Abstract
Background
We aimed to determine sphingolipid levels and examine apoptotic pathways in human retinal pigment epithelial cells (ARPE-19) undergoing endoplasmic reticulum (ER) stress.
Methods
Cells were treated with tunicamycin (TM) to induce ER stress and tauroursodeoxycholic acid (TUDCA), an ER stress inhibitor, was administered to decrease cytotoxicity. Cell viability was measured by MTT assay. Levels of C16–C24 sphingomyelins (SM) and C16–C24 ceramides (CERs) were determined by LC-MS/MS. Glucose-regulated protein 78-kd (GRP78) and nuclear factor kappa-b subunit 1 (NFκB1) gene expressions were evaluated by quantitative PCR analysis, while GRP 78, NF-κB p65, cleaved caspase-3 and caspase-12 protein levels were assesed by immunofluorescence. Ceramide-1-phosphate (C1P) levels were determined by immunoassay, while caspase −3 and −12 activity in cell lysates were measured via a fluorometric method.
Results
Induction of ER stress in TM treated groups were confirmed by significantly increased mRNA and protein levels of GRP78. TM significantly decreased cell viability compared to controls. Treatment with TUDCA along with TM significantly increased cell viability compared to the TM group. A significant increase was observed in C22–C24 CERs, C1P, caspase-3, caspase-12, NFκB1 mRNA and NF-κB p65 protein levels in cells treated with TM compared to controls. Administration of TUDCA lead to a partial decrease in GRP78 expression, NFκB1 mRNA, NF-κB p65 protein, C22–C24 CERs and C1P levels along with a decrease in caspase-3 and -12 activity.
Conclusions
The results of this study reveal the presence of increased long chain CERs, C1P and apoptotic markers in retinal cells undergoing ER stress.
Keywords: Sphingolipid, Tunicamycin, Retinal pigment epithelial cells
List of abbreviations
- AMD
age-related macular degeneration
- Apaf-1
apoptotic protease activating factor-1
- ARPE-19
human retinal pigment epithelial cells
- Bcl-2
B-cell lymphoma/leukemia-2
- C16 CER
N-palmitoyl-D-erythro-sphingosine;
- C16 SM
N-palmitoyl-D-erythro-sphingosylphosphorylcholine
- C18 CER
N-stearoyl-D-erythro-sphingosine;
- C18 SM
N-stearoyl-D-erythro sphingosylphosphorylcholine;
- C1P
ceramide-1-phosphate
- C1P
ceramide-1-phosphate
- C20 CER
N-arachidoyl-D-erythro-sphingosine;
- C22 CER
N-behenoyl-D-erythro-sphingosine;
- C24 CER
N-lignoceroyl-D-erythro-sphingosine;
- C24 SM
N-lignoceroyl-D-erythro sphingosylphosphorylcholine;
- CERs
ceramides
- DMSO
dimethyl sulfoxide;
- ESI
electrospray ionization
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- GRP78
glucose-regulated protein 78-kd
- MRM
multiple reaction monitoring
- MTT
3-(4,5)dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
- NFκB1
nuclear factor kappa-b subunit 1
- N-SMase
neutral SMase
- OMIM
Online Mendelian Inheritance in Man
- PP2A
protein phosphatase 2A
- RPE
retinal pigment epithelium
- SERCA
sarcoplasmic/endoplasmic reticulum Ca2+- ATPase
- SM
sphingomyelins
- SMase
sphingomyelinase
- Tg
thapsigargin
- TM
tunicamycin
- TUDCA
tauroursodeoxycholic acid
- UFLC
ultrafast-liquid chromatography
- UPR
unfolded protein response
1. Introduction
Human retinal pigment epithelium (RPE) is a monolayer of cells found on the outer surface of the neural retina and is vital in preserving retinal function. Closeness of choroidal capillaries facilitates RPE cells to supply nutrients to sustain visual function and to participate in structuring the outer blood-retinal barrier that precludes nonspecific diffusion and transport of material from the choroid [1]. Ischemic and hypoxic injuries as well as many retinal diseases affect RPE cells [2]. Age-related modifications of the RPE comprise the decrease of cell density that can be caused by apoptosis resulting from accumulation of toxic substances [3]. Oxidative stress [4], hyperglycemia [5], mitochondrial dysfunction [6] and ER stress [7] are amongst the numerous investigated proapoptotic factors in RPE cell lines. Endoplasmic reticulum and apoptosis is a recently revealed mechanism of cell death observed in RPE cells [8] and is an important feature of age-related macular degeneration, the most common cause of irreversible vision loss especially in the elderly population over 65 years of age [9].
Basic and clinical research have presented data that ER stress and apoptotic cell death is an important feature in the pathogenesis of age-related macular degeneration (AMD) and other retinal degenerative diseases [10]. However, the mechanism of increased ER stress induced apoptosis in retinal pigment epithelial cells in retinal degeneration remain largely unknown. Understanding the possible mechanisms of dysregulated apoptosis is important because it may initiate novel approaches for the prevention and treatment of retinal degenerative diseases.
Protein-folding, -maturation, -trafficking, lipid synthesis and intracellular calcium homeostasis are several of the key functions of the ER [11]. Endoplasmic reticulum stress results in buildup of unfolded proteins in the ER lumen leading to unstable protein homeostasis. Cells turn on an adaptive system known as the unfolded protein response (UPR) to eradicate toxic protein components which alleviates ER stress and re-establishes protein homeostasis [12]. However, extended periods of ER stress results in failed UPR response and activates the apoptotic cascade [12]. Fundamental mechanisms resulting in the switch of UPR from a prosurvival to a proapoptic stimulus are still not clearly understood.
Alterations in sphingolipid metabolism and buildup of ceramide have been demonstrated to induce ER stress and apoptosis in retinal pigment epithelium subjected to UV and hydrogen peroxide [13]. Ceramide can be generated through de novo synthesis, hydrolysis of sphingomyelin by sphingomyelinase (SMase) and breakdown of glycosphingolipids [14]. Among the different types of SMase enzymes that are present, acid and neutral SMase (N-SMase) have been investigated extensively in response to cellular stress. Endoplasmic reticulum stress response in insulinoma cells was shown to be associated with both increased message and protein levels of N-SMase [15], while on the contrary, ER stress was demonstrated to inhibit N-SMase activity in RPE cells [7].
Localization of ceramide in sphingolipid rich membrane fractions in the mitochondria constitutes the basis for the functional interaction between ceramide and apoptosis [16]. Increased ceramide activates protein phosphatase 2A (PP2A) which in turn leads to dephosphorylation of B-cell lymphoma/leukemia-2 (Bcl-2) protein that causes inactivation of Bcl2's anti-apoptotic function. Ceramide is the principal core for sphingolipid metabolism; its decomposition or alteration leads to the production of several sphingolipids such as C1P that, like ceramide itself, can possiblely serve as both structural and/or cell signaling molecule [17]. Here, we used a cell model to test the hypothesis that ER stress changes sphingolipid levels and induces apoptotic pathways in retinal pigment epithelial cells.
2. Materials and methods
2.1. Cell culture and treatment conditions
The ARPE-19 cell line is a human -RPE-cell line and was obtained from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM): Ham's Nutrient Mixture (F-12), 1:1 (Gibco, Life Technologies Limited, Paisley, UK) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA), 1% penicillin–streptomycin (Gibco, Life Technologies, Grand Island, NY, USA). Cultures were maintained at 37 °C in a humidified atmosphere of 95% air and 5% CO2. After the cell culture reached 80% confluency, the cells were passaged by trypsinization (0.05% Trypsin- EDTA; Gibco, Life Technologies, Paisley, United Kingdom). Cells from passages ten to fifteen were used in the studies.
Tunicamycin (Sigma-Aldrich, St. Louis, MO, USA) was prepared in dimethyl sulfoxide (DMSO; Calbiochem, EMD Bioscience Inc. La Jolla, CA, USA) at a stock concentration of 10 mg/ml. The ER stress inhibitor TUDCA (EMD Millipore Corp. Billerica, MA USA) was dissolved in saline at a stock concentration of 10 mM.
In most of the experiments cells were treated with 10 μg/ml TM for 24 h to induce ER stress. The dose and duration of TM used to treat cells were based on the results of earlier studies [7]. TUDCA was administered as an ER stress inhibitor. In most of the experiments cells were treated with 0.5 mM of TUDCA, administered for 24 h.
2.2. Cell viability assay
Cell viability was measured using the colorimetric 3-(4,5)dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit (Millipore Corporation, Billerica, MA, USA). Living cells reduced MTT to formazan which was quantified by measuring absorbance at 570 nm (MicroQuant Plate Reader, Bio-Tek Instruments Inc. Vermont, USA). Cells were grown to confluence in 96-well plates and incubated with 1 μl/ml DMSO, 1–10 μg/ml TM, 0.1–5 mM TUDCA or 10 μg/ml TM + 0.1–5 mM TUDCA for 18–48 h. Control cells were prepared in plates containing only medium. At the end of the incubation period, MTT was added to each well and incubation was carried out for 4 h at 37 °C. Formazan production was expressed as a percentage of the values obtained from control cells.
2.3. Measurement of sphingomyelins and ceramides
An optimized multiple reaction monitoring (MRM) method was developed using ultrafast-liquid chromatography (UFLC) coupled with MS/MS as previously described [18]. Standards for N-palmitoyl-D-erythro-sphingosylphosphorylcholine (C16 SM), N-stearoyl-D-erythro sphingosylphosphorylcholine (C18 SM), N-lignoceroyl-D-erythro sphingosylphosphorylcholine (C24 SM), N-palmitoyl-D-erythro-sphingosine (C16 CER), N-stearoyl-D-erythro-sphingosine (C18 CER), N-arachidoyl-D-erythro-sphingosine (C20 CER), N-behenoyl-D-erythro-sphingosine (C22 CER) and N-lignoceroyl-D-erythro-sphingosine (C24 CER) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Labeled C16 CER d18:1/16:0 (Palmitoyl-U-13C16) internal standard was obtained from Cambridge Isotope Laboratories (Andover, MA, USA). Solutions of each sphingolipid standard was prepared in methanol (Sigma-Aldrich, St. Louis, MO, USA) at 40 °C with sonication. A UFLC system (LC-20 AD UFLC XR, Shimadzu Corporation, Japan) was coupled to a LCMS-8040 triple quadrupole mass spectrometer (Shimadzu Corporation, Japan). Chromatographic separations were carried out using an HPLC column (XTerra C18, 2.1 mm × 50 mm, Waters, MA, USA) maintained at 60 °C. Sphingolipids were separated using a gradient elution with a flow rate of 0.45 ml/min. Mobile phase solvent A was water–acetonitrile–2-propanol (8:1:1, v/v/v) with 10 mM ammonium formate and solvent B was acetonitrile–2-propanol (9:1, v/v). Gradient program was solvent B, 65% (0–2 min), 90% (2.01–13 min), 100% (13.01–20 min) and 65% (20.1–23 min). MRM transitions and responses were automatically optimized for individual compounds in positive electrospray ionization (ESI). In the positive ESI-MS mode the precursor and product m/z values for all analyzed sphingolipids were as follows: C16 SM, precursor m/z: 703.30, product m/z: 184.20; C18 SM, precursor m/z: 731.40, product m/z: 184.20; C24 SM, precursor m/z: 815.50, product m/z: 184.20; C16 CER, precursor m/z: 538.50, product m/z: 264.40; C16 CER *IS, precursor m/z: 554.30, product m/z: 264.30; C18 CER, precursor m/z: 566.30, product m/z: 264.40; C20 CER, precursor m/z: 594.60, product m/z: 264.50; C22 CER, precursor m/z: 622.60, product m/z: 264.40; C24 Ceramide, precursor m/z: 650.40, product m/z: 264.30. Responses to analyzed sphingolipids were optimized to a linear calibration range from 39 to 625 ng/ml and a sample analysis time of 30 min.
2.4. Preparation of samples for mass spectrometric analysis
Sphingolipid extraction was done as previously described [19]. Two μl of 5 μg/ml internal standard solution was added to 500 μl of cell lysate. Samples were briefly vortexed and 375 μl of chloroform/methanol (1:2, v/v) was added. The samples were sonicated for 30 sn and 100 μl of water was added. The mixtures were vortexed for 5 min and stood at room temperature for 30 min. After centrifugation at 2000g for 5 min, the supernatant was taken and 125 μl of chloroform and 125 μl of water was added. The samples were vortexed and stood for 30 min for phase separation. The upper organic layer was transferred to glass tubes and evaporated at room temperature under a constant stream of nitrogen with height adjustable gas distribution unit (VLM, Bielefeld, Germany). The dried residue was dissolved in 100 μl of methanol and 10 μl was injected into the column.
2.5. Measurement of sphingomyelinase activity
Neutral-SMase activity was measured in cell extracts via a sphingomyelinase assay kit (Abcam, Catalog # ab138876, Cambridge, UK). This assay utilizes sphingomyelin as substrate to specifically monitor SMase activity. First, SMase hydrolyses sphingomyelin to yield ceramide and phosphocholine. The absorbance of the colorimetric probe at 655 nm is proportional to the formation of phosphocholine, therefore to the SMase activity. A standard curve of absorbance values of known amounts of sphingomyelinase standards was generated. Sphingomyelinase activity in the samples (mU/ml) were calculated from their corresponding absorbance values via the standard curve.
2.6. Measurement of ceramide-1-phosphate levels
Ceramide-1-phosphate levels were measured in cell extracts via an ELISA kit (Shanghai YL Biotech Co., Ltd. Catalog # YLA3764HU Shanghai, China). Cellular C1P captured by a solid phase monoclonal antibody was detected with a biotin-labeled polyclonal antibody. A streptavidin-peroxidase conjugate was then added to bind the biotinylated antibody. A TMB substrate was added and the yellow product was measured at 450 nm. A standard curve of absorbance values of known C1P standards was plotted as a function of C1P standard concentrations using the GraphPad Prism Software program for windows version 5.03. (GraphPad Software Inc.). The amount of C1P in the samples were calculated from their corresponding absorbance values via the standard curve.
2.7. RNA extraction
Total RNA was purified from cells using AxyPrep Multisource Total RNA Miniprep Kit (Axygen Biosciences, CA, USA) according to manufacturer instructions. Purified RNA concentration was determined spectrophotometrically at 260 nm. RNA was dissolved with 70 μl TE buffer [10 mM Tris-HCl, 0.1 mM EDTA (pH 7.5)]. 10 μl dissolved RNA was added to 490 μl distilled water (1:50 dilution). Diluted RNA sample was measured at an absorbance of 260 and 280 nm. One absorbance unit at 260 nm was equal to 40 μg/ml of RNA. Prior to quantitative real-time PCR (Q-RT-PCR) analysis, total RNA samples were diluted with RNase-free water to a final concentration of 66 ng/μl.
2.8. Primer and probe design and optimization
Online Mendelian Inheritance in Man (OMIM) database was used as a source for all mRNA sequences. The primers and probes were designed using the Oligoware 1.0 software as previously [20] and were synthesized by Metabion International AG (Steinkirchen, Germany). The primers and probes used in the study are shown in Table 1. Real time PCR was performed using one-run RT PCR kit (SNP Biyoteknoloji, Ankara, Turkey). Primer concentrations were optimized by varing concentrations of both forward and reverse primers in order to determine the minimum primer concentration which would generate the maximum ΔRn (difference between baseline and maximal fluorescence of sample). Optimum probe concentration was determined by running reactions under the optimum primer concentrations and varying the probe concentration. The probe concentration that generated the lowest Ct (threshold cycle defined as the fractional cycle number at which the amount of the amplified target reaches a fixed threshold) was used as the optimum probe concentration. The optimized buffer composition was as follows: 18.3 μl one run miks, 1.1 μl primer mix (10 pmol), 1.1 μl probe (5 pmol) and 0.3 μl RT/hotstart Taq mix. Total RNA was added at 5 μl of volume for measurement of target mRNA in each tissue. The RT-PCR assay was performed using Mx3000p Multiplex Quantitative PCR system (Stratagene, CA, USA). Cycling conditions were: Step 1. 30 min at 50 °C; Step 2. 10 min at 95 °C and then 40 cycles of Step 3. 15 s at 95 °C and then 1 min at 60 °C. Control reactions confirmed that no amplification occurred when “no template” control reactions were performed.
Table 1.
Primers and probes used for real-time-PCR analysis.
| Gene | Accession | ||
|---|---|---|---|
| ACTB | Forward Primer | 5′-CCC AGC ACA ATG AAG ATC AAG ATC-3′ | OMIM -102630 |
| Reverse Primer | 5′-GGGTGTAACGCAACTAAGTCATAGTC-3′ | ||
| Probe | 5′-AGATCATTGCTCCTCCTGAGCGCAAG-3′ | ||
| GRP78 | Forward Primer | 5′-ACAATCAAGGTCTATGAAGGTGAAAGAC-3′ | OMIM -138120 |
| Reverse Primer | 5′-CTCGAAGAATACCATTCACATCTATCTC-3′ | ||
| Probe | 5′-ACTTCAATC TGTGGGACCCCACGAG -3′ | ||
| NFκB1 | Forward Primer | 5′-CCTCCACAAGGCAGCAAATAGACG -3′ | OMIM -164011 |
| Reverse Primer | 5′-AGAGTTAGCAGTGAGGCACCACT -3′ | ||
| Probe | 5′-AGACAGTGA CAGTGTCTGCGACAGC-3′ |
OMIM, Online Mendelian Inheritance in Man; ACTB, actin beta; GRP78, glucose-regulated protein 78-kd.
NFκB1, nuclear factor kappa-b subunit 1.
2.9. Quantitative PCR analysis
Using the MxPro QPCR Software for Mx3000P detection system, the log-linear phase of amplification was monitored to obtain Ct values for each RNA sample. All reactions were run in triplicate and the mRNA levels of each gene normalized to beta actin were expressed as fold change, 2-(ΔΔct) as previously described [21]. The relative change in the mRNA levels of the genes studied were calculated with respect to control operated group by the following equation:
Fold change = 2-(ΔΔct)
| ΔCt= (Ct target-Ct actin beta) |
| ΔΔ Ct = ΔCt treatment group-ΔCt control |
2.10. Caspase activity
Caspase −3 (Catalog #K102-100; BioVision Milpitas, CA, USA) and-12 (Catalog #K139-100; BioVision Milpitas, CA, USA) activity in cell lysates was measured via fluorometric assay kits. ARPE-19 cells were incubated with either 1 μl/ml DMSO; 0.5 mM TUDCA; 10 μg/ml TM and 10 μg/ml TM + 0.5 mM TUDCA for 24 h. Control cells were incubated with only medium. At the end of the incubation period, cells were washed with PBS, 1–5 x106 cells were resuspended in lysis buffer and incubated on ice for 10 min. Caspase-3 and -12 activity were determined by using 50 and 100 μg cell lysate protein, respectively. Caspase activity was measured based on fluorometric detection of the fluorophore 7-amino-4-trifluoromethyl coumarin (AFC) after cleavage from the peptide substrates. Comparison of the fluorescence of AFC in sample groups with control group allowed the determination of fold change in caspase activity.
2.11. Immunofluorescent staining
For immunofluorescence cell-imaging studies, cells were plated at a density of 100,000 cells/chamber in an 8-chamber slide (Merck Millipore, Cork, Ireland) one day prior to staining in order to achieve 70% confluency. Cells were fixed in 4% freshly prepared formaldehyde for 5 min and permeabilized in PBS with %0.1 Triton X-100 for 5 min at room temperature. Cells were then incubated for 1 h in blocking solution (3% bovine serum albumin/0,08% glycine in PBS) and treated with either anti-GRP 78 (1:100, #ab21685 Abcam, Cambridge, MA, USA); anti–NF–κB p65 (1:100, #ab16502, Abcam, Cambridge, MA, USA); anti-cleaved caspase-3 (1:100, #9664, Cell Signaling Technology, Danvers, MA, USA); anti-caspase 12 (1:100, #ab62463 Abcam, Cambridge, MA, USA); overnight. The secondary antibody, Alexa Fluor-488 conjugated goat anti-rabbit (1:300, # A11008 life Technologies, USA), was applied for 1 h at room temperature and nuclei were counterstained with DAPI (Vector Laboratories Inc.,Burlingame, CA, USA) in all experiments. Slides were viewed under a fluorescence microscope (Olympus BX61 fully automated, Tokyo, Japan) and fluorescence intensity was quantified using NIH ImageJ 1.44p software.
2.12. Protein measurements
Protein concentrations were measured at 595 nm by a modified Bradford assay using Coomassie Plus reagent with bovine serum albumin as a standard (Pierce Chemical Company, Rockford, IL).
2.13. Statistical analysis
Statistical analysis was performed using SigmaStat statistical software version 3.5 (Sigma, St. Louis, MO, USA). Statistical analysis for each measurement is described in figure and table legends. To compare the groups via the SigmaStat statistical sofware, we first performed a normality test. A test that passed indicated that the data matched the pattern expected if the data was drawn from a population with a normal distribution. If the sample data were not normally distributed, the normality test failed. In such case the software performed a nonparametric test. The experimental groups were compared by either One Way ANOVA (analysis of variance) or by Kruskal-Wallis One Way ANOVA on Ranks. When there was a statistically significant difference, we used multiple comparison procedures also known as post-hoc tests to determine exactly which groups were different.
3. Results
3.1. Analysis of cell viability
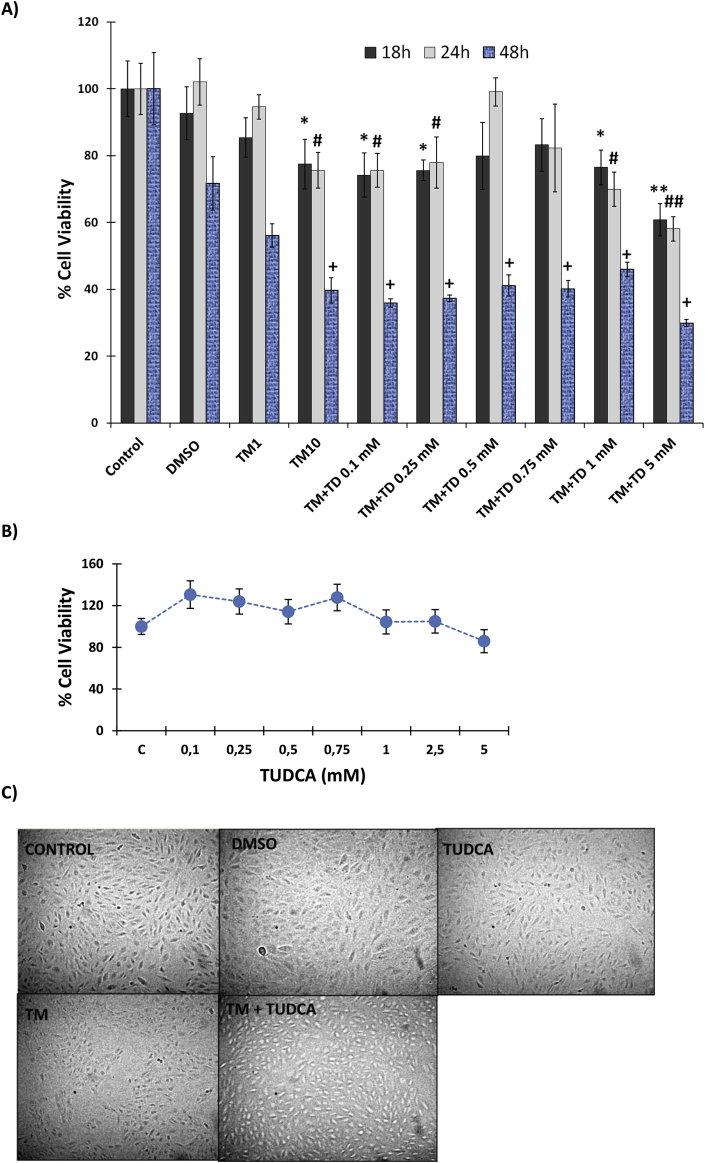
Incubation of human RPE cells with 1 μl/ml DMSO and 1 μg/ml TM showed no cytotoxicity at 18–48h, while 10 μg/ml TM was significantly cytotoxic (Fig. 1A). Incubation of ARPE-19 cells with 10 μg/ml TM, for 24 and 48 h significantly decreased cell viability when compared to control groups (Fig. 1A). Treatment of cells with 0,1–0.25 mM or 1–5 mM TUDCA together with 10 μg/ml TM did not increase cell viability during a 48 h time-course study. Cells treated with 0.5 mM TUDCA for 24 h showed a pronounced increase in cell viability in the presence of 10 μg/ml TM, which was significantly greater than 10 μg/ml TM treatment alone (Fig. 1A). Fig. 1B shows dose-dependent effect of 0.1–5 mM TUDCA on cell viability at 24 h time-course. No significant difference was observed in cell viability compared to non-treated controls. Fig. 1C shows morphological changes that occurred in ARPE-19 cells treated with 1 μl/ml DMSO; 0.5 mM TUDCA; 10 μg/ml TM and 10 μg/ml TM + 0.5 mM TUDCA for 24 h. Cells treated with 10 μg/ml TM showed morphological changes such as cell shrinkage, detachment and rounding. A decrease in cell population was also noted in the TM group.
Fig. 1.
legend: Cell viability analysis. A) Cell viability assessed by MTT assay. DMSO, cells treated with dimethyl sulfoxide (1 μl/ml); TM1, cells treated with 1 μg/ml tunicamycin; TM10, cells treated with 10 μg/ml tunicamycin. TM + TD, cells treated with 10 μg/ml TM and TUDCA at indicated doses. Data shown are representative of 6 separate experiments and values are given as mean ± SD. Statistical analysis was performed by one way analysis of variance with all pairwise multiple comparison procedures done by Tukey test. *, p < 0.001 vs. 18h control. **, p < 0.001 vs. 18h control, 18h DMSO, 18 h TM1, 18 h TM + TD 0.5 mM and 18 h TM + TD 0,75 mM #, p < 0.001 vs. 24h control, 24 h DMSO and 24 h TM + TD 0.5 mM ##, p < 0.001 vs. 24h control, 24h DMSO, 24 h TM1, 24 h TM + TD 0.5 mM and 24 h TM + TD 0,75 mM+, p < 0.001 vs.48h control. B) Dose-dependent effect of TUDCA on cell viability at 24 h time-course. C, control. Data shown are representative of 3 separate experiments and values are given as mean ± SD. No significant difference was observed in cell viability among the different dose groups. C) Morphological changes of ARPE-19 cells observed under an inverted light microscope (20× magnification). Cells were treated with 1 μl/ml DMSO; 0.5 mM taurohyodeoxycholic acid (TUDCA); 10 μg/ml tunicamycin (TM) and 10 μg/ml TM + 0.5 mM TUDCA for 24 h. Cells treated with 10 μg/ml TM showed morphological changes such as cell shrinkage, detachment and rounding. A decrease in cell population was also noted in the TM group.
3.2. Sphingomyelin and ceramide levels
Levels of endogenous sphingolipids measured in ARPE-19 cells are given in Table 2. No significant change was observed in measured sphingomyelins and C16–C20 CERs among the experimental groups. A significant increase was found in C22 and C24 CER levels in ARPE-19 cells treated with 10 μg/ml TM compared to all experimental groups. 0.5 mM TUDCA treatment caused a significant decrease in long chain CER levels in cells treated with 10 μg/ml TM compared to TM treatment alone. However, levels of C22 and C24 CERs were still significantly higher in cells treated with 10 μg/ml TM + 0.5 mM TUDCA compared to control, DMSO and TUDCA groups.
Table 2.
Levels of sphingolipids in ARPE-19 cells.
| Sphingolipids (ng/mg protein) | Control | DMSO | TUDCA | TM | TM + TUDCA |
|---|---|---|---|---|---|
| 16:0 SM (d18:1/16:0) | 847,63 ± 66,99 | 751,87 ± 24,19 | 733,76 ± 39,49 | 739,98 ± 56,56 | 768,16 ± 40,94 |
| 18:0 SM (d18:1/18:0) | 299,77 ± 29,59 | 327,45 ± 29,32 | 297,56 ± 35,94 | 302,22 ± 28,29 | 334,33 ± 55,17 |
| 24:0 SM (d18:1/24:0) | 130,63 ± 12,41 | 133,58 ± 8,52 | 121,65 ± 14,00 | 137,61 ± 8,80 | 127,46 ± 9,00 |
| C16 Ceramide (d18:1/16:0) | 116,86 ± 8,22 | 101,35 ± 2,90 | 103,90 ± 14,12 | 117,10 ± 5,20 | 93,64 ± 8,41 |
| C18 Ceramide (d18:1/18:0) | 37,72 ± 4,06 | 34,69 ± 1,74 | 36,83 ± 5,22 | 33,43 ± 2,87 | 33,12 ± 4,70 |
| C20 Ceramide (d18:1/20:0) | 24,74 ± 4,21 | 27,16 ± 3,45 | 21,40 ± 2,63 | 23,79 ± 1,56 | 20,68 ± 2,45 |
| C22 Ceramide (d18:1/22:0) | 47,06 ± 4,70 | 50,92 ± 3,52 | 38,39 ± 2,91 | 130,21 ± 6,61* | 97,88 ± 1,54** |
| C24 Ceramide (d18:1/24:0) | 267,55 ± 5,82 | 241,14 ± 10,30 | 257,30 ± 7,63 | 402,06 ± 9,01* | 302,03 ± 9,23** |
All values are mean ± SD and n = 3 in each experimental group. SM; sphingomyelin. Cells were treated with 1 μl/ml dimethyl sulfoxide (DMSO); 0.5 mM taurohyodeoxycholic acid (TUDCA); 10 μg/ml tunicamycin (TM) and 10 μg/ml TM + 0.5 mM TUDCA for 24 h. Statistical analysis was performed by one way analysis of variance with all pairwise multiple comparison procedures were done by Tukey test. *, p < 0,001 vs. Control, DMSO, TUDCA and TM + TUDCA. **, p < 0,001 vs. Control, DMSO and TUDCA.
3.3. Neutral sphingomyelinase activity
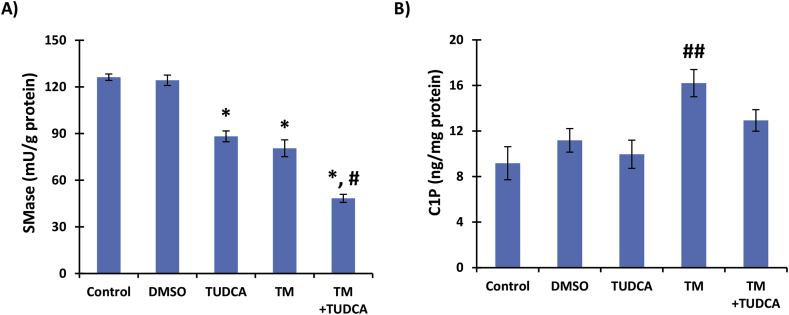
Endoplasmic reticulum stress induced by 24 h treatment with 10 μg/ml TM caused a significant reduction in SMase activity (80,52 ± 5,39 mU/g protein) when compared to control (126,21 ± 2,06 mU/g protein) or DMSO treated cells (124,25 ± 3,36 mU/g protein). This effect was similar with 0.5 mM TUDCA treatment (88,22 ± 3,48 mU/g protein) (Fig. 2A). Cells treated with 10 μg/ml TM + 0.5 mM TUDCA showed a further significant decrease in SMase activity (48,33 ± 2,60 mU/g protein) when compared to 10 μg/ml TM or 0.5 mM TUDCA alone.
Fig. 2.
legend: A) Neutral sphingomyelinase (SMase) activity B) Ceramide-1-phosphate (C1P) levels. ARPE-19 cells treated with 1 μl/ml dimethyl sulfoxide (DMSO); 0.5 mM taurohyodeoxycholic acid (TUDCA); 10 μg/ml tunicamycin (TM) and 10 μg/ml TM + 0.5 mM TUDCA for 24 h. All values are mean ± SD. Data shown are representative of 8 separate experiments. Statistical analysis for SMase measurements were by One Way Analysis of Variance and all pairwise multiple comparison procedures were done by Tukey test.*, p < 0,001 vs. control and DMSO. #, p < 0,001 vs. TUDCA and TM. Statistical analysis for C1P measurements were by Kruskal-Wallis One Way Analysis of Variance and all pairwise multiple comparison procedures were done by Tukey test. ##, p < 0,05 vs. control, DMSO and TUDCA.
3.4. Ceramide-1-phosphate levels
Treatment of ARPE-19 cells with 10 μg/ml TM for 24 h caused a significant increase in C1P levels (mean ± SD) compared to control, DMSO and TUDCA treated groups. Measured C1P levels in control, DMSO, TUDCA, TM and TM + TUDCA groups were 9,17 ± 1,45; 11,18 ± 1,03; 9,96 ± 1,24; 16,20 ± 1,19 and 12,93 ± 0,95 ng/mg protein, respectively (Fig. 2B). Treatment with 0.5 mM TUDCA slightly increased C1P levels in TM treated cells and no significant difference was found between C1P levels between control, DMSO and TUDCA treated groups.
3.5. GRP78 mRNA and GRP78 protein levels
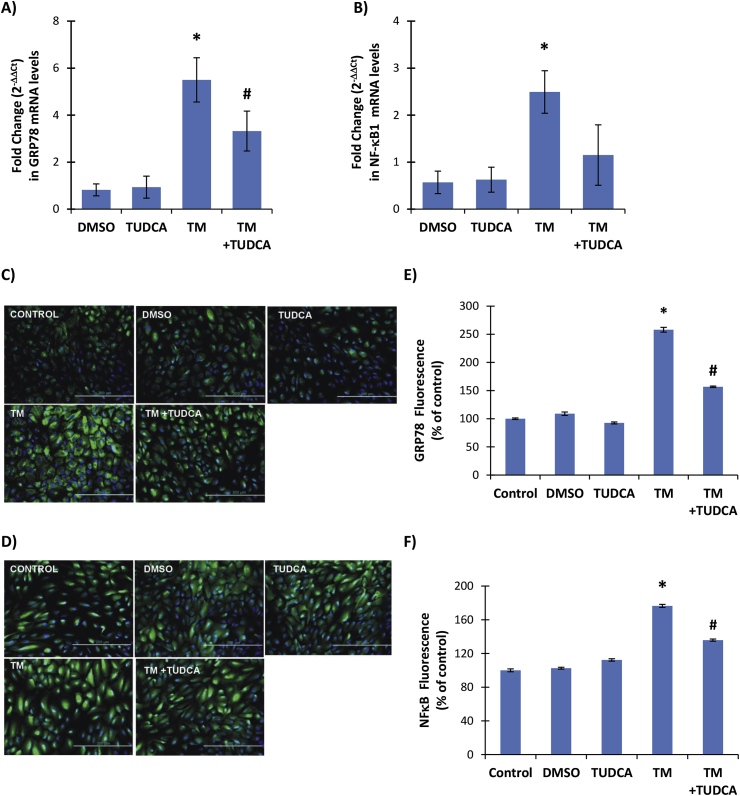
Treatment of ARPE-19 cells with 10 μg/ml TM for 24 h caused a significant increase in ER stress as shown by increased levels of GRP78 mRNA in TM and TM + TUDCA treated cells compared to DMSO and TUDCA groups (Fig. 3A). Treatment with 0.5 mM TUDCA decreased GRP78 mRNA levels in TM treated cells and a significant difference was found between GRP78 mRNA levels between TM and TM + TUDCA treated groups (Fig. 3A). Increased immunostaining of GRP78 was also observed in TM treated cells (Fig. 3C). The immunostaining scores, obtained according to both the percentage and intensity of positive stained cells were statistically increased in TM treated ARPE-19 cells (Fig. 3E).
Fig. 3.
legend: A-B) GRP78 and NF-κB1 mRNA levels. ARPE-19 cells were treated with 1 μl/ml dimethyl sulfoxide (DMSO); 0.5 mM taurohyodeoxycholic acid (TUDCA); 10 μg/ml tunicamycin (TM) and 10 μg/ml TM + 0.5 mM TUDCA for 24 h. Fold change (2−ΔΔCt) in mRNA levels of NFκB1 and GRP78. Values are mean ± SD, n = 3. Statistical analysis was performed by one way analysis of variance with all pairwise multiple comparison procedures done by Holm-Sidak method. *, p < 0.05 vs. DMSO, TUDCA and TM + TUDCA. #, p < 0.05 vs. DMSO and TUDCA. C-D) Representative Immunofluorescent staining of GRP78 and NFκB in ARPE-19 cells, respectively. E-F) Quantitation of fluorescence staining was estimated by ImageJ software. Data shown are representative of 3 separate experiments and values are given as mean ± SD. Statistical analysis was performed by One Way Analysis of Variance and all pairwise multiple comparison procedures done by Tukey test. *, p < 0,001 vs. Control, DMSO, TUDCA and TM + TUDCA. #, p < 0,001 vs. Control, DMSO, TUDCA.
3.6. NF-κB1 mRNA and NF-κB p65 protein levels
Treatment of ARPE-19 cells with 10 μg/ml TM for 24 h caused a significant increase in NF-κB1 mRNA levels in TM treated cells compared to DMSO, TUDCA and TM + TUDCA treated groups (Fig. 3B). Treatment with 0.5 mM TUDCA slightly increased NF-κB1 mRNA levels in TM treated cells and no significant difference was found between NF-κB1 mRNA levels between DMSO and TUDCA treated groups (Fig. 3B). Increased immunostaining of NF-κB p65 protein was also observed in TM treated cells (Fig. 3D). The immunostaining scores, obtained according to both the percentage and intensity of positive stained cells were statistically increased in TM treated ARPE-19 cells (Fig. 3F).
3.7. Caspase activity
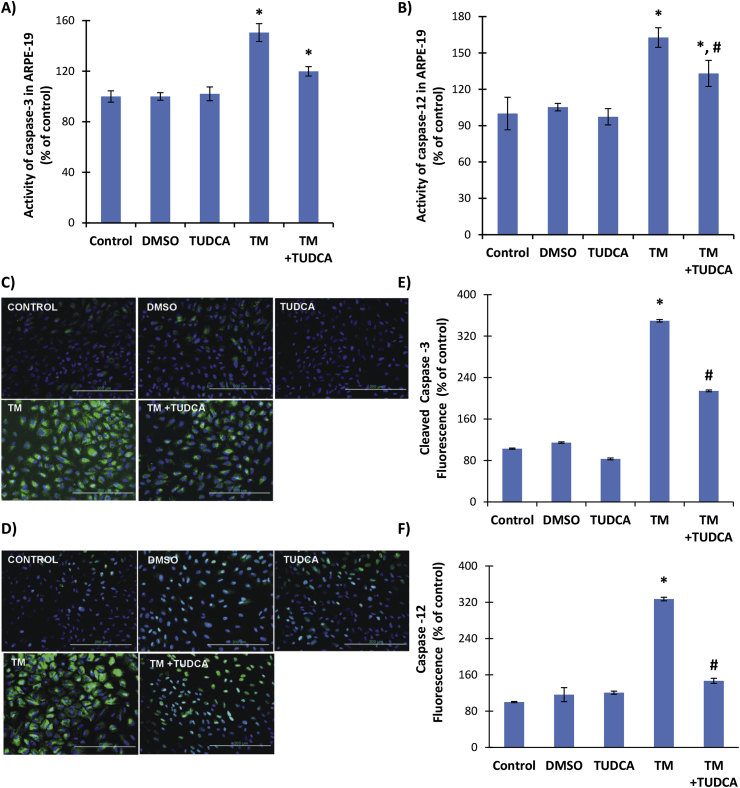
Incubation of ARPE-19 cells with TM for 24 h significantly increased caspase-3 and-12 activity compared to control, DMSO and TUDCA groups (Fig. 4A and B). Treatment with 0.5 mM TUDCA had no effect on caspase-3 activity in TM treated cells (Fig. 4A) but decreased caspase-12 activity in TM treated cells and a significant difference was found between caspase-12 activity between TM and TM + TUDCA treated groups (Fig. 4B). Increased immunostaining of cleaved caspase-3 and caspase 12 protein was also observed in TM treated cells (Fig. 4C and D, respectively). The immunostaining scores, obtained according to both the percentage and intensity of positive stained cells were statistically increased in TM treated ARPE-19 cells (Fig. 4E and F).
Fig. 4.
legend: A) Caspase-3 and B) Caspase-12 activity in ARPE-19 cells. Cells were treated with 1 μl/ml dimethyl sulfoxide (DMSO); 0.5 mM taurohyodeoxycholic acid (TUDCA); 10 μg/ml tunicamycin (TM) and 10 μg/ml TM + 0.5 mM TUDCA for 24 h. All values are mean ± SD. Data shown are representative of 7–8 separate experiments. Statistical analysis was by One Way Analysis of Variance and all pairwise multiple comparison procedures were done by Tukey test. *, p < 0,001 vs. control, DMSO and TUDCA. #p < 0,001 vs. TM. C-D) Representative Immunofluorescent staining of cleaved caspase −3 and caspase −12 in ARPE-19 cells, respectively. E-F) Quantitation of fluorescence staining was estimated by ImageJ software. Data shown are representative of 3 separate experiments and values are given as mean ± SD. Statistical analysis was performed by One Way Analysis of Variance and all pairwise multiple comparison procedures done by Tukey test. *, p < 0,001 vs. Control, DMSO, TUDCA and TM + TUDCA. #, p < 0,001 vs. Control, DMSO, TUDCA.
4. Discussion
This study gives rise to the novel observation that TM treatment in RPE cells leads to increased ceramide levels that can be regulated by an ER stress inhibitor, TUDCA. Information is also presented identifying down regulation of apoptotic response via TUDCA inhibition through a mechanism that involves caspase-12 activity.
Incubation of ARPE-19 cells with 10 μg/ml TM significantly decreased cell viability as previously reported [7]. The results of this study demonstrated that TM directly damaged RPE in cell cultures. These findings are consistint with previous reports showing retinal toxicity of TM both in vivo and in vitro [22,23]. Tunicamycin acts by triggering ER stress through the inhibition of protein glycosylation [24] and the dose of TM administered to RPE cells in this study was determined with reference to previous studies designed to investigate the effects of ER stress on ARPE-19 cell cultures [7]. The generation of cytotoxicity in our experimental cell model could be related to cell death signaling by ER stress response since cells treated with 0.5 mM TUDCA, an ER stress inhibitor, showed a pronounced increase in cell viability in the presence of TM (Fig. 1). TUDCA acts as an effective chemical chaperone in the treatment of ER stress-associated diseases by inhibiting the UPR [25]. TUDCA restrains the initiation of the early signaling steps in the three branches of UPR, preventing the formation and binding of activated transcription factors to the GRP78 promoter for transcriptional activation [25]. The capability of ER stress to induce apoptosis and cytotoxicity in RPE cells have been documented in recent studies [[26], [27], [28]].
A significant increase was observed in cellular levels of C22–C24 CERs in RPE cells treated with TM compared to controls (Table 2). Our results support a previous study that has shown that buildup of ceramide during ER stress induces apoptosis in retinal pigment epithelium subjected to UV and oxidative stress [13]. To our knowledge, this is the first study to report increased levels of long chain fatty acid containing ceramides in ARPE-19 cells treated with TM. It was previously shown that exogenous ceramide led to the inhibition of sarcoplasmic/endoplasmic reticulum Ca2+- ATPase (SERCA) resulting in the induction of [Ca2+] depletion in the ER [29]. This effect of ceramide is similar to thapsigargin, a classic inducer of ER stress [30]. Thapsigargin selectively inhibits SERCA, resulting in Ca2+ depletion from the ER lumen and activation of ER stress. The subsequent increase of Ca2+ concentration in the cytosol and mitochondrial matrix play an important role in exogenous ceramide-induced apoptosis [31]. TUDCA treatment caused a significant decrease in long chain CER levels in cells treated with TM compared to TM treatment alone. The observed decrease in C22–C24 CERs following TUDCA treatment may present a novel mechanism by which TUDCA alleviates ER stress. We have also observed that incubation with TM for 24 h significantly increased C1P levels in RPE cells. To our knowledge, this is the first study to report increased levels of C1P in RPE cells treated with TM. Ceramide-1-phosphate, like ceramide itself, can likely perform as both structural and/or cell signaling molecule [32]. Retinal pigment epithelial cells can generate C1P which acts as an antagonist of ceramide to support cell proliferation and defend cells from diverse injury [32]. Ceramide-1-phosphate promotes photoreceptor survival and differentiation [33]. The growing role for this sphingolipid in the alteration of vital processes in retina cell types and its function in retinal degenerations makes them potential targets for treating these diseases. Endoplasmic reticulum stress was reported to increase the production of C1P in keratinocytes stimulated with a pharmacological ER stressor, thapsigargin (Tg) [34]. Our results support these findings in RPE cells and further shows that inhibition of ER stress via TUDCA treatment can decrease levels of C1P. Latest considerable advance in knowledge of biologically active sphingolipid synthesis, particularly within ceramide and C1P-mediated pathways, has identified central roles for these molecules in cell survival. Ceramide, a chief molecule in sphingolipid metabolism, operationally triggers antiproliferative and apoptotic reactions, while C1P induces responses that make this molecule a cell proliferating lipid [33]. The finding of increased C22–C24 CERs and C1P in our cell model suggests that synthesis of both lipid species are activated in ER stress.
The catalytic role of SMase is to convert sphingomyelin to phosphocholine and ceramide. Ceramide production potentiates apoptotic stimuli in almost any mammalian cell [35] and thus decrease of SMase activity by tunicamycin (Fig. 2) is expected to be an adaptation mechanism to cellular stress. In our study, tunicamycin treatment decreased SMase activity relative to control but this did not prevent RPE cells from caspase activation. This may be due to the fact that ceramide also serves as a substrate for sphingomyelin synthase and decreased ceramide production causes decreased sphingomyelin synthase activity leading to sufficient amount of ceramide that is still present in the milieu to serve in apoptotic cell signaling [36]. However, at greater than 50% inhibition of SMase activity, as observed in TM + TUDCA treated cells, ceramide present in the cell will be used to maintain sphingomyelin synthesis and thus cannot induce apoptotic signaling. The ratio of sphingomyelin:ceramide appears to be sufficiently in favor of sphingomyelin in tunicamycin plus TUDCA treatment to significantly decrease induction of caspase-12 in ARPE-19 cells.
Treatment of ARPE-19 cells with the ER stressor TM for 24 h caused a significant increase in levels of GRP78 mRNA and protein (Fig. 3). Significantly increased mRNA level of GRP78, has become an established indicator for the presence of ER stress [37]. In unstressed cells GRP78 works as a chaperone within the ER and some can be bound to PKR-like ER kinase (PERK). When unfolded proteins are produced, GRP-78 is sequestered away from PERK and binds to unfolded proteins, thereby challenging to maintain homeostasis in the ER. Detachment of GRP78 from PERK, allows the phosphorylation of PERK [37]. We found that inhibition of ER stress in TM + TUDCA treated cells significantly decreased the expression of GRP78, suggesting that ER stress response is altered by TUDCA treatment in RPE cells.
Treatment with TM alone also increased NF-kB1 mRNA and NF-kB protein levels in RPE cell lines (Fig. 3). Obtained data is in agreement with previous studies which demonstrate that TM-induced ER stress triggers activation of transcription factor NF-kB [[38], [39], [40]]. Activation of the IkB/NF-kB transcription pathway is considered to be necessary for cell proliferation [41]. Prolonged ER stress goes through three phases; adaptation, alarm and apoptosis [42]. When adaptation fails, ER stress triggers the activation of NF-kB [40]. The phosphorylation of PERK during prolonged ER stress phosphorylates eukaryotic initiation factor 2 alpha (eIF2α) which activates NF-kB [40]. A significant down regulation of NFkB1 gene transcription was observed in TM + TUDCA treated RPE cells supporting ER stress-mediated activation of the IkB/NF-kB transcription pathway.
Caspase-3 and caspase-12 activity was increased in TM treated RPE cells (Fig. 4). Caspases are key enzymes for apoptosis, which is necessary for preserving homeostasis in eukaryotic. cells. There are two major pathways that induce apoptotic cell death. One is the extrinsic pathway, which involves binding of tumor necrosis factor-α (TNF-α) and Fas ligand to membrane receptors leading to caspase-8 or caspase-10 activation. The other is the intrinsic pathway, in which there is stress-induced mitochondrial cytochrome c release [43]. Released cytochrome c makes a complex with apoptotic protease activating factor-1 (Apaf-1) and procaspase-9, which activates caspase-9. Both pathways converge on caspase-3 activation, resulting in cellular morphological changes such as blebbing and nuclear degradation [43]. C/EBP-homologous protein (CHOP), a transcription factor induced under ER stress, has been reported to be involved in ER stress-induced apoptosis by reducing the expression of Bcl-2 and leading to the release of cytochrome c [44]. In fact, CHOP-deficiency creates resistance to ER stress-induced cell death both in vitro and in vivo [45]. As discussed previously, ER stress also leads to increase cytosolic calcium levels, which leads to the activation of m-calpain [46]. Activated m-calpain cleaves procaspase-12, a caspase localized in the ER, to generate active caspase-12 [47]. Processed-caspase-12 reportedly activates caspase-9 independent of Apaf-1, followed by activation of caspase-3 [48]. Both caspase-3 and -12 enzyme activity was increased in ER stressed RPE cells and TUDCA treatment caused a significant decrease in caaspase-12 activity, supporting the involvement of ER stress in caspase-12 activation.
In conclusion, TM increased ceramide, C1P, GRP78 and NFkB levels and lead to the activation of caspase-3 and -12 in human RPE cell lines. Inhibition of ER stress by TUDCA decreased ceramide/C1P levels, reduced caspase-12 activity, caused a significant reduction of GRP 78 and NF-kB1 expression in RPE cells. Our data suggests that inhibition of ER stress and ceramide formation can potentially be a pharmaceutical target in the prevention and treatment of retinal degenerative diseases associated with apoptosis.
Declaration of competing interest
All authors declare that they have no financial, consulting, and personal relationships with other people or organizations that could influence the presented work.
Acknowledgement
This work was supported by a grant by Akdeniz University Research Foundation (TDK-2018-3040).
Contributor Information
Ebru Afşar, Email: ebrukiracc@gmail.com.
Esma Kırımlıoglu, Email: esma.konuk@gmail.com.
Tuğçe Çeker, Email: tugceker159@gmail.com.
Çağatay Yılmaz, Email: ccagatayyilmaz@gmail.com.
Necdet Demir, Email: necdet.demir@gmail.com.
Mutay Aslan, Email: mutayaslan@akdeniz.edu.tr.
References
- 1.García-Ayuso D., Di Pierdomenico J., Vidal-Sanz M., Villegas-Pérez M.P. Retinal ganglion cell death as a late remodeling effect of photoreceptor degeneration. Int. J. Mol. Sci. 2019 Sep 19;20(18) doi: 10.3390/ijms20184649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H., Chen M. Diabetic retinopathy and dysregulated innate immunity. Vis. Res. 2017 Oct;139:39–46. doi: 10.1016/j.visres.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Hageman G.S., Luthert P.J., Victor Chong N.H., Johnson L.V., Anderson D.H., Mullins R.F. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog. Retin. Eye Res. 2001 Nov;20(6):705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 4.Yin Y., Liu D., Tian D. Salidroside prevents hydroperoxide-induced oxidative stress and apoptosis in retinal pigment epithelium cells. Exp. Ther. Med. 2018 Sep;16(3):2363–2368. doi: 10.3892/etm.2018.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei B., Wang M., Hao W., He X. Mst1 facilitates hyperglycemia-induced retinal pigmented epithelial cell apoptosis by evoking mitochondrial stress and activating the Smad2 signaling pathway. Cell Stress Chaperones. 2019 Jan;24(1):259–272. doi: 10.1007/s12192-018-00963-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Chen M., Wang W., Ma J., Ye P., Wang K. High glucose induces mitochondrial dysfunction and apoptosis in human retinal pigment epithelium cells via promoting SOCS1 and Fas/FasL signaling. Cytokine. 2016 Feb;78:94–102. doi: 10.1016/j.cyto.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Kucuksayan E., Konuk E.K., Demir N., Mutus B., Aslan M. Neutral sphingomyelinase inhibition decreases ER stress-mediated apoptosis and inducible nitric oxide synthase in retinal pigment epithelial cells. Free Radic. Biol. Med. 2014 Jul;72:113–123. doi: 10.1016/j.freeradbiomed.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Jing G., Wang J.J., Zhang S.X. ER stress and apoptosis: a new mechanism for retinal cell death. Exp. Diabetes Res. 2012;2012:589589. doi: 10.1155/2012/589589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kheitan S., Minuchehr Z., Soheili Z.S. Exploring the cross talk between ER stress and inflammation in age-related macular degeneration. PLoS One. 2017 Jul 24;12(7) doi: 10.1371/journal.pone.0181667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenox A.R., Bhootada Y., Gorbatyuk O., Fullard R., Gorbatyuk M. Unfolded protein response is activated in aged retinas. Neurosci. Lett. 2015 Nov 16;609:30–35. doi: 10.1016/j.neulet.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y., Simmen T. Mechanistic connections between endoplasmic reticulum (ER) redox control and mitochondrial metabolism. Cells. 2019 Sep 12;8(9) doi: 10.3390/cells8091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee W.S., Yoo W.H., Chae H.J. ER stress and autophagy. Curr. Mol. Med. 2015;15(8):735–745. doi: 10.2174/1566524015666150921105453. [DOI] [PubMed] [Google Scholar]
- 13.Yao J., Bi H.E., Sheng Y., Cheng L.B., Wendu R.L., Wang C.H., Cao G.F., Jiang Q. Ultraviolet (UV) and hydrogen peroxide activate ceramide-ER stress-AMPK signaling axis to promote retinal pigment epithelium (RPE) cell apoptosis. Int. J. Mol. Sci. 2013 May 17;14(5):10355–10368. doi: 10.3390/ijms140510355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro B.M., Prieto M., Silva L.C. Ceramide: a simple sphingolipid with unique biophysical properties. Prog. Lipid Res. 2014 Apr;54:53–67. doi: 10.1016/j.plipres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Lei X., Zhang S., Bohrer A., Bao S., Song H., Ramanadham S. The group VIA calcium-independent phospholipase A2 participates in ER stress-induced INS-1 insulinoma cell apoptosis by promoting ceramide generation via hydrolysis of sphingomyelins by neutral sphingomyelinase. Biochemistry. 2007 Sep 4;46(35):10170–10185. doi: 10.1021/bi700017z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruvolo P.P. Ceramide regulates cellular homeostasis via diverse stress signaling pathways. Leukemia. 2001 Aug;15(8):1153–1160. doi: 10.1038/sj.leu.2402197. [DOI] [PubMed] [Google Scholar]
- 17.Shaw J., Costa-Pinheiro P., Patterson L., Drews K., Spiegel S., Kester M. Novel sphingolipid-based cancer therapeutics in the personalized medicine era. Adv. Cancer Res. 2018;140:327–366. doi: 10.1016/bs.acr.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Özer H., Aslan İ., Oruç M.T., Çöpelci Y., Afşar E., Kaya S., Aslan M. Early postoperative changes of sphingomyelins and ceramides after laparoscopic sleeve gastrectomy. Lipids Health Dis. 2018 Nov 24;17(1):269. doi: 10.1186/s12944-018-0917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aslan M., Kıraç E., Kaya S., Özcan F., Salim O., Küpesiz O.A. Decreased serum levels of sphingomyelins and ceramides in sickle cell disease patients. Lipids. 2018 Mar;53(3):313–322. doi: 10.1002/lipd.12027. [DOI] [PubMed] [Google Scholar]
- 20.Yapar M., Aydogan H., Pahsa A., Besirbellioglu B.A., Bodur H., Basustaoglu A.C., Guney C., Avci I.Y., Sener K., Setteh M.H., Kubar A. Rapid and quantitative detection of Crimean-Congo hemorrhagic fever virus by one-step real-time reverse transcriptase-PCR. Jpn. J. Infect. Dis. 2005 Dec;58(6):358–362. [PubMed] [Google Scholar]
- 21.Tuzcu H., Unal B., Kırac E., Konuk E., Ozcan F., Elpek G.O., Demir N., Aslan M. Neutral sphingomyelinase inhibition alleviates apoptosis, but not ER stress, in liver ischemia-reperfusion injury. Free Radic. Res. 2017 Mar;51(3):253–268. doi: 10.1080/10715762.2017.1298103. [DOI] [PubMed] [Google Scholar]
- 22.Anderson D.H., Williams D.S., Neitz J., Fariss R.N., Fliesler S.J. Tunicamycin-induced degeneration in cone photoreceptors. Vis. Neurosci. 1988;1(2):153–158. doi: 10.1017/s0952523800001425. [DOI] [PubMed] [Google Scholar]
- 23.Boriushkin E., Wang J.J., Li J., Jing G., Seigel G.M., Zhang S.X. Identification of p58IPK as a novel neuroprotective factor for retinal neurons. Invest. Ophthalmol. Vis. Sci. 2015 Feb 5;56(2):1374–1386. doi: 10.1167/iovs.14-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price N.P., Tsvetanova B. Biosynthesis of the tunicamycins: a review. J. Antibiot. (Tokyo) 2007 Aug;60(8):485–491. doi: 10.1038/ja.2007.62. [DOI] [PubMed] [Google Scholar]
- 25.Berger E., Haller D. Structure-function analysis of the tertiary bile acid TUDCA for the resolution of endoplasmic reticulum stress in intestinal epithelial cells. Biochem. Biophys. Res. Commun. 2011 Jun 17;409(4):610–615. doi: 10.1016/j.bbrc.2011.05.043. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L., Xia Q., Zhou Y., Li J. Endoplasmic reticulum stress and autophagy contribute to cadmium-induced cytotoxicity in retinal pigment epithelial cells. Toxicol. Lett. 2019 Sep 1;311:105–113. doi: 10.1016/j.toxlet.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Weng S., Mao L., Gong Y., Sun T., Gu Q. Role of quercetin in protecting ARPE-19 cells against H2O2-induced injury via nuclear factor erythroid 2 like 2 pathway activation and endoplasmic reticulum stress inhibition. Mol. Med. Rep. 2017 Sep;16(3):3461–3468. doi: 10.3892/mmr.2017.6964. [DOI] [PubMed] [Google Scholar]
- 28.Pesonen M., Pasanen M., Loikkanen J., Naukkarinen A., Hemmilä M., Seulanto H., Kuitunen T., Vähäkangas K. Chloropicrin induces endoplasmic reticulum stress in human retinal pigment epithelial cells. Toxicol. Lett. 2012 Jun 20;211(3):239–245. doi: 10.1016/j.toxlet.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z., Xia Y., Li B., Xu H., Wang C., Liu Y., Li Y., Li C., Gao N., Li L. Induction of ER stress-mediated apoptosis by ceramide via disruption of ER Ca(2+) homeostasis in human adenoid cystic carcinoma cells. Cell Biosci. 2014 Nov 25;4:71. doi: 10.1186/2045-3701-4-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdullahi A., Stanojcic M., Parousis A., Patsouris D., Jeschke M.G. Modeling acute ER stress in vivo and in vitro. Shock. 2017 Apr;47(4):506–513. doi: 10.1097/SHK.0000000000000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinton P., Ferrari D., Rapizzi E., Di Virgilio F., Pozzan T., Rizzuto R. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 2001 Jun 1;20(11):2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simón M.V., Prado Spalm F.H., Vera M.S., Rotstein N.P. Sphingolipids as emerging mediators in retina degeneration. Front. Cell. Neurosci. 2019 Jun 11;13:246. doi: 10.3389/fncel.2019.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miranda G.E., Abrahan C.E., Agnolazza D.L., Politi L.E., Rotstein N.P. Ceramide-1-phosphate, a new mediator of development and survival in retina photoreceptors. Invest. Ophthalmol. Vis. Sci. 2011 Aug 22;52(9):6580–6588. doi: 10.1167/iovs.10-7065. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y.I., Park K., Kim J.Y., Seo H.S., Shin K.O., Lee Y.M., Holleran W.M., Elias P.M., Uchida Y. An endoplasmic reticulum stress-initiated sphingolipid metabolite, ceramide-1-phosphate, regulates epithelial innate immunity by stimulating β-defensin production. Mol. Cell. Biol. 2014 Dec;34(24):4368–4378. doi: 10.1128/MCB.00599-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mullen T.D., Obeid L.M. Ceramide and apoptosis: exploring the enigmatic connections between sphingolipid metabolism and programmed cell death. Anti Cancer Agents Med. Chem. 2012 May;12(4):340–363. doi: 10.2174/187152012800228661. [DOI] [PubMed] [Google Scholar]
- 36.Luberto C., Hannun Y.A. Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. Does sphingomyelin synthase account for the putative phosphatidylcholine-specific phospholipase C? J. Biol. Chem. 1998 Jun 5;273(23):14550–14559. doi: 10.1074/jbc.273.23.14550. [DOI] [PubMed] [Google Scholar]
- 37.Zhu G., Lee A.S. Role of the unfolded protein response, GRP78 and GRP94 in organ homeostasis. J. Cell. Physiol. 2015 Jul;230(7):1413–1420. doi: 10.1002/jcp.24923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pahl H.L., Baeuerle P.A. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-kappa B. EMBO J. 1995 Jun 1;14(11):2580–2588. doi: 10.1002/j.1460-2075.1995.tb07256.x. 31-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung J.H., Su I.J., Lei H.Y., Wang H.C., Lin W.C., Chang W.T., Huang W., Chang W.C., Chang Y.S., Chen C.C., Lai M.D. Endoplasmic reticulum stress stimulates the expression of cyclooxygenase-2 through activation of NF-kappaB and pp38 mitogen-activated protein kinase. J. Biol. Chem. 2004 Nov 5;279(45):46384–46392. doi: 10.1074/jbc.M403568200. [DOI] [PubMed] [Google Scholar]
- 40.Lin W.C., Chuang Y.C., Chang Y.S., Lai M.D., Teng Y.N., Su I.J., Wang C.C., Lee K.H., Hung J.H. Endoplasmic reticulum stress stimulates p53 expression through NF-κB activation. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0039120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oeckinghaus A., Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009 Oct;1(4):a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu C., Bailly-Maitre B., Reed J.C. Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Investig. 2005 Oct;115(10):2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007 Jun;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCullough K.D., Martindale J.L., Klotz L.O., Aw TY., Holbrook N.J. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 2001 Feb;21(4):1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998 Apr 1;12(7):982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muruganandan S., Cribb A.E. Calpain-induced endoplasmic reticulum stress and cell death following cytotoxic damage to renal cells. Toxicol. Sci. 2006 Nov;94(1):118–128. doi: 10.1093/toxsci/kfl084. [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa T., Yuan J. Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J. Cell Biol. 2000 Aug 21;150(4):887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morishima N., Nakanishi K., Takenouchi H., Shibata T., Yasuhiko Y. An endoplasmic reticulum stress-specific caspase cascade in apoptosis. Cytochrome c-independent activation of caspase-9 by caspase-12. J. Biol. Chem. 2002 Sep 13;277(37):34287–34294. doi: 10.1074/jbc.M204973200. [DOI] [PubMed] [Google Scholar]