating in 2018, which previously contained only three non-American sequences. Strains NL-09, DK-09 and four German strains also fell within this American cluster; NL-09 separately from the other 2019 VP1 samples and the other five in the VP4/VP2 region (Figure 2, sequence data not shown). Samples DE-01 and DE-12 formed a pair and were descended from a cluster of B3 strains that circulated in Europe in 2018, including one of the Dutch cases detected in January (NL-02; cluster EU18 in Figures 2 and 3). The other Dutch case from January (NL-07; cluster A2/D in Figures 2 and 3), along with five other strains, belonged to clade A2/D (Figure 4C) and rooted, although separately, in a cluster circulating in Europe in autumn 2018. Five and 21 sequences, respectively, were long enough to be included in the Nextstrain full-length (≥ 6,000 bp) and VP1 (≥ 700 bp) builds; they can be viewed at https://nextstrain.org/enterovirus/d68/genome and https://nextstrain.org/enterovirus/d68/vp1.
Discussion
Here we report the detection of EV-D68 infections associated with both respiratory and neurological manifestations in five European countries in the autumn of 2019. Following its first identification in 1962, only 699 cases of EV-D68 were reported in scientific literature before 2014 [14]. From 2014 onwards, the epidemiology appears to have changed, and EV-D68 has increasingly been associated with outbreaks of respiratory infections and a concurrent upsurge of AFM [15,16] with poor long-term prognosis [17,18]. We identified 15 sequences, collected from five countries, which formed a distinct cluster within subclade B3 not previously described in Europe, illustrating the rapid evolution and spread of EV-D68. The sequences most closely related to this novel B3 cluster were sampled in India in 2017 and from sewage in the United Kingdom in 2018. This would suggest either a very low circulation of strains of this cluster or a lack of sustained global surveillance for EV-D68 and/or submission of sequences in GenBank, or both. The estimated MRCA in mid-2018 suggests that these viruses were already circulating and diversifying during and after the 2018 EV-D68 epidemic, but were not sampled until August 2019. In previous European seasons of EV-D68, Denmark has only reported respiratory infections caused by EV-D68, whereas Germany, France, the Netherlands and Sweden have experienced paralytic cases [11,18,19].
The EV-D68 cases were detected through routine surveillance networks in each country, with alerts to physicians posted in Denmark and the Netherlands. The number of samples processed in each country was within the expected range for the enterovirus season, but the detection rate of EV-D68 was higher than expected in Denmark, Germany and Sweden, where the number of detections for the 2018 season had been unexpectedly low. Detection rates in the other countries were similar to those seen in previous years with low circulation. The rate of hospitalisation among cases was high, however, current surveillance systems primarily allow the detection of severe infections. In this study, 12 of 93 cases were detected through community-based surveillance and a further three cases through sequencing of rhinovirus-positive samples, highlighting the importance of including such surveillance when monitoring infections such as EV-D68. The detection of EV-D68 associated disease indicates a continuous, and most likely underestimated, global circulation of this enterovirus type that was recognised after the large outbreak in the United States in 2014 had sparked increased testing. This justifies the need for reinforced enterovirus surveillance based on a more systematic screening of respiratory samples, collected in hospitalised patients or through the surveillance of respiratory infections (such as influenza community surveillance). Our report also highlights the heterogeneity of the surveillance of EV-D68 in Europe, which may hamper both the comparison of the epidemiological pattern between countries and, more importantly, the early detection of an outbreak. Continuous surveillance will add to our understanding of how EV-D68 evolves outside of outbreak periods and whether low-prevalence years vary in geographic or demographic distribution. The detection of EV-D68 in a presumed year of low activity and the presence of patients with severe neurologic symptoms, stresses the importance of continuous and systematic surveillance and availability of diagnostics for workup of clinical cases.
Acknowledgements
The authors wish to thank the European non-polio Enterovirus Network (ENPEN) for facilitating contact between the countries.
The authors are grateful to all the scientists and researchers who have made their EV-D68 samples and metadata available publicly, often in a very timely manner, which enables the analysis of recent sequences in a global context.
The Danish authors wish to thank the clinical microbiology departments for sending entero- and rhinovirus-positive samples to SSI for further characterisation. We also wish to thank the clinicians in the hospitals for ensuring appropriate sampling and diagnostic testing and providing us with clinical information about enterovirus-positive cases. Finally, we wish to thank Shukriya Barzinci and Mille Weismann Poulsen for carrying out the laboratory analyses at SSI.
The Dutch authors wish to thank the Dutch medical microbiology laboratories for their contribution on the EV surveillance programme and VIRO-TypeNed, in particular thanks to the clinicians, molecular medical microbiologist and technicians in the Dutch hospitals and laboratories for providing us with sequence and clinical information about the EV-D68 cases (Richard Molenkamp (Erasmus Medical Center, Rotterdam), Judith Hoogenboom-Beuving (Reinier Haga, Medical Diagnostic Center, Delft), Hubert GM Niesters, Coretta Van Leer-Buter, Marjolein Knoester (University Medical Center Groningen, Groningen), and Lieuwe Roorda (Maasstad Hospital, Rotterdam)). We would also like to thank Gé A. Donker coordinator of the Nivel Primary Care database – sentinel Practices, Utrecht, The Netherlands and technicians (Bas van der Veer, Anne-Marie van den Brandt, Sharon van den Brink, Lisa Wijsman, Elsa Poorter, Barbara Favier, Jeroen Cremer and Pieter Overduin) running molecular diagnostics and sequencing at RIVM. We also thank Harry Vennema for bioinformatic assistance with analysis of NGS data.
The Swedish authors wish to acknowledge the input of Jan Albert at the Department of Clinical Microbiology, Karolinska University Hospital, and Katherina Zakikhany at the Public Health Agency of Sweden. We also want to thank Lina Thebo at Karolinska Institutet for expert technical assistance, Tanja Normark at SciLifeLab for bioinformatics, and Anders Jonsson, Guillermo Martinez Gonzalez, and Rolf Ingemarsson running diagnostics at the Karolinska University Hospital.
The French authors thank Dr Gisèle Lagathu and Pr Vincent Thibaud (University hospital of Rennes) and Dr Quentin Lepiller (University hospital of Besançon) for taking part in the enterovirus surveillance. We also thank Adeline Duard and Nathalie Rodde for technical assistance.
The German authors wish to thank all clinicians for submitting patient samples for testing. We acknowledge Ortwin Adams (University hospital of Düsseldorf), Christiane Prifert, and Benedikt Weissbrich (University hospital of Würzburg) who substantially contribute to the German EV-D68 surveillance in the context of the RespVir network, as well as Sabine Diedrich (Robert Koch Institute, Rolf Kaiser (University hospital of Cologne), Souhaib Aldabbagh (Institute of Virology, University of Bonn Medical Center) and all technicians in the labs.
Analyses were run using sciCORE (http://scicore.unibas.ch/) scientific computing core facility at the University of Basel.
Conflict of interest: None declared.
Authors’ contributions: SEM: Initiated the study, wrote the first draft, edited subsequent drafts and prepared the final manuscript.
KB: Provided data, critically reviewed and edited all drafts of the manuscript.
RD: Provided data, critically reviewed and edited all drafts of the manuscript.
AMir: Provided data, critically reviewed and edited all drafts of the manuscript.
JLB: Critically reviewed and edited the manuscript.
SibB: Provided data, critically reviewed and edited the manuscript.
SteB: Provided data, critically reviewed and edited the manuscript.
SinB: Provided data, critically reviewed and edited the manuscript.
AMEH: Provided data, critically reviewed and edited the manuscript.
MH: Provided data, critically reviewed and edited the manuscript.
VVJ: Provided data, critically reviewed and edited the manuscript.
UBH: Provided data, critically reviewed and edited the manuscript.
CH: Critically reviewed and edited the manuscript.
MP: Provided data, critically reviewed and edited the manuscript.
MKT: Provided data, critically reviewed and edited the manuscript.
EBH: Carried out the phylogenetic analysis, critically reviewed and edited all drafts of the manuscript.
AMeij: Provided data, critically reviewed and edited all drafts of the manuscript.
References
- 1.Kramer R, Sabatier M, Wirth T, Pichon M, Lina B, Schuffenecker I, et al. Molecular diversity and biennial circulation of enterovirus D68: a systematic screening study in Lyon, France, 2010 to 2016. Euro Surveill. 2018;23(37):1700711. 10.2807/1560-7917.ES.2018.23.37.1700711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Messacar K, Pretty K, Reno S, Dominguez SR. Continued biennial circulation of enterovirus D68 in Colorado. J Clin Virol. 2019;113:24-6. 10.1016/j.jcv.2019.01.008 [DOI] [PubMed] [Google Scholar]
- 3.Uprety P, Curtis D, Elkan M, Fink J, Rajagopalan R, Zhao C, et al. Association of enterovirus D68 with acute flaccid myelitis, Philadelphia, Pennsylvania, USA, 2009-2018. Emerg Infect Dis. 2019;25(9):1676-82. 10.3201/eid2509.190468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cottrell S, Moore C, Perry M, Hilvers E, Williams C, Shankar AG. Prospective enterovirus D68 (EV-D68) surveillance from September 2015 to November 2018 indicates a current wave of activity in Wales. Euro Surveill. 2018;23(46):1800578. 10.2807/1560-7917.ES.2018.23.46.1800578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bal A, Sabatier M, Wirth T, Coste-Burel M, Lazrek M, Stefic K, et al. Emergence of enterovirus D68 clade D1, France, August to November 2018. Euro Surveill. 2019;24(3):1800699. 10.2807/1560-7917.ES.2019.24.3.1800699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pellegrinelli L, Giardina F, Lunghi G, Uceda Renteria SC, Greco L, Fratini A, et al. Emergence of divergent enterovirus (EV) D68 sub-clade D1 strains, northern Italy, September to October 2018. Euro Surveill. 2019;24(7):1900090. 10.2807/1560-7917.ES.2018.24.7.1900090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The United Kingdom Acute Flaccid Paralysis Afp Task Force An increase in reports of acute flaccid paralysis (AFP) in the United Kingdom, 1 January 2018-21 January 2019: early findings. Euro Surveill. 2019;24(6):1900093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González-Sanz R, Taravillo I, Reina J, Navascués A, Moreno-Docón A, Aranzamendi M, et al. Enterovirus D68-associated respiratory and neurological illness in Spain, 2014-2018. Emerg Microbes Infect. 2019;8(1):1438-44. 10.1080/22221751.2019.1668243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Midgley SE, Christiansen CB, Poulsen MW, Hansen CH, Fischer TK. Emergence of enterovirus D68 in Denmark, June 2014 to February 2015. Euro Surveill. 2015;20(17):21105. 10.2807/1560-7917.ES2015.20.17.21105 [DOI] [PubMed] [Google Scholar]
- 10.Meijer A, van der Sanden S, Snijders BE, Jaramillo-Gutierrez G, Bont L, van der Ent CK, et al. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology. 2012;423(1):49-57. 10.1016/j.virol.2011.11.021 [DOI] [PubMed] [Google Scholar]
- 11.Dyrdak R, Grabbe M, Hammas B, Ekwall J, Hansson KE, Luthander J, et al. Outbreak of enterovirus D68 of the new B3 lineage in Stockholm, Sweden, August to September 2016. Euro Surveill. 2016;21(46):30403. 10.2807/1560-7917.ES.2016.21.46.30403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyrdak R, Mastafa M, Hodcroft EB, Neher RA, Albert J. Intra- and interpatient evolution of enterovirus D68 analyzed by whole-genome deep sequencing. Virus Evol. 2019;5(1):vez007. 10.1093/ve/vez007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121-3. 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm-Hansen CC, Midgley SE, Fischer TK. Global emergence of enterovirus D68: a systematic review. Lancet Infect Dis. 2016;16(5):e64-75. 10.1016/S1473-3099(15)00543-5 [DOI] [PubMed] [Google Scholar]
- 15.Dyda A, Stelzer-Braid S, Adam D, Chughtai AA, MacIntyre CR. The association between acute flaccid myelitis (AFM) and Enterovirus D68 (EV-D68) - what is the evidence for causation? Euro Surveill. 2018;23(3):e. 10.2807/1560-7917.ES.2018.23.3.17-00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Messacar K, Asturias EJ, Hixon AM, Van Leer-Buter C, Niesters HGM, Tyler KL, et al. Enterovirus D68 and acute flaccid myelitis-evaluating the evidence for causality. Lancet Infect Dis. 2018;18(8):e239-47. 10.1016/S1473-3099(18)30094-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yea C, Bitnun A, Robinson J, Mineyko A, Barton M, Mah JK, et al. Longitudinal outcomes in the 2014 acute flaccid paralysis cluster in Canada. J Child Neurol. 2017;32(3):301-7. 10.1177/0883073816680770 [DOI] [PubMed] [Google Scholar]
- 18.Knoester M, Helfferich J, Poelman R, Van Leer-Buter C, Brouwer OF, Niesters HGM, et al. Twenty-nine cases of enterovirus-D68-associated acute flaccid myelitis in Europe 2016: a case series and epidemiologic overview. Pediatr Infect Dis J. 2019;38(1):16-21. 10.1097/INF.0000000000002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antona D, Kossorotoff M, Schuffenecker I, Mirand A, Leruez-Ville M, Bassi C, et al. Severe paediatric conditions linked with EV-A71 and EV-D68, France, May to October 2016. Euro Surveill. 2016;21(46):30402. 10.2807/1560-7917.ES.2016.21.46.30402 [DOI] [PMC free article] [PubMed] [Google Scholar]