Abstract
Background
Mycoplasma pneumoniae is a leading cause of community-acquired pneumonia, with large epidemics previously described to occur every 4 to 7 years.
Aim
To better understand the diagnostic methods used to detect M. pneumoniae; to better understand M. pneumoniae testing and surveillance in use; to identify epidemics; to determine detection number per age group, age demographics for positive detections, concurrence of epidemics and annual peaks across geographical areas; and to determine the effect of geographical location on the timing of epidemics.
Methods
A questionnaire was sent in May 2016 to Mycoplasma experts with national or regional responsibility within the ESCMID Study Group for Mycoplasma and Chlamydia Infections in 17 countries across Europe and Israel, retrospectively requesting details on M. pneumoniae-positive samples from January 2011 to April 2016. The Moving Epidemic Method was used to determine epidemic periods and effect of country latitude across the countries for the five periods under investigation.
Results
Representatives from 12 countries provided data on M. pneumoniae infections, accounting for 95,666 positive samples. Two laboratories initiated routine macrolide resistance testing since 2013. Between 2011 and 2016, three epidemics were identified: 2011/12, 2014/15 and 2015/16. The distribution of patient ages for M. pneumoniae-positive samples showed three patterns. During epidemic years, an association between country latitude and calendar week when epidemic periods began was noted.
Conclusions
An association between epidemics and latitude was observed. Differences were noted in the age distribution of positive cases and detection methods used and practice. A lack of macrolide resistance monitoring was noted.
Keywords: Mycoplasma pneumoniae, epidemiology, diagnostics, surveillance
Introduction
Mycoplasma pneumoniae is a major cause of respiratory infection in humans and macrolide antibiotics, such as azithromycin, are used as the first-line of treatment in many countries. The bacterium is transmitted from person-to-person by respiratory droplets with the incubation period ranging from 4 days to 3 weeks [1]. Because of M. pneumoniae’s intrinsic resistance to many antibiotics, including all cell wall inhibitors, macrolide antibiotics such as azithromycin and clarithromycin are the drug of choice for treatment. In cases of suspected infection in immunocompromised individuals, bactericidal fluoroquinolones may be considered. Tetracyclines are an alternative for treatment of adults with possible macrolide-resistant M. pneumoniae infections. Prudent use of antibiotics has been urged for all cases of M. pneumoniae infection because of worldwide reports of macrolide resistance, which have been reported as ranging from 0.2% in Sweden to more than 90% in China [2-5].
M. pneumoniae infections show seasonal variation. In temperate climates, the number of infections peak during the latter months of the years, with epidemic periods every 4 to 7 years on average [6-8]. The most recent survey in 2012 by Lenglet et al indicated that some countries in the European Union and European Economic Area experienced an increase in M. pneumoniae cases in 2011 whereas others did not, indicating that a universal geographic increase had not occurred [5]. Little is understood about the transmission of M. pneumoniae within populations and several factors have been postulated to account for transmission dynamics, including the immunity level of the population, the bacterial population based on the P1 adhesin type, the age and extent of mixing of children in educational settings.
Methodologies for detection of M. pneumoniae include nucleic acid amplification tests (NAAT), serology and culture with varying sensitivities and specificities. There is no international standard material for quality control detection in assays, although external quality control schema exist for some methodologies (NAAT). There are no internationally defined guidelines on the requirements for surveillance of M. pneumoniae, macrolide resistance testing and surveillance, reference system structure, routine testing and bacterial strain discrimination. However, a few countries such as France and the United States (US) have surveillance within specific regions and national surveillance is seen in countries such as Denmark [9] and Japan, the latter of which has maintained an active surveillance system for this pathogen for some time [10]. Overall, laboratory confirmed cases and surveillance data regarding the number of cases and reported cases of macrolide resistance are likely to be underestimated. This is further confounded because an undefined proportion of patients will have mild disease or may be carriers within community settings, without active testing to confirm the infection. Further underestimation is likely to occur from patients receiving empirical treatment in the absence of laboratory-confirmed infection with M. pneumoniae.
In response to an increase in infection seen in several countries in 2016, the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group for Mycoplasma Infections (ESGMI), now called the Study Group for Mycoplasma and Chlamydia Infections (ESGMAC), established this study [9,11]. The purpose of the study was to gain a greater understanding of the diagnostic methods used to detect M. pneumoniae; gain a greater understanding of the testing and surveillance in use for M. pneumoniae (macrolide resistance, seasonality); to identify epidemics; to determine detections per age group, age demographics for positive detections and concurrence of epidemics and annual peaks across geographical areas; and to determine the effect of geographical location on timing of epidemics.
Methods
Study type, data collection and analysis
ESGMI conducted a retrospective email-based survey in May 2016 of ESGMI members in countries in Europe and Israel, asking members to describe existing laboratory-confirmed case data for M. pneumoniae infection. This retrospective study involved sending an email-based survey to 18 experts collating laboratory-confirmed documented detections of M. pneumoniae from national laboratory and surveillance institutions or, if not available, other regional laboratory and surveillance institutions. Mycoplasma experts invited to participate in the study were either active members of ESGMI or authors listed in the previous study by Lenglet et al [5]. Participants were invited to join the study and provide the number of detections confirmed by nucleic acid amplification test (NAAT), serology, culture and total overall between weeks commencing 3 January 2011 to 24 April 2016. Positive results and, if available, negative results were also collated. For Germany and France, only regional data were available.
Additional information was requested, including what diagnostic methods were used to detect M. pneumoniae; whether surveillance for M. pneumoniae was in use; if macrolide resistance was being monitored by countries; and M. pneumoniae detection number per age group.
Data from each participating country was collated and aggregated to give total number of detections per age group and four weekly moving averages of detections per country and overall where possible. We did not request information on the sex of patient from which detections were made. Total weekly data were subcategorised by age group: 0 to 4 years, 5 to 9 years, 10 to 14 years, 15 to 24 years, 25 to 44 years, 45 to 64 years, ≥ 65 years or unknown.
Case definition
Cases of M. pneumoniae infection were defined by local practice. Because of local variation, this study collated information on M. pneumoniae detections, not cases.
De-duplication and exclusion criteria
Because of the heterogeneous nature of M. pneumoniae data collection from each country, defining study-wide de-duplication criteria was not feasible. Participants were therefore asked to detail if data with duplicate samples from the same patient (e.g. with NAAT and serology) was included as a single category and if possible, to include as serology. Specific exclusion criteria were also not set for similar reasons stated above. Responses for de-duplication and exclusion criteria are listed in Supplementary Table S1.
Characteristics of epidemics using the Moving Epidemic Method
To determine the characteristic properties of M. pneumoniae epidemics across the 12 countries for which data were provided on M. pneumoniae positivity, the Moving Epidemic Method (MEM) was used [12]. An epidemic slope threshold of 2% was chosen and used to determine the pre-epidemic period, epidemic period and post-epidemic period for the five periods coinciding with annual peaks spanning week 19 of the first year through week 18 of the following year. These data were used to calculate the epidemic duration for each country, as well as the percentage of positive samples that were identified within this period. Data generated from the MEM model, such as week number in which epidemics began, were used to correlate the onset of epidemics with the geographical location of each country. Statistical analysis for generating p values and calculation of r2 were performed with GraphPad Prism 5.0.
Ethics statement
This retrospective study involved collation of anonymised surveillance data for epidemiological analysis. Ethical approval was not required and no patient identifiers were included in the study.
Results
Of the 18 countries approached, 11 countries across Europe and Israel, provided information regarding M. pneumoniae. For Cyprus, Malta, the Netherlands, Poland, Slovakia and Spain, M. pneumoniae national testing and surveillance systems were not in place or a response with data was not received within the timeframe of the study.
Mycoplasma pneumoniae detection methods
During the study period, a total of 95,666 detections of M. pneumoniae were confirmed from participating countries (Table 1). The method of M. pneumoniae detection varied between the 12 countries. Two countries, Denmark and Israel, reported exclusively NAAT use; two countries, Greece and Ireland, reported serology exclusively; five countries, Germany, Hungary, Norway, Sweden and the United Kingdom (excluding Northern Ireland) used a combination of NAAT and serology; one country, Slovenia, used NAAT in combination with culture; and two countries, Belgium and France, used all three methods. No country relied on culture alone (Table 1).
Table 1. Mycoplasma pneumoniae detection methods, percent of positive samples and macrolide resistance monitoring by country, 11 countries in Europe and Israel, 2011–2016.
| Country | Nucleic acid amplification test | Culture | Serology | Total number of positive samples | Total number of negative samples | Percent of positive samples (%) | Macrolide resistance monitoring | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Performed | Number of positive detections | Performed | Number of positive detections | Performed | Number of positive detections | |||||
| Belgium | Yes | 894 | Yes | 49 | Yes | 12,047 | 21,094a | ND | NA | Only monitored when samples test positive at the NRC. No testing in sentinel laboratories. |
| Denmark | Yes | 20,081 | No | NA | No | NA | 20,081 | 264,770 | 7.0 | No routine surveillance system in place. In 2010/11, 2011/12 and 2015/16, 809 samples were examined identifying 13 macrolide resistance-associated mutations (1.5%). Samples are investigated upon physician request. |
| France | Yes | 92 | Yes | 53b | Yes | 298 | 390 | 7,463 | 5.0 | Performed on all clinical specimens detected as M. pneumoniae-positive by NAAT since 2013 [13]. |
| Germany | Yes | 127 | No | NA | Yes | 316 | 443 | 10,143 | 4.2 | No comment |
| Greece | No | NA | No | NA | Yes | 140 | 140 | 1,498 | 8.5 | Information not provided |
| Hungary | Yes | 17 | No | NA | Yes | 1,117 | 1,134 | 6,109 | 15.7 | Information not provided |
| Ireland | No | NA | No | NA | Yes | 535 | 535 | 2,853 | 15.8 | Information not provided |
| Israel | Yes | 848 | No | NA | No | NA | 848 | 5,309 | 13.8 | No active monitoring. |
| Norway | Yes | 13,980 | No | NA | Yes | 10,678 | 24,658 | ND | NA | Information not provided |
| Slovenia | Yes | 1,172 | Yes | 827 | No | NA | 1,172 | 8,872 | 11.7 | Only upon physician request |
| Sweden | Yes | 9,499 | No | NA | Yes | 10,024 | 19,523 | 169,501 | 10.3 | No active monitoring. |
| United Kingdom (excluding Northern Ireland) | Yes | 385 | No | NA | Yes | 5,263 | 5,648 | ND | NA | No national system. All positive samples referred to PHE are tested. |
| Total | NA | 47,095 | NA | 876 | NA | 40,418 | 95,666 | 476,518 | NA | NA |
NA: not applicable; NAAT: Nucleic acid amplification test; ND: no data available; NRC: National Reference Centre; PHE: Public Health England.
a Method of detection not known for sentinel laboratories.
b Not included in the overall total for the purpose of de-duplication as these 53 samples were already listed in NAAT or serological counts.
Responses were received to state M. pneumoniae testing and surveillance is not in place on a national scale or case data were not available within the timespan of the survey from European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group for Mycoplasma Infections (ESGMI) members from Cyprus, Malta, the Netherlands, Poland, Slovakia or Spain.
The greatest number of positive samples were reported using NAAT (47,095; 49%) followed by serology (40,418; 42%). Only 876 (1%) samples were reported positive by culture and 7,277 (8%) of tests were not specified. Norway contributed the greatest number of M. pneumoniae-positive detections to the total figure (24,658; 26%) and Greece the lowest (140; 0.1%). De-duplication data were determined at country level (Supplementary Table S1).
Macrolide resistance monitoring
With regards to active monitoring of macrolide resistance, five countries did not comment; Belgium noted that monitoring was only carried out on positive samples identified at the National Reference Laboratory, but not at sentinel laboratories; Slovenia noted that macrolide resistance determination was carried out only upon physician request; in this study, Denmark stated that NAAT-positive samples from three recent M. pneumoniae epidemic periods were investigated, finding a low level (1.5%) of macrolide resistance, and that clinical samples are investigated on request from physicians; the Mycoplasmal and Chlamydia Infections in Humans Research Department, University of Bordeaux, France initiated national systematic monitoring of all NAAT-positive clinical specimens in 2013 using an in-house published method [13]. In 2017, England and Wales introduced monitoring of all positive NAAT samples referred to the National Reference Laboratory. Two countries stated that no monitoring for resistance was in place (Table 1).
Total number of detections and seasonality
The distribution and seasonality of the 95,666 detections from the 12 countries across the study period was determined. To account for weekly bias in reporting, data were converted to four weekly moving average. The greatest number of positive samples from the four weekly moving average data was 1,759 positive detections during week 48 of 2011.
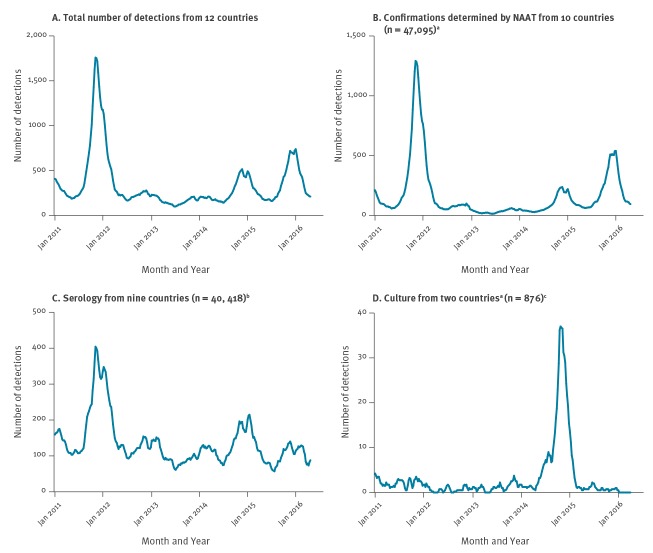
Detection of M. pneumoniae by NAAT correlated with the overall detections (Figure 1A and Figure 1B). Detection of M. pneumoniae by culture accounted for the lowest number of positive samples per methodology; 1% of the total positive samples. Detection using serology was the second most common method for detecting M. pneumoniae-positive patients (Figure 1C). The four weekly moving average for detection by culture (Figure 1D) was less than five positive samples for all reporting weeks with the exception of Slovenia in the 2015 season when a maximum average of 52 positive samples was identified.
Figure 1.
Four weekly moving average data on Mycoplasma pneumoniae infections by detection methods, 11 countries in Europe and Israel, 2011–2016
NAAT: nucleic acid amplification tests.
a Ten countries: Belgium, Denmark, France, Germany, Hungary, Israel, Norway, Slovenia, Sweden and the United Kingdom (excluding Northern Ireland).
b Nine countries: Belgium, France, Germany, Greece, Hungary, Ireland, Norway and Sweden and the United Kingdom (excluding Northern Ireland).
c Two countries: Belgium and Slovenia. Culture data from France not included as it was duplicated in NAAT and serological data.
Epidemic periods based on the Moving Epidemic Method
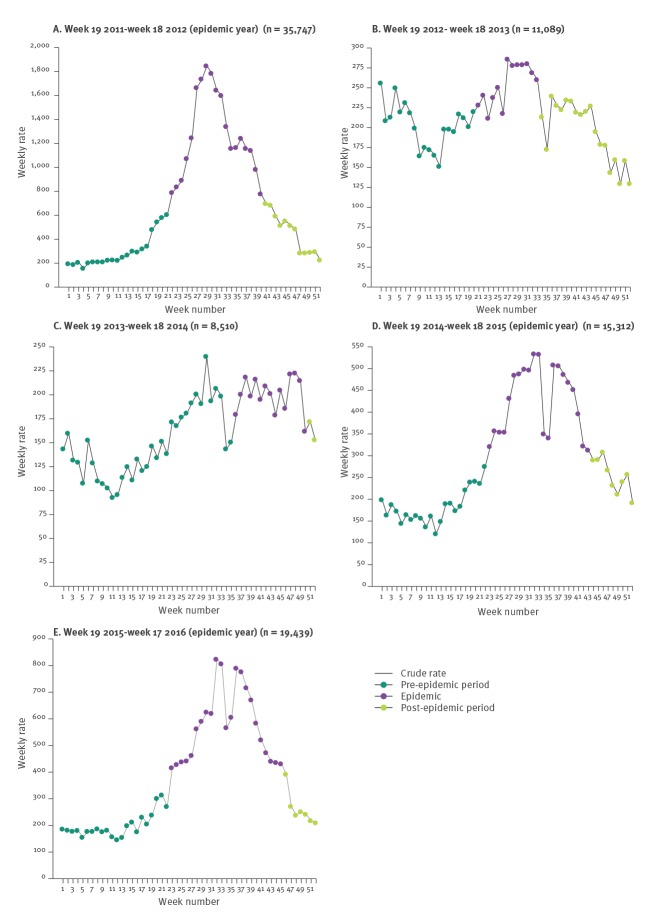
Analysis of detections during the annual periods was carried out using MEM (Figure 2). For the five annual periods described, we noted 35,747; 11,089; 8,510; 15,312; 19,439 detections for the periods 2011/12, 2012/13, 2013/14, 2014/15 and 2015/16, respectively. Three epidemics were detected between 2011/12, 2014/15 and 2015/16, in which 67%, 59% and 68% of each period’s detections were identified during the calculated epidemic period, respectively. Epidemics had longer duration, 19, 21 and 23 weeks, respectively, compared with the duration observed during annual seasonal peaks of infection (non-epidemic periods). In 2012/13, the duration was 13 weeks with 30% of total detections and in 2013/14, 15 weeks with 35% of total detections. For countries providing the total number of negative samples, the percentage of positive samples identified during the pre-epidemic, epidemic and post-epidemic period was calculated for the epidemic periods of 2011/12, 2014/15 and 2015/16 (Table 2). For all periods, there was a greater percentage of positive samples during the epidemic period than in the pre-epidemic period.
Figure 2.
Analysis of Mycoplasma pneumoniae epidemic periods using the Moving Epidemic Method, 11 countries in Europe and Israel, 2011–2016
MEM: Moving Epidemic Method.
Week numbers represent epidemic week period (week 1 represents calendar week 19). Green dots represent pre-epidemic period, purple dots represent epidemic period and yellow dots represent post-epidemic period, as calculated by the MEM.
Table 2. Proportion of total samples positive for Mycoplasma pneumoniae during the pre-epidemic, epidemic and post-epidemic periods, 8 countries in Europe and Israel, epidemic periods 2011/12, 2014/15 and 2015/16a.
| Country | 2011/12 | 2014/15 | 2015/16 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre-epidemic | Epidemic | Post-epidemic | Total across all periods | Detection per peak week during epidemic period | Pre-epidemic | Epidemic | Post-epidemic | Total across all periods | Detection per peak week during epidemic period | Pre-epidemic | Epidemic | Post-epidemic | Total across all periods | Detection per peak week during epidemic period | ||||||||||||||||
| n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | n/N | % | |
| Denmark | 920/7,784 | 12 | 6,932/47,522 | 15 | 804/15760 | 5 | 8,656/71,066 | 12 | 185/879 | 21 | 291/12,171 | 2 | 1,206/23,252 | 5 | 204/7,876 | 3 | 1,701/43,299 | 4 | 90/1238 | 7 | 1,120/18,340 | 6 | 5,751/52,827 | 11 | 712/16,139 | 4 | 7,583/87,306 | 9 | 546/3, 457 | 16 |
| France | 71/862 | 8 | 41/410 | 10 | 27/583 | 5 | 139/1,855 | 7 | 7/37 | 19 | 32/786 | 4 | 17/208 | 8 | 19/381 | 5 | 68/1,375 | 5 | 6/44 | 14 | 28/690 | 4 | 8/86 | 9 | 22/506 | 4 | 58/1,282 | 5 | 4/29 | 14 |
| Germany | 38/833 | 5 | 33/360 | 9 | 37/669 | 6 | 108/1,862 | 6 | 7/44 | 16 | 8/447 | 2 | 77/1,373 | 6 | 4/248 | 2 | 89/2,068 | 4 | 7/39 | 18 | 22/917 | 2 | 13/293 | 4 | 19/1,639 | 1 | 54/2,849 | 2 | 4/46 | 9 |
| Greece | NA | NA | 18/110 | 16 | 16/224 | 7 | 34/334 | 10 | 5/8 | 63 | NA | NA | 9/27 | 33 | 28/304 | 9 | 37/331 | 11 | 3/6 | 50 | 0/15 | 0 | 2/11 | 18 | 6/179 | 3 | 8/205 | 2 | 2/3 | 67 |
| Hungary | 15/154 | 10 | 32/169 | 19 | 105/942 | 11 | 152/1,265 | 12 | 7/20 | 35 | 75/442 | 17 | 186/760 | 24 | 54/367 | 15 | 315/1,569 | 20 | 5/12 | 42 | 96/570 | 17 | 50/220 | 23 | 59/544 | 11 | 205/1,334 | 15 | 10/30 | 33 |
| Ireland | 53/290 | 18 | 77/261 | 30 | 15/78 | 19 | 145/629 | 23 | 3/6 | 50 | 28/263 | 11 | 31/153 | 20 | 17/144 | 12 | 76/560 | 14 | 4/10 | 40 | 66/374 | 18 | 39/153 | 25 | 17/73 | 23 | 122/600 | 20 | 8/12 | 67 |
| Israel | 84/572 | 15 | 81/346 | 23 | NA | NA | 165/918 | 18 | 19/55 | 35 | 57/499 | 11 | 87/597 | 15 | NA | NA | 144/1,096 | 13 | 5/17 | 29 | 106/1052 | 10 | 82/687 | 12 | 3/36 | 8 | 191/1,775 | 11 | 11/45 | 24 |
| Slovenia | 32/389 | 8 | 31/349 | 9 | 12/413 | 3 | 75/1,151 | 7 | 4/18 | 22 | 166/775 | 21 | 583/1,866 | 31 | 83/1,037 | 8 | 832/3,678 | 23 | 38/78 | 49 | NA | NA | 29/469 | 6 | 28/1,447 | 2 | 57/1,916 | 3 | 5/27 | 19 |
| Sweden | 1,389/7,389 | 19 | 5,356/28,226 | 19 | 859/10,034 | 9 | 7,604/45,649 | 17 | 186/686 | 27 | 863/10,734 | 8 | 1,447/17,242 | 8 | 501/8,729 | 6 | 2,811/36,705 | 8 | 100/950 | 11 | 500/7,955 | 6 | 1,356/12,414 | 11 | 967/14,066 | 7 | 2,823/34,435 | 8 | 98/731 | 13 |
| Total | 2,602/18,273 | 14 | 12,601/77,753 | 16 | 1,875/28,703 | 7 | 17,078/124,729 | 14 | 423/1,753 | 24 | 1,520/26,117 | 6 | 3,643/45,478 | 8 | 910/19,086 | 5 | 6,073/90,681 | 7 | 258/2,394 | 11 | 1,938/29,913 | 6 | 7,330/67,160 | 13 | 1,833/34,629 | 5 | 11,101/13,1702 | 8 | 688/4,380 | 16 |
NA: not applicable.
a Only those countries reporting both positive and negative sample data are included.
Distribution of Mycoplasma pneumoniae positivity by age
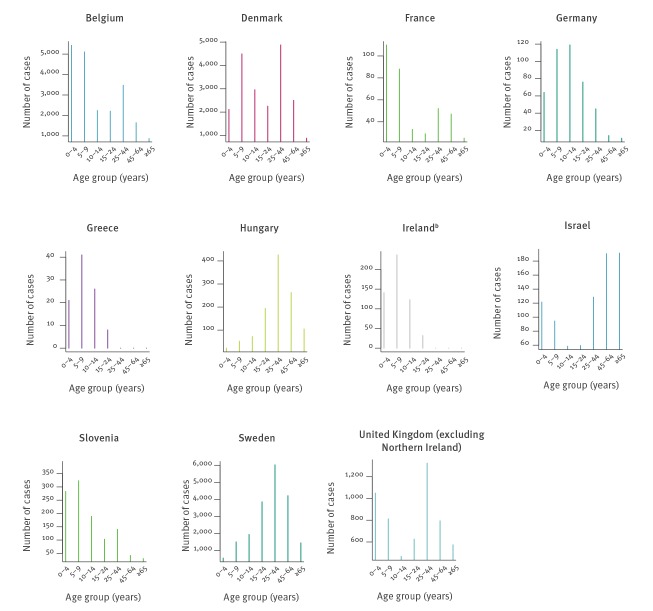
The distribution of M. pneumoniae positivity by age group was determined for 11 countries (Figure 3) as data obtained from Norway was not subcategorised by age. Detections per age group varied, and infections were noted in all age groups (0–4 years of age 9,832; 5–9 years of age 12,957; 10–14 years of age 8,144; 15–24 years of age 9,311; 25–44 years of age 16,281; 45–64 years of age 9,586; ≥ 65 years of age 4,080; total with specified age group 70,191). Even distribution across all age groups was not noted in the data from any country. Country-specific age data revealed three distinct patterns. Four countries, Germany, Greece, Ireland and Slovenia, showed skewing of positive samples to younger patients (< 10 years of age), whereas two countries, Hungary and Sweden, showed skewing towards older patients (> 25 years of age). Five countries reported a bimodal distribution: Belgium, Denmark, France, Israel and the United Kingdom (excluding Northern Ireland), reported a bimodal distribution. Of note, data for Ireland was not available for cases 25 years of age and above.
Figure 3.
Number of Mycoplasma pneumoniae detections by age group, 10 countries in Europea and Israel, 2011–2016 (n = 70,191)
a Age-specific data were not available for Norway.
b Ireland did not test for M. pneumoniae in patients 25 years of age and above.
Correlation between latitude and epidemic period onset
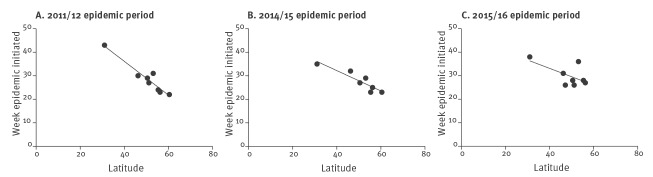
When examining the epidemic periods of 2011/12, 2014/15 and 2015/16, a clear association between the country latitude and beginning of the national epidemic period was observed (Figure 4). This was statistically significant for the 2011/12 period (p < 0.005; r2 = 0.92) and the 2014/15 period (p < 0.005; r2 = 0.84). However, significance was not achieved during the 2015/16 period (p = 0.1; r2 = 0.38). The association was most apparent during the major epidemic period of 2011/12 when the epidemic period was first noted in Norway (60.4oN) during epidemic week 22 (calendar week 40 of 2011) and was then observed to start in Israel (31oN) during epidemic week 43 (calendar week 9 of 2012). There was a lack of an association during the non-epidemic periods of 2012/13 and 2013/14 (data not shown).
Figure 4.
Correlation between country latitude and epidemic week for Mycoplasma pneumoniae infections, 7 European countries and Israel, epidemic periods 2011/12, 2014/15 and 2015/16
Countries in which epidemic periods began prior to that predicted by the Moving Epidemic Method were removed from the analysis. A statistically significant association between the week in which the country epidemic began and epidemic week number was seen in 2011/12 (p < 0.005; r2 = 0.92) and 2014/15 (p < 0.005; r2 = 0.84). No significant association was seen in 2015/16 (p = 0.1; r2 = 0.38).
Discussion
Our study involved assembling the largest dataset thus far of M. pneumoniae-positive samples and associated data on the methods used to detect M. pneumoniae per country. NAATs were the most common method used among the 12 countries. A variety of commercial and in-house methodologies were used in the detection of M. pneumoniae in our study. However, a recent study of 13 assays used across Europe, Israel and the US demonstrated comparable levels of detection of 20 M. pneumoniae genomes per reaction [14]. Serological methods were also commonly used to detect M. pneumoniae infections. The presence of an IgM response may indicate recent acquisition of infection, but it may be an unreliable marker because of documented long-term persistence of antibodies [15]. Culture-dependent methods were used by three countries. Culture has the benefit of confirming the presence of viable M. pneumoniae and may permit phenotypic antimicrobial susceptibility testing. However, because of the considerable time required for growth, up to 4 weeks, it does not provide results within a clinically relevant time period.
Currently, no single method is recommended for routine detection of M. pneumoniae, and international guidelines or control materials do not exist. Real-time PCR and serology have previously shown agreement in their ability to detect M. pneumoniae among samples (90% agreement), however, 7% of patients were PCR positive, with limited evidence of seroconversion [16]. No single test can reliably detect all infections. Therefore, a combined approach utilising both NAAT and serology may help to identify any potential false-negative results, with NAAT used for acute phase detection [17].
In addition to method of detection, we sought to identify if surveillance for macrolide resistance was routinely undertaken. Routine macrolide resistance monitoring was not systematically in place. This may contribute to the under-detection of resistance or reflect low levels of macrolide resistance reported across Europe [18-20]. High levels of resistance have been noted, for example, in Israel (30%) [21] and China wherein reported macrolide resistance levels are 90 to 100% [22,23]. High incidence of macrolide resistance necessitates co-ordinated surveillance across Europe. It would be of benefit to increase physicians’ knowledge of macrolide resistance rates and acquisition, as well as an understanding of the patient’s recent travel history when considering therapy. In the authors’ opinion, co-ordinated international surveillance for macrolide resistance should be considered as macrolides are the current treatment of choice in Europe.
Regardless of the methodology used to detect M. pneumoniae, clear seasonal trends were apparent between January 2011 and April 2016, with peaks in infection occurring between the fourth quarter of a year and first quarter of the next. To calculate the epidemic period for each season, the MEM, as described by Vega et al, was used [12]. Three clear epidemics were noted in 2011/12, 2014/15 and 2015/16. M. pneumoniae epidemics have been suggested to occur every 4 to 7 years, lasting for longer than one annual season in some cases [8,24]. However, in this study, based on a very large data set, the interval between epidemic occurrence was found to be 3 years from 2011/12 to 2014/15 and 1 year from 2014/15 to 2015/16. The latter epidemic period may reflect a secondary peak of cases within a 2-year epidemic span as previous epidemics have been shown to persist for some time [5,24]. Confirmation of circulating genotypes of M. pneumoniae from large geographical areas would be of interest to determine the microbiological nature of strains within these and epidemic periods.
For the major epidemic periods, we sought to determine any changes between the pre-epidemic, epidemic and post-epidemic periods in the reported number of detections in countries reporting both positive and negative data. The greatest rise was seen in Ireland where 18% of samples were positive for M. pneumoniae in the pre-epidemic period, while 30% were positive during the epidemic period in 2011/12. In Denmark, France, Slovenia and Sweden, the prevalence in the post-epidemic period was lower than that of the pre-epidemic period in 2011/12. This lower prevalence in the post-epidemic period may reflect over-sampling as a bias resulting from higher prevalence during the epidemic. This may also reflect an increase in the population burden of infection prior to an epidemic. Additional analysis is required to understand if this is the case and if monitoring of levels can be used to predict imminent epidemics.
A notable observation was the pattern of positive detections stratified by age, with detections in all age groups. M. pneumoniae cases are classically seen among children 5 to 14 years of age, with those under 5 years experiencing milder disease [17]. This trend was seen in Germany, Greece, Ireland and Slovenia which skew towards the younger age groups. For Greece, this can be attributed to the acquisition of samples from a tertiary children’s hospital in Athens. It should be noted that the skew towards younger age groups seen in Ireland may reflect the nature of only investigating patients up to 25 years of age. An observed peak in infection among the under 14 years age groups was followed by a second peak in the 25 to 44 years age group giving a bimodal distribution in five countries. Finally, a skew to the older age groups was seen in Hungary and Sweden. This observation is not likely to be an artefact of testing methodology whereby older people may be more likely to have existing IgM levels as both countries do not solely rely on serology. The reason for this increased detection in people above 44 years of age in two countries is unknown; it may simply reflect differences in local testing guidelines and routine practice for respiratory screening, or reflect age-based screening practices (e.g. the majority of Hungarian and Swedish samples derived from a population > 25 years of age).
The final analysis examined the association between the start of epidemic periods as calculated by the MEM model and the geographical location of the country as determined by latitude. Geographical location has not been thought to be of importance in the progression of M. pneumoniae infection, but our data suggests that more northern countries experience the start of epidemic periods earlier than those in the south. This association was most clear during the 2011/12 epidemic period, but also held true for the subsequent epidemic periods of 2014/15 and 2015/16. Previous national-based studies have shown epidemics to be polyclonal in nature [25-30]. Establishing whether the microbiological nature of the epidemics across Europe are clonal or not, may be beneficial and influence future sentinel surveillance design. The impact of climatic factors on M. pneumoniae infections has yet to be investigated.
Limitations
A number of limitations are apparent. First, because of the variable reporting methods of each country, specific case definitions were not considered and de-duplication methodologies were not imposed but rather set at submitter-specific level. Overall, reported testing activity or testing-incidence was very different between countries (regions), and conclusions based on analysis across countries must be considered with caution. Ireland did not provide a complete dataset because they only investigated M. pneumoniae in patients who were 24 years of age and under. The true number of positive individuals from this population is therefore likely to be underestimated. Second, the data from some countries may not be fully representative of the country as a whole. For example, data from Germany and France was obtained from a single region within each country and how representative this is of national coverage is unknown. Third, the data examining the distribution of detections stratified by age group should be interpreted with a level of caution. The age categories did not contain equal weighting of age groups; for example, there were fewer ages encompassed in the 0 to 4 years group compared with older age groups such as the 25 to 44 years group. Finally, this data did not take the subtypes of M. pneumoniae that have been described into account.
Possible clinical impact
The comparative nature of this study has highlighted a number of interesting points with regards to trends in the testing and epidemiology of M. pneumoniae infections. First, cases may be under-detected in countries because of limitations in the age groups examined for the infection. A number of countries showed a skew towards patients > 25 years of age and Belgium, Denmark, France, the United Kingdom (excluding Northern Ireland) and Israel showed a bimodal distribution, suggesting investigations for M. pneumoniae, although of importance in young children, should not be restrictive and that consistent testing guidelines are required. It would be beneficial to have an agreed international case definition for infection with M. pneumoniae.
This study also highlights a lack of antimicrobial resistance testing and surveillance of M. pneumoniae, resulting in a limited evidence base on resistance to guide therapy. Without active coordinated monitoring, it will not be possible to track changes in resistance profiles and the emergence of high-level macrolide resistant clones. There is an absence of a structured European level surveillance and resistance monitoring for this infection, despite the high levels of resistance in some global areas. In the authors’ opinion, structured surveillance should be implemented in Europe.
Finally, the observation relating to association between northern latitudes and earlier epidemic start week may suggest the need for another more focused study. It would be interesting to assess the potential value of a rapid, real-time reporting system of M. pneumoniae infections across Europe. Such a system could possibly aid in future epidemiological studies and resistance monitoring, and help to predict the M. pneumoniae epidemic season throughout the continent.
Conclusion
As this large study demonstrates, there is currently no standardised method for detecting M. pneumoniae infection among patients and macrolide resistance screening is sporadic in European countries despite high levels in some areas globally. A wave of epidemics from more northern latitudes to the more southern ones occurs during epidemic years and the reason for this is not known. There is a need for testing guidelines and standardised international control material for use in testing. The potential value of a co-ordinated international surveillance and macrolide resistance monitoring system needs to be further addressed.
Acknowledgements
We are very grateful to all those who provided the data or nil returns. We would specifically like to thank the following contributors:
Belgium: Sophie Quoilin; Croatia: Sanja Kurečić Filipović and Iva Pem Novosel; Czech Republic: Jitka Castkova and Vratislav Nemecek; Denmark: Sofie Gillesberg Raiser; the Danish Microbiology Data Base (MiBa) Board of representatives for their improvement of the study, the Danish Clinical Microbiology Departments for providing data to MiBa and samples for macrolide resistance examination; staff of the Bacterial PCR Laboratory Statens Serum Institut for examination of samples. England and Wales: Elaine Stanford (Public Health England); Estonia: Irina Filippova; Finland: Ruska Rimhanen-Finne, Markku Kuusi (National Institute for Health and Welfare, Finland); France: Anne-Marie Roque-Afonso; Greece: Kassiani Gkolfinopoulou; Ireland: Niamh Murphy; Italy: Stefania D’Amato and Anna Rita Ciccaglione; Malta: Tanya Melillo; Netherlands: Petra Brandsema (Epidemiology and Surveillance Unit), Marit de Lange (Centre Infectious Disease Control Netherlands (RIVM), Bilthoven), Daan Notermans (RIVM - Centre Infectious Disease Control Netherlands); Norway: Bernardo Guzmán Herrador; Poland: Piotr Polanski and Anna Baumann-Popczyk; Sweden: Andreas Edberg (Klinisk Mikrobiologi Karlstad), Arne Kötz (Klinisk Mikrobiologi Halmstad), Annika Tiveljung Lindell (Karolinska Universitetslaboratoriet Solna), Hilde Riedel ( Klinisk mikrobiologi Uppsala), Kåre Bondeson (Klinisk mikrobiologi Uppsala); Lena Jakobsson (Klinisk mikrobiologi Örebro), Sara Mernelius (Klinisk mikrobiologi Jönköping), Michael Toepfer (Unilabs Skövde), Marianne Bäckman (Klinisk mikrobiologi Aleris), Nina Kamenska (Klinisk mikrobiologi Trollhättan), Claudia Popa (Laboratoriemedicin Västernorrland), Ann-Chatrine Petersson (Klinisk mikrobiologi Skåne), Camilla Norling (Klinisk mikrobiologi Falun), Baqir Haitham (Klinisk mikrobiologi Linköping); Scotland: Kate Templeton (ESVC) and Rory Gunson (WOSSVC); Slovenia: Sabina Pervić, Ines B. Šimaga, Rok Kogoj; Spain: Elena Vanessa Martínez and Carmen Varela.
Supplementary Data
ESCMID Study Group for Mycoplasma and Chlamydia Infections (ESGMAC) Mycoplasma pneumoniae subgroup members not listed as an individual author
Belgium: Nathalie Bossuyt (Service Epidemiology of Infectious Diseases, Sciensano, Brussels); Katrien Lagrou (National Reference Centre for Respiratory Pathogens, University Hospitals Leuven, Leuven)
Cyprus: George Mitis (Immunology Department, Nicosia General Hospital Nicosia; Maria Koliou (Department of Paediatrics, Archbishop Makarios III Hospital, Nicosia)
Czech Republic: Martina Havlickova, Jan Kyncl (National Institute of Public Health, Prague)
Denmark: Hanne-Dorthe Emborg, Marianne Voldstedlund (Statens Serum Institut, Copenhagen)
Germany: Colin Rae MacKenzie (Institute of Medical Microbiology and Hospital Hygiene, Heinrich-Heine-University, Duesseldorf)
Greece: Helena C. Maltezou, Theano Georgakopoulou (National Public Health Organization, Athens); Evangelina Petridou, Kirkira Banou (Aghia Sophia Children Hospital, Athens)
Hungary: Eszter Balla (Department of Reference Laboratories, National Public Health Center, Budapest)
Ireland: Jeff Connell, Zoe Yandle, Joanne Moran (University College Dublin, National Virus Reference Laboratory, Dublin); Karen Burns (Health Protection Surveillance Centre, Dublin)
Israel: Ayelet Michael-Gayego, Allon E. Moses (Department of Clinical Microbiology and infectious Diseases, Hadassah-Hebrew University Medical Centre, Jerusalem)
Malta: Tanya Melillo Fenech (Ministry of Health, Valletta)
Norway: Gabriel Ånestad, Hans Blystad, Didrik F. Vestrheim (Norwegian Institute of Public Health, Oslo)
Portugal: Filipe Froes (Hospital Pulido Valente and General Directorate of Health Consultant for Pneumology, Lisbon)
Slovenia: Darja Kese (University of Ljubljana, Faculty of Medicine
Institute of Microbiology and Immunology, Ljubljana) and Maja Socan (National Institute of Public Health, Ljubljana)
Spain: Rosa Cano Portero (National Center for Epidemiology, Instituto de Salud Carlos III. CIBERESP), Madrid); Sara Santos Sanz and Berta Suárez Rodríguez (Coordination for Alerts and Public Health Emergencies, Directorate General of Public Health, Ministry of Health, Social Affairs and Equality, Madrid)
Sweden: Anders Ternhag (Karolinska University Hospital and Public Health Agency of Sweden, Stockholm); Christian Giske (Karolinska University Hospital, Stockholm); Karin Tegmark Wisell (Public Health Agency of Sweden, Stockholm)
United Kingdom: Diogo Pereira Marques, Arlene Reynolds, Jim McMenamin (Health Protection Scotland, Glasgow)
Conflict of interest: None declared.
Authors’ contributions: VC, SU, MB, MI, KL, RD, CB, RNP, SP and OBS conceived the study. VC designed, issued and collated results from the questionnaire. VC, MB and XZ undertook data analysis. XZ and MB undertook modelling and statistics. MB, XZ, VC and SU drafted the manuscript. The ESCMID Study Group for Mycoplasma and Chlamydia Infections (ESGMAC) Mycoplasma pneumoniae subgroup members contributed information on country practice or data for evaluation; with some stating that they have no system or that they have a system but cannot return data. All authors contributed to the manuscript and draft and approved study design.
References
- 1.Sánchez-Vargas FM, Gómez-Duarte OG. Mycoplasma pneumoniae-an emerging extra-pulmonary pathogen. Clin Microbiol Infect. 2008;14(2):105-17. 10.1111/j.1469-0691.2007.01834.x [DOI] [PubMed] [Google Scholar]
- 2.Xin D, Mi Z, Han X, Qin L, Li J, Wei T, et al. Molecular mechanisms of macrolide resistance in clinical isolates of Mycoplasma pneumoniae from China. Antimicrob Agents Chemother. 2009;53(5):2158-9. 10.1128/AAC.01563-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao F, Lv M, Tao X, Huang H, Zhang B, Zhang Z, et al. Antibiotic sensitivity of 40 Mycoplasma pneumoniae isolates and molecular analysis of macrolide-resistant isolates from Beijing, China. Antimicrob Agents Chemother. 2012;56(2):1108-9. 10.1128/AAC.05627-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gullsby K, Olsen B, Bondeson K. Molecular Typing of Mycoplasma pneumoniae Strains in Sweden from 1996 to 2017 and the Emergence of a New P1 Cytadhesin Gene, Variant 2e. J Clin Microbiol. 2019;57(6):e00049-19. 10.1128/JCM.00049-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenglet A, Herrador Z, Magiorakos AP, Leitmeyer K, Coulombier D, European Working Group on Mycoplasm C, European Working Group on Mycoplasma pneumoniae surveillance Surveillance status and recent data for Mycoplasma pneumoniae infections in the European Union and European Economic Area, January 2012. Euro Surveill. 2012;17(5):20075. 10.2807/ese.17.05.20075-en [DOI] [PubMed] [Google Scholar]
- 6.Rasmussen JN, Voldstedlund M, Andersen RL, Ellermann-Eriksen S, Jensen TG, Johansen HK, et al. Increased incidence of Mycoplasma pneumoniae infections detected by laboratory-based surveillance in Denmark in 2010. Euro Surveill. 2010;15(45):19708. [PubMed] [Google Scholar]
- 7.Rastawicki W, Kaluzewski S, Jagielski M, Gierczyski R. Epidemiology of Mycoplasma pneumoniae infections in Poland : 28 years of surveillance in Warsaw 1970-1997. Euro Surveill. 1998;3(10):99-100. 10.2807/esm.03.10.00095-en [DOI] [PubMed] [Google Scholar]
- 8.Zhang XS, Zhao H, Vynnycky E, Chalker V. Positively interacting strains that co-circulate within a network structured population induce cycling epidemics of Mycoplasma pneumoniae. Sci Rep. 2019;9(1):541. 10.1038/s41598-018-36325-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Statens Serum Institut (SSI). Epidemi med Mycoplasma pneumoniae. [Epidemic with Mycoplasma pneumoniae]. Copenhagen; SSI: October 2016. Danish. Available from: https://www.ssi.dk/aktuelt/nyhedsbreve/epi-nyt/2016/uge-41---2016
- 10.Kawai Y, Miyashita N, Kubo M, Akaike H, Kato A, Nishizawa Y, et al. Nationwide surveillance of macrolide-resistant Mycoplasma pneumoniae infection in pediatric patients. Antimicrob Agents Chemother. 2013;57(8):4046-9. 10.1128/AAC.00663-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Public Health England (PHE). Annual summary of Mycoplasma pneumoniae laboratory surveillance data, 2016, England and Wales. London: PHE; January 2017. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/817798/MycoplasmaAnnualSummary2016.pdf
- 12.Vega T, Lozano JE, Meerhoff T, Snacken R, Mott J, Ortiz de Lejarazu R, et al. Influenza surveillance in Europe: establishing epidemic thresholds by the moving epidemic method. Influenza Other Respir Viruses. 2013;7(4):546-58. 10.1111/j.1750-2659.2012.00422.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peuchant O, Ménard A, Renaudin H, Morozumi M, Ubukata K, Bébéar CM, et al. Increased macrolide resistance of Mycoplasma pneumoniae in France directly detected in clinical specimens by real-time PCR and melting curve analysis. J Antimicrob Chemother. 2009;64(1):52-8. 10.1093/jac/dkp160 [DOI] [PubMed] [Google Scholar]
- 14.Dumke R, Benitez AJ, Chalker V, Gullsby K, Henrich B, Hidalgo-Grass C, et al. Multi-center evaluation of one commercial and 12 in-house real-time PCR assays for detection of Mycoplasma pneumoniae. Diagn Microbiol Infect Dis. 2017;88(2):111-4. 10.1016/j.diagmicrobio.2017.03.004 [DOI] [PubMed] [Google Scholar]
- 15.Nir-Paz R, Michael-Gayego A, Ron M, Block C. Evaluation of eight commercial tests for Mycoplasma pneumoniae antibodies in the absence of acute infection. Clin Microbiol Infect. 2006;12(7):685-8. 10.1111/j.1469-0691.2006.01469.x [DOI] [PubMed] [Google Scholar]
- 16.He XY, Wang XB, Zhang R, Yuan ZJ, Tan JJ, Peng B, et al. Investigation of Mycoplasma pneumoniae infection in pediatric population from 12,025 cases with respiratory infection. Diagn Microbiol Infect Dis. 2013;75(1):22-7. 10.1016/j.diagmicrobio.2012.08.027 [DOI] [PubMed] [Google Scholar]
- 17.Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. Mycoplasma pneumoniae from the Respiratory Tract and Beyond. Clin Microbiol Rev. 2017;30(3):747-809. 10.1128/CMR.00114-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown RJ, Macfarlane-Smith L, Phillips S, Chalker VJ. Detection of macrolide resistant Mycoplasma pneumoniae in England, September 2014 to September 2015. Euro Surveill. 2015;20(48):30078. 10.2807/1560-7917.ES.2015.20.48.30078 [DOI] [PubMed] [Google Scholar]
- 19.Dumke R, Lück C, Jacobs E. Low rate of macrolide resistance in Mycoplasma pneumoniae strains in Germany between 2009 and 2012. Antimicrob Agents Chemother. 2013;57(7):3460. 10.1128/AAC.00706-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereyre S, Touati A, Petitjean-Lecherbonnier J, Charron A, Vabret A, Bébéar C. The increased incidence of Mycoplasma pneumoniae in France in 2011 was polyclonal, mainly involving M. pneumoniae type 1 strains. Clin Microbiol Infect. 2013;19(4):E212-7. 10.1111/1469-0691.12107 [DOI] [PubMed] [Google Scholar]
- 21.Averbuch D, Hidalgo-Grass C, Moses AE, Engelhard D, Nir-Paz R. Macrolide resistance in Mycoplasma pneumoniae, Israel, 2010. Emerg Infect Dis. 2011;17(6):1079-82. 10.3201/eid/1706.101558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao F, Qu JX, Liu ZJ, Qin XG, Cao B. [The clinical characteristics, treatment and outcome of macrolide-resistant Mycoplasma pneumoniae pneumonia in children]. Zhonghua Jie He He Hu Xi Za Zhi. 2013;36(10):756-61. [PubMed] [Google Scholar]
- 23.Zhou Z, Li X, Chen X, Luo F, Pan C, Zheng X, et al. Macrolide-resistant Mycoplasma pneumoniae in adults in Zhejiang, China. Antimicrob Agents Chemother. 2015;59(2):1048-51. 10.1128/AAC.04308-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalker VJ, Stocki T, Mentasti M, Fleming D, Sadler C, Ellis J, et al. Mycoplasma pneumoniae infection in primary care investigated by real-time PCR in England and Wales. Eur J Clin Microbiol Infect Dis. 2011;30(7):915-21. 10.1007/s10096-011-1176-3 [DOI] [PubMed] [Google Scholar]
- 25.Kenri T, Okazaki N, Yamazaki T, Narita M, Izumikawa K, Matsuoka M, et al. Genotyping analysis of Mycoplasma pneumoniae clinical strains in Japan between 1995 and 2005: type shift phenomenon of M. pneumoniae clinical strains. J Med Microbiol. 2008;57(Pt 4):469-75. 10.1099/jmm.0.47634-0 [DOI] [PubMed] [Google Scholar]
- 26.Kogoj R, Praprotnik M, Mrvič T, Korva M, Keše D. Genetic diversity and macrolide resistance of Mycoplasma pneumoniae isolates from two consecutive epidemics in Slovenia. Eur J Clin Microbiol Infect Dis. 2018;37(1):99-107. 10.1007/s10096-017-3106-5 [DOI] [PubMed] [Google Scholar]
- 27.Dumke R, Von Baum H, Lück PC, Jacobs E. Subtypes and variants of Mycoplasma pneumoniae: local and temporal changes in Germany 2003-2006 and absence of a correlation between the genotype in the respiratory tract and the occurrence of genotype-specific antibodies in the sera of infected patients. Epidemiol Infect. 2010;138(12):1829-37. 10.1017/S0950268810000622 [DOI] [PubMed] [Google Scholar]
- 28.Martínez MA, Ruiz M, Zunino E, Luchsinger V, Aguirre R, Avendaño LF. Identification of P1 types and variants of Mycoplasma pneumoniae during an epidemic in Chile. J Med Microbiol. 2010;59(Pt 8):925-9. 10.1099/jmm.0.018333-0 [DOI] [PubMed] [Google Scholar]
- 29.Zhao F, Liu L, Tao X, He L, Meng F, Zhang J. Culture-Independent Detection and Genotyping of Mycoplasma pneumoniae in Clinical Specimens from Beijing, China. PLoS One. 2015;10(10):e0141702. 10.1371/journal.pone.0141702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown RJ, Nguipdop-Djomo P, Zhao H, Stanford E, Spiller OB, Chalker VJ. Mycoplasma pneumoniae Epidemiology in England and Wales: A National Perspective. Front Microbiol. 2016;7:157. 10.3389/fmicb.2016.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.