Abstract
Pregnane X receptor (PXR) is the major regulator of xenobiotic metabolism. PXR itself is controlled by various signaling molecules including glucocorticoids. Moreover, negative feed-back regulation has been proposed at the transcriptional level. We examined the involvement of the 3′-untranslated region (3′-UTR) of NR1I2 mRNA and microRNAs in PXR- and glucocorticoid receptor (GR)-mediated regulation of NR1I2 gene expression. PXR ligands were found to significantly downregulate NR1I2 mRNA expression in a set of 14 human hepatocyte cultures. Similarly, PXR was downregulated by PCN in the C57/BL6 mice liver. In mechanistic studies with the full-length 3′-UTR cloned into luciferase reporter or expression vectors, we showed that the 3′-UTR reduces PXR expression. From the miRNAs tested, miR-18a-5p inhibited both NR1I2 expression and CYP3A4 gene induction. Importantly, we observed significant upregulation of miR-18a-5p expression 6 h after treatment with the PXR ligand rifampicin, which indicates a putative mechanism underlying NR1I2 negative feed-back regulation in hepatic cells. Additionally, glucocorticoids upregulated NR1I2 expression not only through the promoter region but also via 3′-UTR regulation, which likely involves downregulation of miR-18a-5p. We conclude that miR-18a-5p is involved in the down-regulation of NR1I2 expression by its ligands and in the upregulation of NR1I2 mRNA expression by glucocorticoids in hepatic cells.
KEY WORDS: Gene expression, MicroRNA, Glucocorticoid, Regulation, Pregnane X receptor, Cytochrome P450 3A4
Abbreviations: 3′-UTR, 3′-untranslated region; CAR, constitutive androstane receptor; CYP3A4, cytochrome P450 3A4; DEX, dexamethasone; DMEs, drug metabolizing enzymes; DMSO, dimethyl sulfoxide; ER, estrogen receptor; Gluc, Gaussia luciferase; GRα, glucocorticoid receptor α; LBD, ligand binding domain; miRNA, microRNA; MRE, miRNA-response element; NR, nuclear receptor; PB, phenobarbital; PCN, pregnenolone 16α-carbonitrile; PHHs, primary human hepatocytes; PPARα, peroxisome proliferator-activated receptor α; PXR, pregnane X receptor; Rif, rifampicin; RXRα, retinoid X receptor α; SEAP, secreted alkaline phosphatase
Graphical abstract
miR-18a-5p is involved in the down-regulation of pregnane X receptor (PXR) gene (NR1I2) expression by its ligands and in the upregulation NR1I2 mRNA expression by glucocorticoids in hepatic cells. Glucocorticoids and PXR ligands thus regulate NR1I2 expression not only through the promoter region but also via 3ʹ-UTR regulation.
1. Introduction
Pregnane X receptor (PXR, encoded by NR1I2 gene) is a member of the nuclear receptor (NR) superfamily, which includes the steroid, retinoid, and thyroid hormone receptors. PXR is the major xenobiotic receptor that coordinately regulates transcription of key phase I and II biotransformation enzymes, as well as some drug transporters, to detoxify and eliminate xenobiotics and endotoxins from the body. As a ligand-activated nuclear receptor, PXR triggers the transcription of various target genes, such as CYP3A4 and CYP2C9, conjugation enzymes, such as UGT1A1, and transporters, such as ABCB11, 2, 3. In some cases, however, activation of PXR has been associated with downregulation of gene expression, as exemplified by the OCT1 gene4,5. In addition to its canonical xenobiotic sensing function, PXR has many other cellular and physiological roles and has been implicated in glucose, lipid and bile acid metabolism, energy homeostasis, inflammatory response, cell proliferation, apoptosis, and cell migration1,2. PXR agonists include a broad spectrum of structurally diverse compounds, such as rifampicin, phenobarbital, SR-12813, and hyperforin, due to the large and considerably flexible ligand-binding domain (LBD) cavity of PXR3.
Although tremendous effort has been spent studying the function of PXR in both exogenous and endogenous metabolism, less is known about the exact mechanisms that are responsible for its regulation. NR1I2 expression and transcriptional activity are coordinately regulated at the transcriptional, posttranscriptional, and posttranslational levels, and numerous signaling cascades have been implicated in modulating PXR activity6,7. As a consequence of this multifactorial regulation, human hepatic NR1I2 mRNA is substantially variable in its expression8.
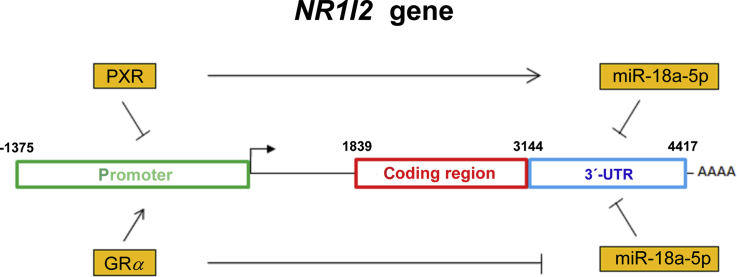
Recent in silico predictions identified a number of potential binding sites for other NRs within the human NR1I2 gene proximal promoter (∼2.2 kb) suggesting hormonal or endobiotic regulation of PXR. Forced overexpression of estrogen receptor (ER), glucocorticoid receptor α (GRα), and peroxisome proliferator-activated receptor α (PPARα) exert positive effects on NR1I2 transcription, but PXR and constitutive androstane receptor (CAR) have negative effects on PXR expression in hepatic cells7,9. However, no putative binding sites for either PXR or CAR have been identified within the NR1I2 promoter by detailed bioinformatic analysis9. Glucocorticoids have been shown to regulate hepatic NR1I2 expression, and the crosstalk between PXR and GRα has been reported in detail in primary human hepatocytes with dexamethasone at submicromolar concentrations10,11. Additionally, positive transactivation of NR1I2 by HNF4α through the direct repeat 1 (DR1) element located in the –88 to –76 bp region or by PPARα via a binding site located in the –1514 to –1321 bp region has been described9,12. Nevertheless, NR1I2 mRNA expression does not correlate with PXR protein levels in the liver, which is an organ where NR1I2 is abundantly expressed; this suggests that PXR is subject to posttranscriptional regulation13.
MicroRNAs (miRNAs) are small noncoding RNAs approximately 22 nt in length that are considered the principal posttranscriptional regulators14. To date, more than 2600 mature human miRNAs have been annotated in the miRBase repository (v. 22.1, October 2018)15. Primarily but not exclusively, miRNAs bind to responsive elements within the 3ʹ-untranslated region (3ʹ-UTR) of a target gene mRNA, resulting in degradation of the mRNA or translational repression. Interestingly, individual mRNAs can be targeted by many diverse miRNAs, leading to fine-tuning of gene expression. In contrast, specific miRNAs can simultaneously control the expression of many target mRNAs14,16. It is currently supposed that most human genes are under the control of miRNAs. Indeed, a growing body of evidence has also confirmed that miRNAs play a significant role in the regulation of genes involved in xenobiotic metabolism. In addition, drugs may regulate their pharmacokinetics and response via miRNAs, which is a topic that has been reviewed elsewhere17, 18, 19. In line with this assumption, it has been demonstrated that miRNAs may interfere with NR1I2 expression, as suggested for miR-18a-5p20, miR-148a13, miR-30c-1-3p21, miR-34a-5p8,22, miR-449a22, and miR-140-3p23. Importantly, negative regulation of NR1I2 expression via miR-18a-5p by PXR ligands has been shown recently in LS180 cells20.
Since ligand-activated NRs are subjects of positive and negative regulation24 and keeping in mind the complexity of transcriptional and posttranscriptional regulation of NR1I2 expression, we hypothesized for the first time in this work that miRNAs may be involved in a NR1I2 autoregulatory feedback loop. To test this hypothesis, we analyzed changes in NR1I2 expression after PXR activation, employing both primary human hepatocytes and mice. Furthermore, we evaluated the mechanisms underlying such changes with a focus on transcriptional and posttranscriptional regulation. Subsequently, we examined the impact of activated GR on posttranscriptional stabilization of NR1I2 transcripts. By employing various constructs with the 3ʹ-UTR of NR1I2 mRNA, we deciphered a key role for miRNA-18a-5p in PXR-mediated and GR-induced regulation of PXR expression through the full-length 3ʹ-UTR of the NR1I2 gene.
2. Materials and methods
2.1. Chemicals
Rifampicin (a prototypical ligand of the human PXR, Rif), SR-12813 (a human PXR ligand), and phenobarbital (PB) were purchased from Sigma–Aldrich (St. Louis, MO, USA). Dexamethasone (DEX) (a prototypical ligand of GR), RU486 (mifepristone, GR antagonist), DMSO (dimethyl sulfoxide), pregnenolone 16α-carbonitrile (PCN, an agonist of mouse PXR), hydrocortisone acetate, corticosterone, triamcinolone acetonide, and 6α-methylprednisolone were purchased from Sigma–Aldrich. Stock solutions (1000×) were prepared in DMSO prior to dilution in cell culture medium. The final concentration of DMSO in the media did not exceed 0.2% (v/v) in any experiments. SPA70, an PXR inhibitor, has been purchased from abcr GmbH Im (Karlsruhe, Germany)25.
miRCURY LNA microRNA mimics cel-miR-39-3p (mimic control, 479902-001), hsa-miR-18a-5p (471717-001), hsa-miR-148a-3p (339173 YM00472598-ADA), hsa-miR-34a-5p (339173 YM00473212-ADA), hsa-miR-449a (339173 YM00473262-ADA), and miRCURY LNA inhibitors, such as negative control A (inhibitor control, 199006-001), hsa-miR-18a-5p (4100723-001), and hsa-miR-34a-5p (339121 YI04100982-ADA), were obtained from Exiqon (now part of QIAGEN, Germantown, MD, USA).
2.2. Plasmids
The p3A4-luc plasmid bears a distal XREM (–7836/–7208) and a basal promoter sequence (prPXRE, –362/+53) from the CYP3A4 gene promoter region. The pER6-3A4-tat-luc plasmid carries three copies of the proximal CYP3A4 promoter ER6 response element cloned into the NheI and BglII sites of the pGL4.23 (Promega, Madison, WI, USA) upstream of the SV40 minimal promoter.
The pmiRGLO Dual-Luciferase miRNA target expression vector (Promega) is designed for an insertion miRNA target site downstream of the firefly luciferase gene. The pmiRGLO-UTR reporter plasmid, containing the 3ʹ-UTR of the NR1I2 gene sequence (1–1273 nt downstream the coding region), was prepared by cloning the 3ʹ-UTR sequence into XhoI and SaII restriction sites of the pmiRGLO vector. The pmiRGLO-18a-5p-Compl construct was generated by insertion of a complementary sequence for miR-18a-5p (5ʹ-ctatctgcactagatgcacctta-3ʹ) into XhoI and SaII restriction sites of the pmiRGLO vector. The pmiRGLO-MRE-1041-1064 (5ʹ-aagaaccatttacatgcacctta-3ʹ), pmiRGLO-MRE-1041-1064-REV, and pmiRGLO-MRE-69-91 (5ʹ-gccaagacagatggacactgcca-3ʹ) constructs contain either a putative responsive element for miR-18a-5p (1041-1064 nt from the stop codon), a reverse sequence of the putative responsive element for miR-18a-5p, or a putative shared responsive sequence of the 3ʹ-UTR of the NR1I2 gene for miR-34a-5p and miR-449a (69-91 nt from the stop codon) in the pmiRGLO vector. All constructed vectors were sequence verified.
pMIR-luc-control (LR-1000) and pMIR-luc-34a-5p-Compl (LR-0014) constructs carrying a complementary binding site for miR-34a-5p were purchased from BioCat (Heidelberg, Germany).
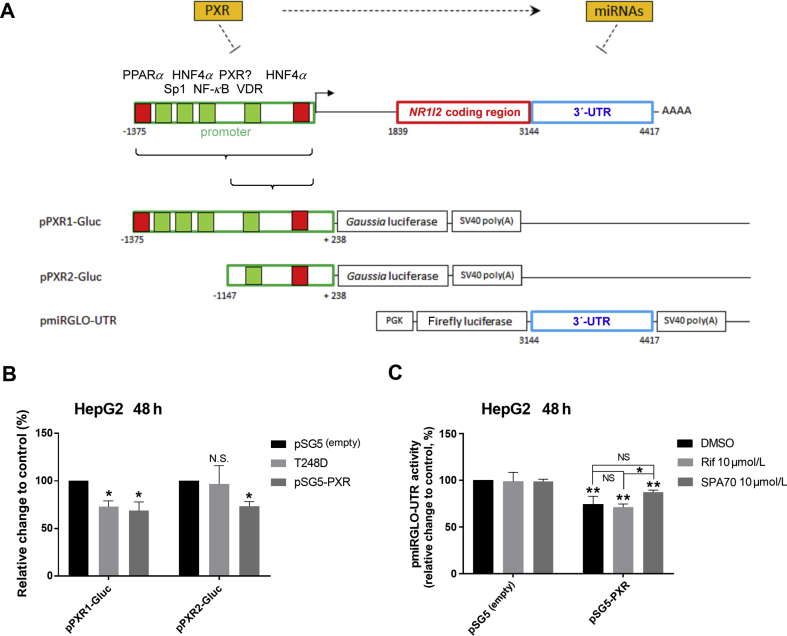
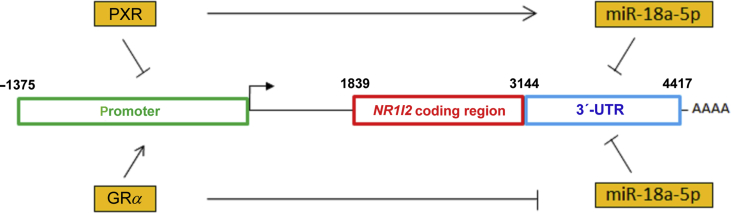
pPXR1-Gluc or pPXR2-Gluc reporter vectors (HPRM45199-PG04 and HPRM23633-PG04) with secreted Gaussia luciferase (Gluc) were purchased from GeneCopoeia (Rockville, MD, USA) together with a luciferase detection kit. pPXR1-Gluc contains a sequence from –1375 to +238 and pPXR2-Gluc sequence from –1147 to +238 of the promoter region upstream of the human NR1I2 gene. The sequence from –1375 to –1147 (228 nt) in pPXR1-Gluc, but not in pPXR2-Gluc, contains putative response elements for HNF4α, PPARα, Sp1 and NF-κB transcription factors9 (see schematic in Fig. 3A). Both reporter constructs contain cDNA for secreted alkaline phosphatase (SEAP) as an internal control for transfection normalization.
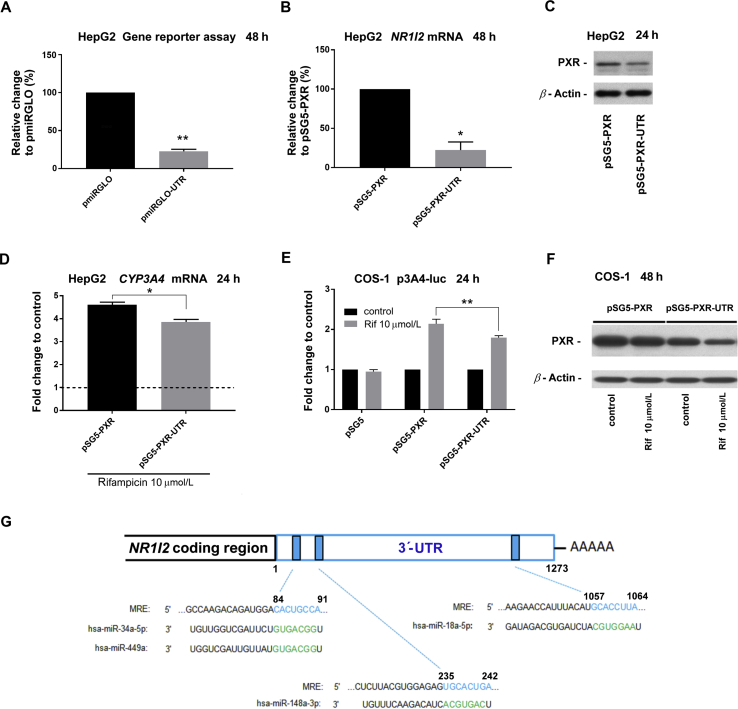
Figure 3.
Implication of the NR1I2 promoter region and 3ʹ-UTR in PXR regulation. (A) Scheme depicting the NR1I2 gene with its regulatory regions. Suggested or confirmed transcription binding sites within the promoter region are shown as green and red boxes, respectively9,12. A schematic overview of reporter plasmids alignment under the scheme. The numbers indicate nucleotide positions either in the promoter region or in mRNA of the NR1I2 gene. (B) HepG2 cells were transiently transfected with either the pPXR1-Gluc or pPXR2-Gluc Gaussia luciferase reporter construct (150 ng/well) concomitantly with the pSG5-RXRα expression vector (50 ng) and with either pSG5-PXR, pSG5-T248D or pSG5 (empty) control vector (100 ng). The next day, the medium was changed and after 24 h, Gaussia luciferase and SEAP activities were assessed. The results are presented as relative change in SEAP-normalized Gaussia luciferase activities to control experiments cotransfected with empty pSG5 vector (100%). The data are shown as the mean ± SD (n = 3). (C) HepG2 cells were transiently transfected with either pmiRGLO-UTR or pmiRGLO (empty) vector (50 ng) together with either pSG5-PXR expression vector or pSG5 (empty) control vector (100 ng). The next day, cells were treated with rifampicin (Rif, 10 μmol/L), SPA70 (a PXR antagonist, 10 μmol/L) or vehicle (DMSO; 0.1%) for 24 h. Samples were subsequently analyzed by Dual-Luciferase Reporter Assay System. Firefly luciferase activities were normalized to Renilla luciferase activities and further normalized to the corresponding data obtained from experiments with the empty pmiRGLO vector. The data are shown as the mean ± SD (n = 3) and are expressed as relative change to DMSO-treated controls cotransfected with the empty pSG5 vector defined as 100%. *P < 0.05 or indicates a statistically significant effect of PXR activation compared to empty pSG5 vector-transfected controls or to rifampicin-treated pSG-PXR cells; **P < 0.01 indicates a statistically significant suppression of pmiRGLO-UTR vector activity; N.S. indicates a statistically insignificant effect.
The expression plasmid for human PXR receptor pSG5-hPXRΔATG (further abbreviated as pSG5-PXR) was kindly provided by Dr. S. Kliewer (University of Texas, Dallas, TX, USA). The constitutively active T248D mutant of PXR was described in our previous work26. The pSG5-PXR-UTR expression vector, including both open reading frames encoding human wild-type PXR and the 3ʹ-UTR of the NR1I2 gene mRNA, was constructed by insertion of the 3ʹ-UTR region into the BamHI restriction site downstream of the cDNA for PXR. Orientation of the 3ʹ-UTR insert in pSG5-PXR-UTR was verified by sequencing. The expression plasmid encoding human GRα (pSG5-hGRα) was a generous gift from Dr. J. Palvimo (University of Helsinki, Helsinki, Finland), and the pSG5-RXRα construct came from Dr. C. Carlberg (University of Kuopio, Kuopio, Finland). pRL-TK was acquired from Promega. The empty expression vector pSG5 was obtained from Agilent (Santa Clara, CA, USA).
2.3. Cell lines
Human hepatoblastoma-derived (HepG2), human hepatocellular carcinoma (Huh-7), and African green monkey kidney-derived (COS-1) cells were obtained from the European Collection of Authenticated Cell Cultures (ECACC) and cultured in antibiotic-free Dulbecco's modified Eagle's medium (DMEM, 10569010, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, F2442, Sigma–Aldrich) at 37 °C in a humidified incubator under 5% CO2.
2.4. Primary human hepatocytes
Primary human hepatocytes (PHHs) were provided by QPS Hepatic Biosciences (Barcelona, Spain) or were isolated, as described previously27, from donor organs unsuitable for transplantation or from liver resections performed in adult patients for medical reasons unrelated to our research program (only noncancerous liver tissue was collected). Liver samples were obtained from the Biological Resource Centre of Montpellier University Hospital (CRB-CHUM; http://www.chu-montpellier.fr; Biobank ID: BB-0033-00031), and this study benefitted from the expertise of Dr. Jeanne Ramos (hepatogastroenterology sample collection) and Prof. Sylvain Lehmann (CRB-CHUM manager). Patient clinical characteristics are presented in Table 1. The procedures were approved by the French Ethics Committee, and written or oral consent was obtained from all patients or their families.
Table 1.
Donor information and culture conditions of isolated primary human hepatocytes.
| Liver | Origin | Age (years) | Sex | Culture medium | Pathology |
|---|---|---|---|---|---|
| Liv1 | QPS | 61 | M | ISOM | Cholangiocarcinoma |
| Liv2 | Resection | 52 | F | HGM | Cyst adenoma |
| Liv3 | QPS | 66 | F | ISOM | Colorectal cancer |
| Liv4 | Resection | 70 | F | HGM | Cholangiocarcinoma |
| Liv5 | Donor | 73 | M | HGM | Stroke |
| Liv6 | Resection | 76 | M | ISOM, HGM | Metastasis from colon cancer |
| Liv7 | Resection | 70 | M | ISOM | Metastasis from colon cancer |
| Liv8 | Resection | 46 | F | ISOM | Cholangiocarcinoma |
| Liv9 | Resection | 64 | M | ISOM | Hepatocellular carcinoma |
| Liv10 | Resection | 46 | M | LNF | Angioma |
| Liv11 | Resection | 62 | M | LNF | Metastasis from colon cancer |
| Liv12 | Resection | 70 | M | LNF | Metastasis of GIST |
| Liv13 | Resection | 80 | F | ISOM | Metastasis of colon cancer |
Primary human hepatocytes were seeded at 1.5 × 105 cells/cm2 in collagen-coated plates and cultured in a 5% CO2 humidified atmosphere at 37 °C in serum-free long-term (Lanford medium, LNF) or short-term (ISOM medium) culture medium27 or hepatocyte growth medium (HGM: WME medium supplemented with 5 μg/mL insulin, 0.1 μmol/L hydrocortisone, 10 μg/mL transferrin, 250 μg/mL ascorbic acid, 3.75 mg/mL fatty acid-free bovine serum albumin, 2 mmol/L glutamine, penicillin and streptomycin). Primary human hepatocytes were incubated with rifampicin (Rif, 5 or 10 μmol/L), SR-12813 (3 μmol/L) or phenobarbital (500 μmol/L) for 24 or 48 h two days postseeding.
Information about the 13 donors of freshly isolated Primary human hepatocytes is shown. Details are included for each donor regarding age, sex, and pathology. Additionally, origin and cultivation medium are indicated for each individual culture of Primary human hepatocytes. Gender: M (male), F (female). ISOM medium28; HGM, hepatocyte growth medium; LNF, Lanford medium.
2.5. Commercial human hepatocytes
Commercial primary human hepatocytes (HEP220A0, Long-term human hepatocytes, batch Hep220980-MA96, a sample Liv14) were purchased from Biopredic International (Saint Gregoire, France). Prior to treatment, commercial Primary human hepatocytes were cultivated in a serum-free medium composed of basal hepatic cell medium (Biopredic, MIL600C, batch MIL600052) and additives for hepatocyte culture medium (Biopredic, ADD222C, batch ADD222039) overnight at 37 °C in a humidified incubator with 5% CO2.
2.6. Animal experiments
Twelve-week-old male C57/BL6 mice (Velaz, Czech Republic) were housed in a temperature- and light-controlled facility with 12 h light–dark cycling. All mice were fed commercially available laboratory chow diet (Velaz) ad libitum. Mice were randomly divided into 12 groups consisting of 5 animals each and were all administered either a single i.p. injection of mouse PXR ligand pregnenolone 16α-carbonitrile (PCN, 50 mg/kg) dissolved in corn oil plus 30% DMSO or vehicle (olive oil plus 30% DMSO).
Individual groups of mice were then sacrificed by exsanguination under general anesthesia induced by isoflurane after 3, 6, 24, 48, 72, or 168 h postadministration of vehicle or PCN. Livers were immediately weighted, frozen in liquid nitrogen and subsequently stored at −80 °C until Western blot and RT-qPCR analysis. All animals received humane care in accordance with the EU Directive 2010/63/EU and guidelines for animal experiments set by the Institutional Animal Use and Care Committee of Charles University, Faculty of Medicine in Hradec Kralove, Czech Republic. The protocol for all experiments was approved by the same committee.
2.7. Luciferase reporter gene assays
All reporter gene assays were performed in HepG2, Huh-7 or COS-1 cells using Lipofectamine 3000 Reagent (Thermo Fisher Scientific) following the manufacturer's protocol, as described previously4,29,30. Briefly, cells were seeded into 48-well plates overnight and transfected with different luciferase reporter constructs either alone or in combination with appropriate expression plasmids, miRNA mimics or miRNA inhibitors at indicated concentrations. If necessary, an equivalent amount of an empty plasmid, such as pmiRGLO, pMIR-luc-control, or pSG5, was transfected into the cells to maintain equal quantities of DNA in respective controls. After stabilization, cells were, if needed, further treated with PXR or GR ligands at indicated times and concentrations. Compounds were diluted in Opti-MEM I Reduced Serum Medium (11058021, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 5% FBS. Then, cells were lysed and measured for both firefly and Renilla luciferase activities by a plate reader, Synergy2 (BioTek, Winooski, VT, USA), employing a Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activities were normalized to Renilla luciferase activities to reduce variability in transfection efficiency. Since the pmiRGLO vector contains a Renilla luciferase coding sequence for transfection normalization, cotransfection with pRL-TK was performed only in experiments with different reporter vectors than those with pmiRGLO. In the indicated cases, measured firefly luciferase activities were double normalized according to the published protocol to eliminate the background effect of tested ligand or miRNA reagent on activity of the empty parent vector31. According to this procedure, the Renilla-normalized firefly luciferase activities obtained from different reporter vectors were further normalized to the corresponding data that were obtained under identical conditions with control (empty) reporter vectors. The results are presented as the relative change or fold change in normalized firefly luciferase activities to the control activities (defined as 100% or 1). The data are presented as the mean ± SD acquired from biological replicates performed in at least technical triplicates (n ≥ 3).
2.8. Dual secreted Gaussia luciferase and SEAP reporter assays
HepG2 cells were seeded into 48-well plates and transiently transfected with a reporter gene construct (either pPXR1-Gluc or pPXR2-Gluc) together with expression vectors using Lipofectamine 3000 Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The next day, the medium was changed based on the experimental setting to either conditioned or nonconditioned Opti-MEM I Reduced Serum Medium supplemented with 5% FBS. After 24 h, medium was analyzed for activities of both SEAP, with Ready-To-Glow Secreted Luciferase Reporter Systems (Clontech, Palo Alto, CA, USA), and Gaussia luciferase, with Secrete-Pair Gaussia Luciferase Assay Kit (GeneCopoeia, Rockville, MD) and GL-S buffer for more stable activity. The Gaussia luciferase and SEAP signals were detected by the plate reader Synergy2 (BioTek, Winooski, VT), and Gaussia luciferase activities were normalized to SEAP activities. The data are presented as the relative change in normalized Gaussia luciferase activities to control activities, which were defined as 100%. The data are presented as the mean ± SD (n = 3).
2.9. RT-qPCR analysis
HepG2 or Huh-7 cells seeded onto 12-well plates were transfected with expression plasmids and, if needed, further treated with PXR or GR ligands at indicated times and concentrations. Primary human hepatocytes (Liv14) were treated with Rif (10 μmol/L) for 24 h. RT-qPCR analyses (performed at the Faculty of Pharmacy, Charles University) were performed as described previously with gene specific assays for CYP3A4 (hCYP3A4_Q2), hypoxanthine phosphoribosyltransferase (HPRT, 3033-F), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 3058-F) and NR1I2 (hNR1I2_Q8) (Generi Biotech, Hradec Kralove, Czech Republic)4,29. The results are presented as fold or relative change in gene expression compared to corresponding controls (defined as 1 or 100%). The data are presented as the mean ± SD (n = 3).
For mouse experiments (performed at the Medical Faculty, Charles University), total RNA from liver samples was isolated using TRI reagent (Sigma–Aldrich), converted into cDNA by High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA) and subsequently analyzed on a QuantStudio 7.0 PCR machine (Thermo Fisher Scientific) by using TaqMan Fast Universal PCR Master Mix and TaqMan Gene Expression Assays for detection of Nr1i2 (Mm01344139_m1) or Cyp3a11 (Mm00731567_m1) mRNA (Thermo Fisher Scientific). The results were calculated using threshold cycle values and normalized to Gapdh mRNA levels. The results are presented as relative change in gene expression compared to corresponding controls. The data are presented as the mean ± SD acquired from five biological samples per group.
RT-qPCR experiments (performed at INSERM) with Primary human hepatocytes were performed after total RNA extraction with TRIzol Reagent and reverse transcription of 500 ng total RNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qPCR was performed using the Roche SYBR Green reagent and a LightCycler 480 apparatus (Roche Diagnostics, Meylan, France) with the following program: one step at 95 °C for 10 min followed by 50 cycles of denaturation at 95 °C for 10 s, annealing at 65 °C for 15 s, and elongation at 72 °C for 15 s. Primer sequences are listed in Table 2. Relative quantification was performed delta-delta method and accounting for reaction efficiencies. Ct values of genes of interest were normalized to Ct values for the reference gene RPLP0 (Liv3-13) or GAPDH (Liv1-2). The results are presented as relative change in gene expression compared to vehicle control (DMSO), which was defined as 1. The data are shown as the mean±SD obtained from biological samples. RTs were run in biological duplicates, and qPCR reactions were run in technical duplicates.
Table 2.
Primer sequences for RT-qPCR with SYBR green chemistry.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| CYP3A4 | GCCTGGTGCTCCTCTATCTA | GGTGTTGACCATCATAAAG |
| NR1I2 | GGACCAGCTGCAGGAGCAAT | CATGAGGGGCGTAGCAAAGG |
| RPLP0 | TCGACAATGGCAGCATCTAC | GCCTTGACCTTTTCAGCAAG |
| GAPDH | GGTCGGAGTCAACGGATTTGGTCG | CAAAGTTGTCATGGATGACC |
2.10. Expression analysis of miRNAs
HepG2 cells that were seeded into 12-well plates were transfected with miRNA inhibitor (50 ng)/mimic (25 ng) reagents or pSG5-PXR expression vector (400 ng) for 24 or 48 h. If needed, the cells were further treated with Rif (10 μmol/L) at the indicated times. Total RNA was isolated with spin column chromatography using a miRCURY RNA Isolation Kit (Cell & Plant; now part of QIAGEN, Germantown, MD, USA). Concentration and purity of total RNA were determined by a NanoDrop spectrophotometer (Thermo Fisher Scientific). Total RNA was reverse-transcribed to miR-18a-5p specific cDNA using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) at 16 °C for 30 min followed by incubation at 42 °C for 30 min with subsequent 85 °C for 5 min. cDNA samples were subsequently analyzed with TaqMan assays for miR-18a-5p (Assay ID 002422) and U6 snRNA (Assay ID 001973) using a TaqMan Fast Advanced Master Mix on the StepOnePlus Real-Time PCR System (all Thermo Fisher Scientific). The Ct values for miR-18a-5p were normalized to the Ct values of the reference gene U6. The results are presented as fold or relative change in gene expression compared to corresponding controls (defined as 1 or 100%). The data are shown as the mean±SD acquired from biological replicates performed in technical triplicates (n = 3).
2.11. miRNA expression profiling
Huh-7 cells were seeded into 12-well plates and transfected with pSG5-GRα (600 ng per well) overnight followed by treatment with DMSO (0.1%, v/v) or DEX (100 nmol/L) for 24 h. Cells were lysed in QIAzol Lysis Reagent (QIAGEN, Germantown, MD, USA), and total RNA, enriched for small RNAs, was isolated using the miRNeasy Mini Kit (QIAGEN, Germantown, MD, USA). MiRNA expression profiling of 754 miRNAs was performed using TaqMan Array Human MicroRNA Card Set v3.0 (Thermo Fisher Scientific). All procedures followed standard manufacturers' recommendations. Briefly, total RNA (100 ng) was reverse-transcribed into cDNA using TaqMan MicroRNA Reverse Transcription Kit together with Megaplex RT Primers (Thermo Fisher Scientific). cDNA samples were further analyzed on the 7900HT Real-Time PCR System (Thermo Fisher Scientific) using TaqMan Universal Master Mix II, no UNG (Thermo Fisher Scientific). Raw data were processed using SDS software version 2.4 (Thermo Fisher Scientific). The primary data were further processed using ExpressionSuite Software v1.1 (Thermo Fisher Scientific). The relative quantification was performed by delta-delta method assuming a PCR efficiency of 100%. RNU48 was determined to be the most stable reference gene and was thus used for normalization of miRNA expression levels. The results are presented as relative change in miRNA expression compared to DMSO control of a representative experiment. Experiments have been repeated with the same results.
2.12. Western blotting assays
Western blotting experiments were performed using SDS-PAGE electrophoresis for total protein fractions of mouse liver samples as described elsewhere32. Primary antibodies used for detection were anti-PXR (PA5-41170) and anti-CYP3A11 (PA1-343, Thermo Fisher Scientific). Protein band intensity on PVDF membranes was quantified using Quantity One imaging software (Bio-Rad, Hercules, CA, USA). Protein expression was normalized to expression of β-actin (A5316, Sigma–Aldrich). The results are presented as relative change in protein expression compared to corresponding controls in the same interval after PCN application. The data are shown as the mean ± SD acquired from five biological samples per group. For Western blotting experiments with primary human hepatocytes and cancer cell lines lysates, β-actin (A5316, Sigma–Aldrich), PXR (PA5-41170, Thermo Fisher Scientific), CYP3A4 (PA1-343, Thermo Fisher Scientific), and RXRα (5388, Cell Signaling Technology, Danvers, MA, USA) primary antibodies were used.
2.13. Statistical analysis
Statistical analyses were performed using GraphPad PRISM 7 software (GraphPad Software Inc., San Diego, CA, USA). Differences between groups were compared using Student's paired two-tailed t test. One-way analysis of variance (ANOVA) with Dunnett's test was applied to the data if more than two groups were analyzed. Nonparametric Wilcoxon matched-pairs tests and parametric paired t test were used to statistically compare paired expression data between control and PXR ligand-treated samples in human hepatocytes. Nonparametric Mann–Whitney test was used to compare unpaired data in mice.
3. Results
3.1. PXR ligands downregulate PXR expression in human hepatocytes
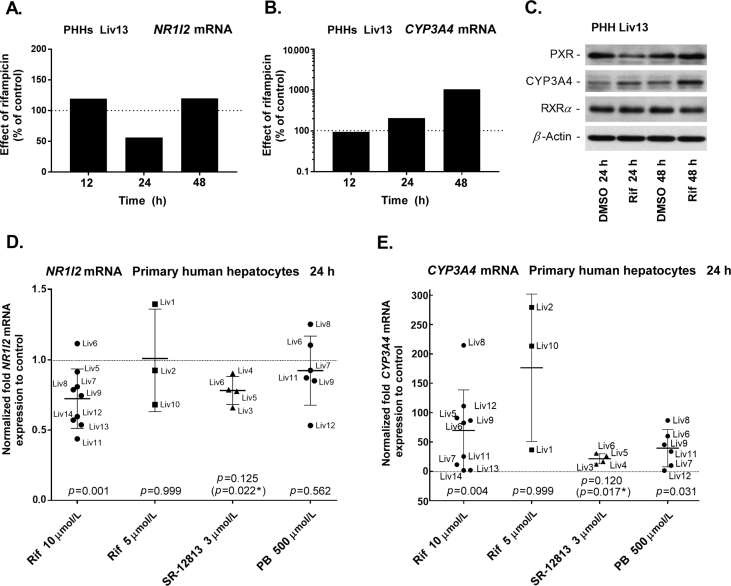
It has been shown that feed-back regulation is a common phenomenon among nuclear receptors24. Therefore, in our preliminary experiment, we explored whether treatment with a model PXR ligand, rifampicin, resulted in pronounced downregulation of NR1I2 transcripts in one culture of Primary human hepatocytes (Liv13). We observed a time-dependent effect of rifampicin on NR1I2 gene mRNA expression (Fig. 1A). Correspondingly, rifampicin induced expression of CYP3A4, the dominant PXR target gene, in the same hepatocyte culture (Fig. 1B). At the protein level, CYP3A4 was consistently upregulated by rifampicin at both 24 and 48 h intervals, however, PXR was apparently downregulated after 24 h treatment but not after 48 h. Expression of the PXR heterodimerzation partner, RXRα, was not affected by rifampicin treatment (Fig. 1C). In a follow-up study with PHH cultures, we confirmed that rifampicin (10 μmol/L) (P = 0.001) and another PXR activator, SR-12813 (3 μmol/L) (P = 0.022, paired t test), significantly suppress NR1I2 mRNA expression 24 h after treatment (Fig. 1D), but the expression of CYP3A4 mRNA was significantly upregulated in cultures of human hepatocytes (Fig. 1E). Phenobarbital (500 μmol/L) also significantly upregulated CYP3A4 mRNA expression even though the expression of NR1I2 mRNA was not significantly affected in six hepatocyte cultures (Fig. 1D and E). The expression of both genes was highly variable among liver donors, reflecting known interindividual variability in hepatic NR1I2 expression. The genetic variation in 3′UTR may also contribute to the variability.
Figure 1.
Ligand-dependent activation of PXR downregulates NR1I2 expression in primary human hepatocytes. NR1I2 mRNA (A) or CYP3A4 mRNA (B) expression in primary human hepatocytes (donor Liv13) treated with vehicle (DMSO; 0.1%, v/v) or rifampicin (Rif, 10 μmol/L) for 12, 24, and 48 h, respectively. The expression data were normalized to the RPLP0 reference gene, and data are expressed as relative change to corresponding control (DMSO-treated) experiments defined as 100%. (C) Western blotting analysis of PXR, CYP3A4 and RXRα expression in primary human hepatocytes (Liv13) treated with vehicle (DMSO; 0.1%) or Rif (10 μmol/L) for 24 or 48 h. β-Actin was used as a loading control. (D) NR1I2 mRNA and (E) CYP3A4 mRNA expression. Primary human hepatocytes (Liv1–13) and commercial (Liv14) human hepatocyte cultures were treated with vehicle (DMSO; 0.1%), Rif (10 or 5 μmol/L), SR-12813 (3 μmol/L) or phenobarbital (500 μmol/L) for 24 h. The data are presented as fold change in expression after PXR ligand treatment versus vehicle (DMSO)-treated controls defined as 1. Nonparametric Wilcoxon matched-pairs tests or paired t test were used to statistically compare paired expression data between control and PXR ligand-treated samples in human hepatocytes. *P < 0.05, statistically significant effect of PXR ligand (paired t test, P-value in parentheses).
3.2. PCN significantly downregulates mouse PXR in mice livers
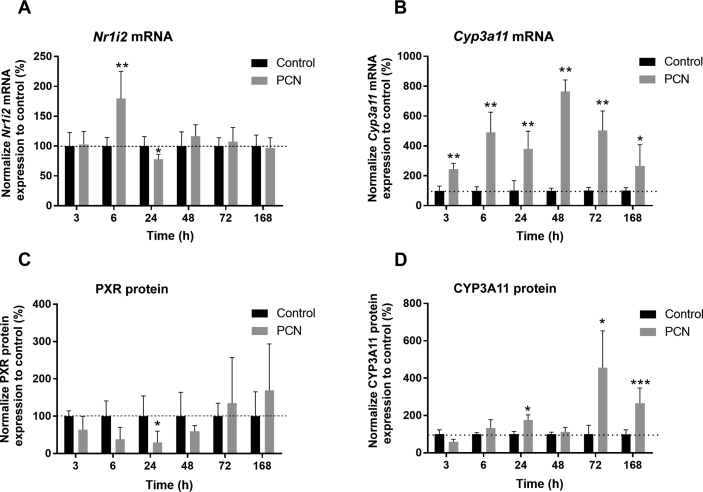
A similar pattern of PXR expression was demonstrated in mice administered a single dose of PCN, the agonist of mouse PXR. Decreased expression of Nr1i2 gene mRNA was observed 24 h after PCN exposure but was preceded by upregulation of Nr1i2 transcript at 6 h (Fig. 2A). As expected, the prototypical murine PXR target gene Cyp3a11 was significantly upregulated in response to PCN in murine livers (Fig. 2B). In agreement with these findings, the protein levels of PXR were significantly decreased at early intervals after PCN administration (Fig. 2C), but CYP3A11 protein was increased after 24, 72, and 168 h post-PCN application (Fig. 2D).
Figure 2.
PCN-mediated activation of mouse PXR affects Nr1i2 expression in mouse liver. Mouse PXR (A) or CYP3A11 (B) mRNA expression in mice that were administered a single i.p. injection of vehicle or PCN (50 mg/kg) and sacrificed after 3, 6, 24, 48, 72, or 168 h postapplication. In addition, liver sample homogenates were analyzed for mouse PXR (C) and CYP3A11 (D) protein expression employing Western blotting. Gapdh was used as a reference gene for RT-qPCR expression data normalization, and β-actin was used for Western blotting data normalization. The data are shown as the mean ± SD (n = 5 per group) and are expressed as change relative to vehicle-treated control samples defined as 100%. Mann–Whitney test was used to compare unpaired data in mice. *P < 0.05, **P < 0.01, and ***P < 0.001 indicate a statistically significant effect of PCN treatment.
Herein, we can conclude that activation of PXR or its murine orthologue leads to time-dependent downregulation of NR1I2 expression, which indicates feedback regulation of PXR.
3.3. Confirmation of NR1I2 promoter region and 3′-UTR in PXR regulation
To further examine PXR regulation, we used several luciferase reporter gene vectors with different promoter regions or the full-length 3ʹ-UTR sequence of NR1I2 gene mRNA (Fig. 3A). Our findings revealed that forced overexpression of PXR itself led to decreased activity of the pPXR1-Gluc vector bearing the promoter sequence from –1375 to +238 upstream of the human NR1I2 gene. Similar findings were shown for the mutant T248D, which produces a constitutively active form of PXR26 (Fig. 3B). The activity of the pPXR2-Gluc reporter vector bearing a shorter promoter region (–1147 to +238) of the NR1I2 gene was statistically significantly downregulated with wild-type PXR construct. The sequence from –1375 to –1147 (228 nt) in pPXR1-Gluc, but not in pPXR2-Gluc, containing numerous putative response elements for HNF4α, PPARα, Sp1 and NF-κB transcription factors9 (see schematic in Fig. 3A) thus does not seem to be important for the regulation although further detail experiments are needed. In subsequent experiments, we proved that the pmiRGLO-UTR construct harboring the 3ʹ-UTR of the NR1I2 mRNA was inhibited by PXR overexpression in HepG2 cells (Fig. 3C). Rifampicin treatment had no further significant effect on the observed inhibition. However, we observed significantly different activation of pmiRGLO-UTR after treatment with rifampicin, an agonist of PXR, or SPA70, a PXR antagonist (Fig. 3C). We thus suppose that ectopic PXR may be activated by endogenous ligands in HepG2 cells and that the activation can be abrogated by PXR antagonist SPA70.
These data demonstrate that both promoter and 3ʹ-UTR regions of the NR1I2 mRNA are important for PXR feed-back regulation (Fig. 3A).
3.4. The 3ʹ-UTR of NR1I2 mRNA is involved in the suppression of PXR expression and the reduction of rifampicin-mediated upregulation of CYP3A4
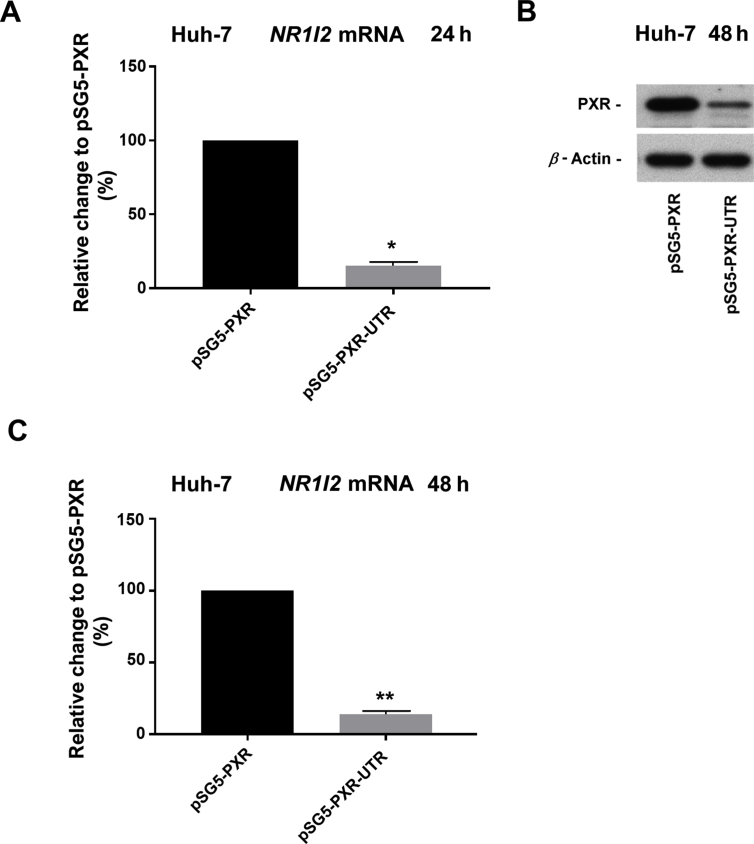
It is widely recognized that miRNAs predominantly bind to responsive elements within the 3ʹ-UTR of their target genes. Therefore, to address whether cellular miRNAs may affect the posttranscriptional regulation of PXR, HepG2 cells were transfected with either pmiRGLO (empty) or pmiRGLO-UTR vector. Detected luciferase activity was higher in cells transfected with the control empty vector (pmiRGLO) compared to cells transfected with the luciferase reporter construct bearing the 3ʹ-UTR of the NR1I2 gene mRNA (Fig. 4A). Consistently, transfection with the PXR expression vector harboring the 3ʹ-UTR resulted in lower PXR expression at both the mRNA and protein levels in HepG2 cells (Fig. 4B and C). The same trend was also observed in undifferentiated Huh-7 cells, which are a near PXR-null cellular model (Supporting Information Figs. S1A–C). Taken together, it is plausible that some miRNAs in hepatic cell lines may cause general repressive effects on the 3ʹ-UTR of NR1I2 mRNA.
Figure 4.
The 3ʹ-UTR is involved in downregulation of PXR expression and decreased rifampicin-mediated upregulation of CYP3A4. (A) HepG2 cells were transiently transfected with either pmiRGLO-UTR or empty pmiRGLO vector (50 ng). After 48 h, the samples were analyzed by the Dual-Luciferase Reporter Assay kit. Firefly luciferase activities were normalized to Renilla luciferase activities. The data are shown as the mean ± SD (n = 3) and are expressed as relative change to an empty pmiRGLO vector defined as 100%. HepG2 cells were transiently transfected with either pSG5-PXR or pSG5-PXR-UTR vector (200 ng) and cotransfected with pSG5-RXRα (200 ng) for 24 h. After another 24 or 48 h in fresh medium, total RNA was isolated and analyzed for basal levels of NR1I2 mRNA by RT-qPCR (B) or for PXR protein expression (C). The HPRT reference gene-normalized data are shown as the mean ± SD (n = 3) and are expressed as relative change to values gained from pSG5-PXR/pSG5-RXRα experiments (100%). Cell lysates were analyzed for expression of PXR protein by Western blotting. β-Actin was used as a loading control. Representative Western blotting results are shown. (D) HepG2 cells were transiently transfected with either pSG5-PXR or pSG5-PXR-UTR vector (200 ng) and cotransfected with pSG5-RXRα (200 ng). The next day, cells were treated with rifampicin (Rif, 10 μmol/L) for 24 h. Total RNA was isolated and analyzed for induction of CYP3A4 mRNA by RT-qPCR. The HPRT reference gene-normalized data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to vehicle-treated controls (defined as 1). (E) COS-1 cells were transiently transfected with a CYP3A4 promoter luciferase reporter vector (p3A4-luc, 150 ng), pRL-TK (30 ng), and pSG5-RXRα (50 ng) together with either pSG5-PXR, pSG5-PXR-UTR or empty pSG5 vector (50 ng). The next day, cells were treated with rifampicin (Rif, 10 μmol/L) or vehicle (DMSO; 0.1%) for another 24 h. Then, samples were analyzed using a Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla activities. The data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to respective DMSO-treated controls. (F) COS-1 cells were transiently transfected with either a pSG5-PXR or pSG5-PXR-UTR construct (200 ng) and cotransfected with pSG5-RXRα (200 ng). The next day, cells were treated with vehicle (DMSO; 0.1%) or rifampicin (Rif, 10 μmol/L) for 48 h. Samples were analyzed for expression of PXR protein by Western blotting. β-Actin was used as a loading control. Representative Western blotting results are shown. *P < 0.05, **P < 0.01 indicate statistically significant effects of the 3ʹ-UTR region of NR1I2 mRNA. (G). Schematic overview showing sequence complementarity between particular miRNAs and in silico predicted or experimentally confirmed miRNA-response elements (MREs) within the 3ʹ-UTR of human NR1I2 mRNA. Green letters depict seed regions of miRNAs. Blue letters refer to sequences within MREs complementary to miRNA seed regions. Numbers above sequences indicate a position of nucleotides downstream from the coding region of NR1I2 mRNA.
Next, we investigated whether this suppressive effect may affect the inducibility of the PXR target gene CYP3A4 in HepG2 and COS-1 cells. Indeed, rifampicin-mediated upregulation of CYP3A4 mRNA was mildly decreased in cells transfected with pSG5-PXR-UTR relative to cells transfected with the pSG5-PXR vector (Fig. 4D). This difference was also pronounced in the CYP3A4 promoter luciferase reporter gene assay using COS-1 cells (Fig. 4E). Of particular interest, rifampicin enhanced the suppressive role of the 3ʹ-UTR on PXR protein expression in COS-1 cells transfected with pSG5-PXR-UTR. However, rifampicin did not affect PXR protein expression in COS-1 cells transfected with pSG5-PXR (Fig. 4F). For this reason, we can assume that rifampicin-induced changes in the cellular miRNA expression profile may result in the suppression of NR1I2 expression through its 3ʹ-UTR region of NR1I2 mRNA.
3.5. MiR-18a-5p is a relevant regulator of NR1I2 expression and transcriptional activity
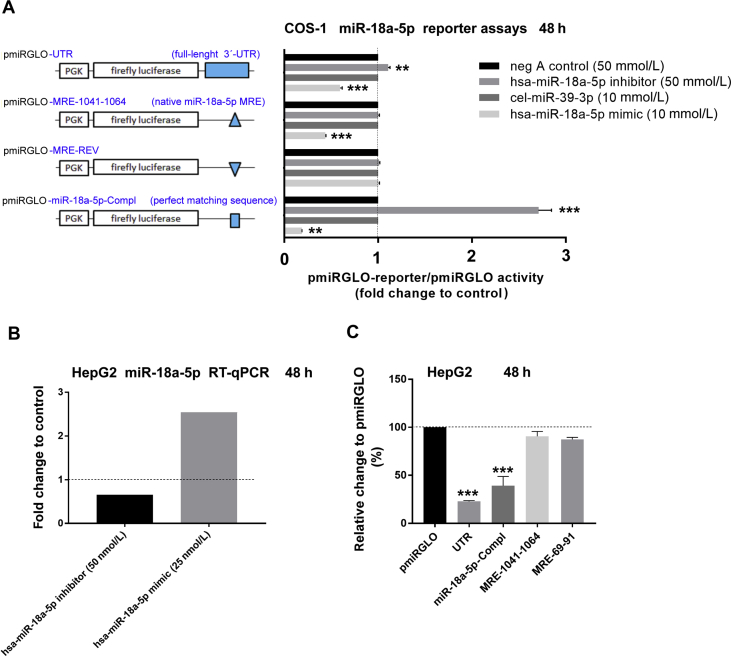
To reveal which miRNAs might be implicated in rifampicin-induced NR1I2 mRNA destabilization via its full-length 3ʹ-UTR, we first screened the effect of a set of miRNA mimics on the pmiRGLO-UTR luciferase reporter vector. The tested miRNAs were selected based on previous literature reports and on our recent bioinformatics study33. An overview of sequence complementarity between candidate miRNAs and their putative miRNA-response elements (MREs) within 1273-bp-long 3ʹ-UTR of NR1I2 mRNA is shown in Fig. 4G.
Gene reporter assays performed in HepG2 cells indicated that all selected miRNA mimics, such as hsa-miR-18a-5p, hsa-miR-449a, hsa-miR-34a-5p, and hsa-miR-148a-3p, were able to attenuate pmiRGLO-UTR activity compared to the nonspecific control (cel-miR-39-3p) mimic (Fig. 5A). However, only miR-18a-5p and miR-148a-3p were potent negative regulators of rifampicin-induced and PXR-mediated activation of the CYP3A4-based luciferase reporter construct pER6-3A4-tat-luc in HepG2 cells (Fig. 5B). In contrast, hsa-miR-449a enhanced the effect of rifampicin on the PXR-responsive reporter construct. Hsa-miR-34a-5p had no effect on the activation of the reporter construct by rifampicin (Fig. 5B). The latter experiments were performed on the pER6-3A4-tat-luc plasmid, which carries three copies of the proximal CYP3A4 promoter ER6 response element and displays minimal basal PXR-non-specific activation in HepG2 cells. Of note, miRNA mimics at a concentration of 25 nmol/L were used in these experiments in accordance with manufacturer's recommendation.
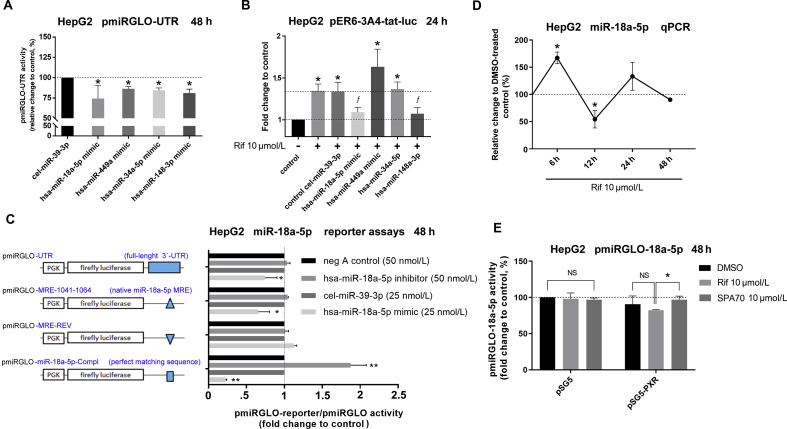
Figure 5.
MiR-18a-5p is a relevant regulator of NR1I2 expression and transcriptional activity. (A) Dual-Luciferase Reporter Assay in HepG2 cells transiently transfected with either pmiRGLO or pmiRGLO-UTR construct (50 ng) with one of the following miRNA mimic reagents such as cel-miR-39-3p (mimic control), hsa-miR-18a-5p mimic, hsa-miR-449a mimic, hsa-miR-34a-5p mimic or hsa-miR-148a-3p mimic (25 nmol/L) for 48 h. Firefly luciferase activities were normalized to Renilla activities and further normalized to the corresponding data with the empty pmiRGLO vector. The data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to the miRNA mimic control cel-miR-39-3p (100%). (B) Dual-Luciferase Reporter Assay in HepG2 cells transiently transfected with pER6-3A4-tat-luc PXR-responsive luciferase reporter vector (150 ng) and pRL-TK (30 ng) together with either cel-miR-39-3p mimic control, hsa-miR-18a-5p mimic, hsa-miR-449a mimic, hsa-miR-34a-5p mimic or hsa-miR-148a-3p mimic (25 nmol/L) for 48 h. Cells were treated with rifampicin (Rif, 10 μmol/L) or vehicle (DMSO; 0.1%) for a further 24 h. Firefly luciferase activities were normalized to Renilla activities. The data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to DMSO-treated control (defined as 1). (C) Dual-Luciferase Reporter Assay in HepG2 cells transiently transfected with either pmiRGLO, pmiRGLO-UTR, pmiRGLO-MRE-1041-1064, pmiRGLO-MRE-1041-1064-REV or pmiRGLO-18a-5p-Compl construct (50 ng) together with one of the miRNA inhibitor/mimic reagents, such as negative control A (inhibitor control), hsa-miR-18a-5p inhibitor (50 nmol/L), cel-miR-39-3p (mimic control) or hsa-miR-18a-5p mimic (25 nmol/L) for 48 h. Firefly luciferase activities were normalized to Renilla activities and further normalized to corresponding firefly luciferase activities obtained from empty pmiRGLO vector experiments. The data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to inhibitor miRNA or mimic controls (defined as 1). (D) HepG2 cells were transiently transfected with the pSG5-PXR expression vector (400 ng) for 24 h. Then, cells were treated with vehicle (DMSO; 0.1%) or rifampicin (10 μmol/L) for 6, 12, 24, or 48 h, respectively. Total RNA was isolated and analyzed for miR-18a-5p expression by RT-qPCR. U6 reference gene-normalized data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to corresponding vehicle-treated controls (defined to be 100%). (E) HepG2 cells were transiently transfected with either pmiRGLO or pmiRGLO-18a-5p (50 ng) together with either the pSG5-PXR expression vector or empty pSG5 control vector (100 ng). The next day, cells were treated with rifampicin (Rif, 10 μmol/L), SPA70 (an PXR antagonist, 10 μmol/L) or vehicle (DMSO; 0.1%) for 48 h. Samples were subsequently analyzed by Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla activities and further normalized to corresponding firefly luciferase activities obtained from the empty pmiRGLO vector. The data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to DMSO-treated control transfected with empty pSG5 vector (defined as 100%). *P < 0.05 or **P < 0.01 indicates a statistically significant effect to vehicle-treated, negative controls or empty vector controls. fP < 0.05 indicates a statistically significant effect of miRNA mimic on Rif-induced action.
To further characterize the role of miR-18a-5p in this phenomenon, we performed a reporter assay with a set of luciferase reporter constructs containing the full-length 3′-UTR or native, reversed or miR-18a-5p complementary MREs (Fig. 5C, left panel). We found that the miR-18a-5p mimic has the ability to significantly decrease activation of constructs generated with both the full-length 3ʹ-UTR and those with one copy of the native MRE for miR-18a-5p. The construct with the reversed MRE exerted no effects on miR-18a-5p in HepG2 (Fig. 5C, right panel) or COS-1 cells (Supporting Information Fig. S2A). In control experiments, we demonstrated functional transfection of the miR-18a-5p mimic and inhibitor into HepG2 cells (Fig. S2B) and significant endogenous activity of miR-18a-5p by employing the miR-18a-5p-Compl reporter vector (Fig. S2C). Additionally, treatment with rifampicin led to time-dependent changes in miR-18a-5p expression in HepG2 cells, with significant upregulation of miR-18a-5p at 6 h, suppression at 12 h and normalization to original levels between 24 and 48 h posttreatment (Fig. 5D). Moreover, HepG2 cells transiently transfected with the miR-18a-5p-Compl luciferase construct with the pSG5-PXR expression vector that were treated with rifampicin showed inhibited reporter construct activity, even though the effect was not statistically significant. Treatment with SPA70, a PXR antagonist, reversed the suppressive effect of PXR on pmiRGLO-18a-5p activation, but the effect was statistically significant only in comparison with rifampicin effect (Fig. 5E). These data confirm augmented miR-18a-5p functional expression in HepG2 cells in response to both overexpressed and activated PXR (Fig. 5E). Finally, miR-18a-5p mimic down-regulated PXR expression in HepG2 cells (data not shown), which is in agreement with published data in LS180 cells20. Taken together, these data suggest that miR-18a-5p is a direct regulator of NR1I2 gene expression and that rifampicin-mediated changes in miR-18a-5p expression may help to explain the negative regulation of activated PXR.
3.6. Activators of GR upregulate NR1I2 mRNA levels by both increasing NR1I2 mRNA stability via the 3ʹ-UTR and by activating the NR1I2 promoter
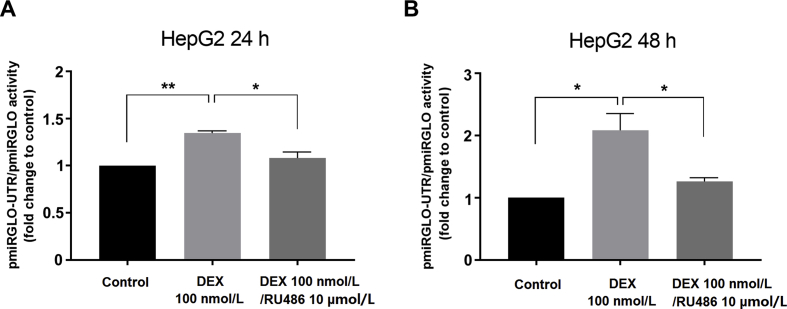
In the next series of experiments, we attempted to decipher the role of the 3ʹ-UTR region of NR1I2 mRNA in GR-mediated PXR expression regulation. To address this aim, pSG5-PXR-UTR was transfected into Huh-7 cells to explore the effect of subsequent treatment with dexamethasone. Huh-7 cells were selected due to their functional activity of GR signaling34. We observed significantly higher expression of NR1I2 mRNA after treatment with dexamethasone (100 nmol/L) than in control experiments with the PXR expression vector lacking the 3ʹ-UTR (Fig. 6A). Since dexamethasone has been demonstrated to stimulate PXR-mediated CYP3A4 expression10, we investigated whether the 3ʹ-UTR of NR1I2 mRNA may have any impacts on CYP3A4 mRNA induction. Indeed, dexamethasone induced CYP3A4 mRNA in the presence of pSG5-PXR expression vector, but CYP3A4 mRNA upregulation was even greater in the vector with the 3ʹ-UTR (Fig. 6B). We further showed that dexamethasone enhanced luciferase activity in response to the pmiRGLO-UTR vector, which was reversed by the GR antagonist RU486 in Huh-7 cells (Fig. 6C) and HepG2 cells (Supporting Information Figs. S3A and B). Finally, we found that the observed 3ʹ-UTR stabilization in the pmiRGLO-UTR construct was not selective for dexamethasone but was also present in response to other glucocorticoids (Fig. 6D). These luciferase gene reporter experiments were performed in parallel with pmiRGLO-UTR and pmiRGLO (empty) controls, and the final activation of the 3ʹ-UTR was calculated as normalized activation (ratio).
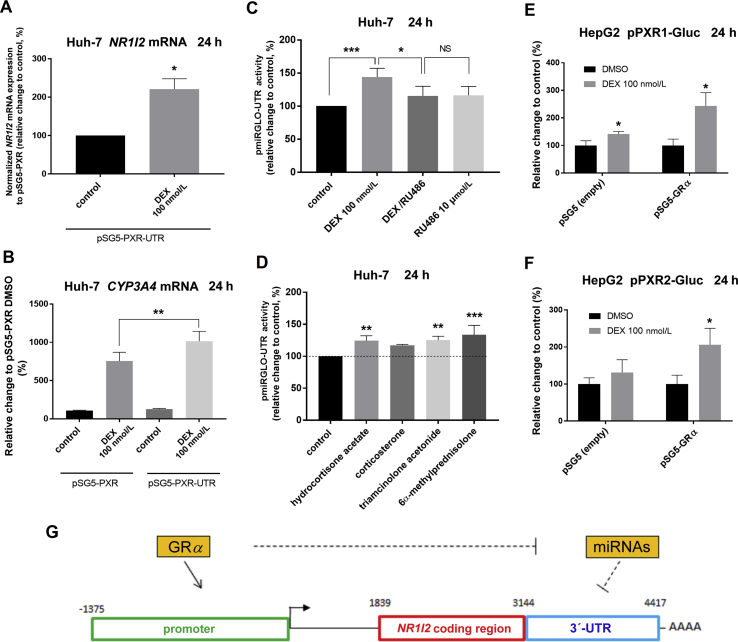
Figure 6.
Activators of the glucocorticoid receptor upregulate NR1I2 mRNA levels both by increasing PXR stability via the 3ʹ-UTR and by NR1I2 promoter activation. (A) Huh-7 cells were transiently transfected with pSG5-PXR or pSG5-PXR-UTR (200 ng) and cotransfected with the glucocorticoid receptor expression vector (pSG5-GRα, 600 ng). The next day, cells were treated with vehicle (DMSO; 0.1%) or dexamethasone (DEX, 100 nmol/L) for 24 h. Total RNA was isolated and analyzed for NR1I2 mRNA expression by RT-qPCR. The data were first normalized to the GAPDH reference gene and were acquired from pSG5-PXR-UTR transfected samples, which were further normalized to corresponding experiments transfected with the PXR expression vector lacking the 3ʹ-UTR. The data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to DMSO-treated controls (defined as 100%). (B) Huh-7 cells were transiently transfected with either pSG5-PXR or pSG5-PXR-UTR (200 ng) and were cotransfected with the pSG5-GRα expression vector (600 ng). The next day, cells were treated with vehicle (DMSO; 0.1%) or DEX (100 nmol/L) for 24 h. Total RNA was isolated and analyzed for CYP3A4 mRNA expression by RT-qPCR. GAPDH reference gene-normalized data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to the mean of vehicle-treated pSG5-PXR/pSG5-GRα-transfected experiments (defined as 100%). *P < 0.05, **P < 0.01 indicates a statistically significant effect of the 3ʹ-UTR region of NR1I2 mRNA. Huh-7 cells were transiently transfected with pmiRGLO or pmiRGLO-UTR vector (50 ng) together with pSG5-GRα expression vector (150 ng). The next day, cells were treated with DEX (100 nmol/L), RU486 (a GR antagonist, 10 μmol/L), a combination of DEX (100 nmol/L)/RU486 (10 μmol/L) or vehicle (DMSO; 0.2%, v/v) (C) or with glucocorticoids hydrocortisone acetate, corticosterone, triamcinolone acetonide, and 6α-methylprednisolone (100 nmol/L) (D) for 24 h. After treatment, samples were analyzed by Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla activities and further normalized to activation of the empty pmiRGLO vector. The data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to vehicle-treated control (defined as 100%). *P < 0.05, **P < 0.01, ***P < 0.001 indicates a statistically significant effect compared to vehicle-treated controls. (E) and (F) HepG2 cells were transiently transfected with either a pPXR1-Gluc or pPXR2-Gluc (150 ng) NR1I2 promoter luciferase reporter construct concomitantly with pSG5-GRα expression vector or pSG5 empty control (100 ng). The next day, cells were treated with dexamethasone (DEX, 100 nmol/L) or vehicle (DMSO; 0.1%) for 24 h. Then, Gaussia luciferase and SEAP activities were assessed. The results are presented as relative change in SEAP-normalized Gaussia luciferase activities compared to DMSO-treated controls (defined as 100%). The data are shown as the mean ± SD (n = 3). *P < 0.05 indicates a statistically significant effect of DEX. (G) The scheme summarizes hypothetical mechanisms for how activated GR enhances NR1I2 mRNA expression. On one side, GR stabilizes the 3ʹ-UTR of NR1I2 mRNA and on the other hand, GR triggers NR1I2 gene transcription.
In additional experiments, we confirmed previously reported findings that ligand-activated GR triggers promoter-dependent transcription of NR1I2 gene by employing NR1I2 promoter Gaussia reporter constructs in HepG2 cells (Fig. 6E and F). In summary, we propose that activated GR may have a dual role in PXR upregulation, as it stabilizes NR1I2 mRNA via the 3ʹ-UTR potentially by suppression of targeting miRNA(s) and activates NR1I2 gene transcription from the NR1I2 promoter (Fig. 6G).
3.7. Dexamethasone decreases miR-18a-5p expression and activity
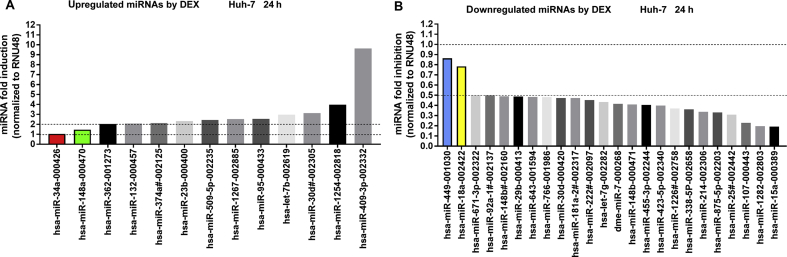
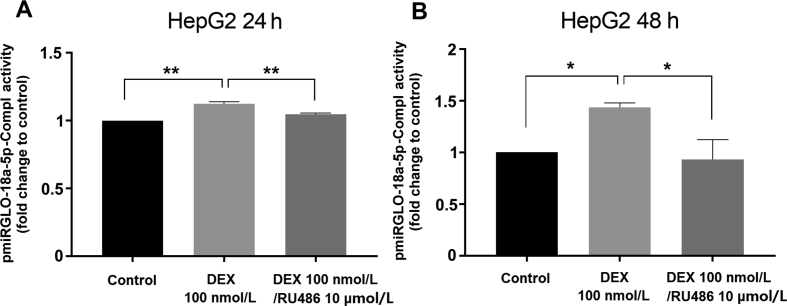
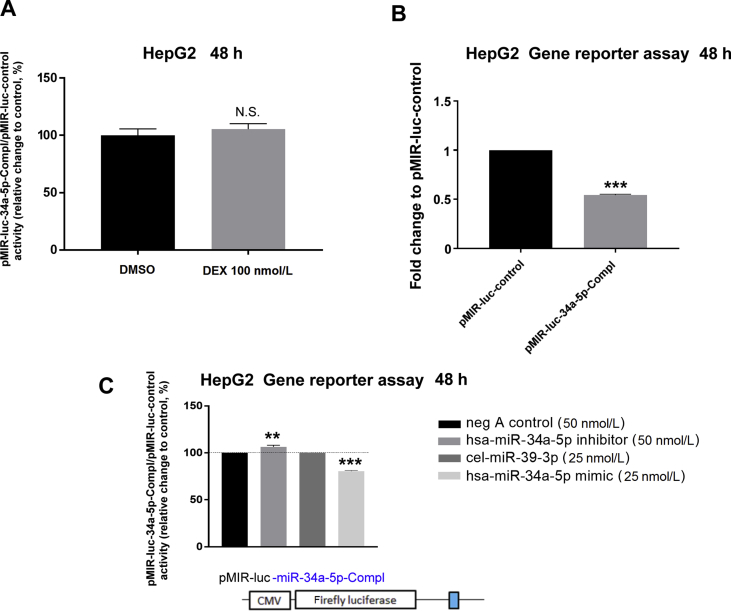
To determine which miRNAs demonstrate altered expression in response to dexamethasone treatment, we performed screening analysis of 754 miRNAs in response to either dexamethasone- or control-treated samples of Huh-7 cells using a TaqMan Array Human MicroRNA Card. Profiling showed 11 upregulated (≥2) and 22 downregulated (≤0.5) miRNAs after 24 h treatment with dexamethasone (Fig. 7A and B). Closer inspection also revealed that hsa-miR-148a-3p was slightly upregulated, while hsa-miR-34a-5p was unchanged (Fig. 7A). In contrast, hsa-miR-18a-5p and hsa-miR-449a were downregulated 24 h posttreatment with dexamethasone (Fig. 7B). Since hsa-miR-18a-5p was suppressed by dexamethasone and the functional miR-18a-5p responsive element was confirmed to be within the 3ʹ-UTR of NR1I2 mRNA (Fig. 5C, Fig. S2A, and reported in20), miR-18a-5p may be the candidate miRNA responsible for GR-mediated posttranscriptional stabilization of NR1I2 mRNA. To examine this hypothesis, we transiently transfected HepG2 cells with the pmiRGLO-18a-5p-Compl and pSG5-GRα expression vectors followed by treatment with dexamethasone for either 24 or 48 h. As expected, dexamethasone increased normalized activity of the miR-18a-5p-sensitive luciferase reporter compared to vehicle, and this effect was reversed by treatment with RU486 (Fig. 8A and B). This was not the case for the pMIR-luc-34a-5p-Compl luciferase construct with a perfect matching sequence for miR-34a-5p (Supporting Information Fig. S4A). This finding is in agreement with the TaqMan Array analysis data, where dexamethasone did not alter hsa-miR-34a-5p expression (Fig. 7A), although miR-34a-5p appears to exhibit potent endogenous activity in HepG2 cells (Figs. S4B and C). Altogether, these data provide evidence that dexamethasone treatment in hepatic cell lines downregulates expression of hsa-miR-18a-5p, subsequently resulting in stabilization of the 3ʹ-UTR of NR1I2 mRNA and NR1I2 mRNA upregulation (Fig. 9).
Figure 7.
Dexamethasone-induced changes in miRNA expression profiles in Huh-7 cells. Huh-7 cells were transfected with pSG5-GRα (600 ng) overnight and then treated with DMSO (0.1%) or dexamethasone (DEX, 100 nmol/L) for 24 h. Total RNA was isolated and analyzed using a miRNA expression TaqMan array card. RNU48 reference gene-normalized data are expressed as fold change compared to vehicle-treated experiments defined as 1. Only miRNAs with upregulated (≥2) (A) or downregulated (≤0.5) (B) expression in response to DEX treatment are shown. Additionally, the color bars indicate other miRNAs of interest potentially involved in PXR 3ʹ-UTR regulation. Data of a representative experiments are shown.
Figure 8.
Dexamethasone treatment stabilizes miR-18a-5p-responsive luciferase construct. HepG2 cells were transiently transfected with pmiRGLO or pmiRGLO-18a-5p-Compl (50 ng) together with the glucocorticoid receptor expression vector (pSG5-GRα, 150 ng). The next day, cells were treated with dexamethasone (DEX, 100 nmol/L), a combination of DEX (100 nmol/L)/RU486 (10 μmol/L) or vehicle (DMSO; 0.2%) for 24 (A) or 48 h (B). After treatment, samples were analyzed with Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla activities and were further normalized to the corresponding activity of the empty pmiRGLO vector. The data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to DMSO-treated controls (defined as 1). *P < 0.05, **P < 0.01 indicate a statistically significant effect compared to vehicle-treated or DEX-treated samples.
Figure 9.
Summary of PXR expression regulation. PXR suppresses its own expression via a dual mechanism including promoter suppression and mRNA destabilization. The latter may be mediated at least partially via upregulation of miR-18a-5p expression. GR receptor triggers transcription of the NR1I2 promoter and stabilizes NR1I2 mRNA posttranscriptionally through downregulation of miR-18a-5p.
4. Discussion
Drug-metabolizing enzymes (DMEs) are involved in removal of xenobiotics, and maintain the homeostasis of certain endogenous compounds. To achieve balance between these metabolic processes, DMEs must be tightly regulated. Several nuclear receptors have been shown to be involved in mutual feed-back and feed-forward loops that are responsible for orchestrating these metabolic pathways.
In this study, we demonstrated that NR1I2 expression may be under feed-back loop regulation through dual mechanisms involving both transcriptional and posttranscriptional regulation based on the following observations: (i) rifampicin represses PXR transcript and protein expression in a time-dependent manner in primary human hepatocytes (Fig. 1A and C); (ii) SR-12813, another PXR activator, downregulated NR1I2 mRNA expression 24 h posttreatment (Fig. 1D); (iii) PCN, an agonist of mouse PXR, suppressed both mRNA and protein levels of PXR in mice 24 h after administration (Fig. 2A and C); and (iv) both the promoter and 3ʹ-UTR regions of the NR1I2 gene suppress PXR expression (Fig. 3B and C).
Our results are consistent with another study that used primary rat hepatocytes, where PCN-activated PXR attenuated its own expression at both the mRNA and protein levels after 48 h35. Negative PXR feed-back regulation was further supported in NR1I2 promoter reporter assays using Huh-7 cells35. However, discordant data have also been reported in intestinal cell lines20,36, in an immortalized human hepatocyte cell line37 and in a few isolated human hepatocyte experiments38,39. Nevertheless, the extent of changes in NR1I2 transcript was highly variable among human liver donors used in this study after treatment with PXR ligands. This is not surprising, since donor-dependent differences in the inducibility of PXR target genes have also been previously described38,40. Thus, different tissue models and small sample sizes in studies with human hepatocytes may be the reason for this discrepancy. Moreover, detailed time course analysis is missing in these reports.
In this study, we also confirmed that the overexpression of ectopic PXR suppresses promoter-mediated transcription or NR1I2 gene. The mechanism explaining this phenomenon remains unclear since no putative binding site for PXR was found within its promoter in in silico analysis or in mechanistic studies either for human or mouse PXR9. It is interesting that PXR downregulates its own transcription both via the promoter and 3ʹ-UTR, regardless of whether PXR is activated by a ligand. We suppose that forced expression of exogenous PXR in the absence of a ligand may mimic activated PXR, which was previously reported10. One explanation may be that overexpression of PXR causes increased sensitivity to endogenous ligands. This hypothesis is further confirmed by no subsequent activation of PXR by rifampicin and by reversal of the effect by the PXR antagonist, SPA70, in the experiments (Fig. 3C).
In addition, we showed for the first time that PXR negatively affects the luciferase gene reporter construct carrying the full-length 3ʹ-UTR of NR1I2 mRNA. As a result of this finding, we aimed to uncover the precise role of the 3ʹ-UTR in PXR regulation. It is known that the 3ʹ-UTR of NR1I2 mRNA is the primary site to which miRNAs bind and exert their negative posttranscriptional effects. Although more than one hundred unique miRNAs have been predicted to potentially target the 3ʹ-UTR of NR1I2 mRNA33, there is still relatively scarce information on posttranscriptional regulation of PXR. As shown by Takagi et al.13, levels of NR1I2 mRNA expression does not correlate with PXR protein levels in human liver samples (n = 25), suggesting that PXR is subject to posttranscriptional regulation. However, in a follow-up correlation study, a linear relationship was observed between NR1I2 mRNA and protein levels in liver samples (n = 24) obtained from a Chinese Han population, raising the question of potential ethnic differences in PXR expression regulation41. However, relatively small sample sizes have been enrolled in both these studies, impairing adequate statistical power. In a pioneering report, a functional responsive element for miR-148a was found in the 3ʹ-UTR of NR1I2 mRNA, and an inverse correlation of miR-148a expression with PXR protein/mRNA ratio has been reported13. However, this observation was not confirmed by others41,42, and Lamba et al.8 even showed a positive correlation.
In our experiments, we revealed that the 3ʹ-UTR of NR1I2 mRNA attenuates PXR expression (Fig. 4A–C) and suppresses rifampicin-mediated upregulation of CYP3A4 mRNA in HepG2 cells (Fig. 4D). It is therefore plausible that endogenous miRNAs in these cells may result in a general repressive effect on the 3ʹ-UTR. An analogous observation was reported for HNF4α22. Moreover, rifampicin enhanced the repressive role of the 3ʹ-UTR in PXR protein expression (Fig. 4F), thus indicating that rifampicin-induced changes in cellular miRNAs profile may inhibit PXR expression via its 3ʹ-UTR. In agreement with this hypothesis, a modulatory effect of rifampicin on miRNA expression profiles has been recently reported in primary human hepatocytes43,44.
To shed light on the missing link between rifampicin and miRNA regulation of the PXR 3ʹ-UTR, we decided to include miR-148a13 and miR-18a-5p20 in our subsequent analysis, as their binding sites were experimentally confirmed within the 3ʹ-UTR of NR1I2 mRNA. Additionally, miR-449a and miR-34a-5p, which share the same seed sequences (Fig. 4G), were also used as they were both predicted in our in silico analysis33. In addition, miR-34a-5p8,22 and miR-449a22 have been reported to be negatively associated with NR1I2 mRNA expression. Both miR-34a-5p and miR-449a are also detectable in human hepatic cell line22,45, which is in agreement with our experiments where endogenous miR-34a-5p downregulated a luciferase reporter construct with a cloned complementary sequence for miR-34a-5p in HepG2 (Fig. S4B). Among the tested miRNAs, we found that only miR-18a-5p and miR-148a mimics attenuated rifampicin-induced PXR-mediated transactivation of CYP3A4-derived luciferase reporter construct (Fig. 5B), which is consistent with previous findings13,20. Notably, miR-449a even enhanced the activation of the CYP3A4-derived luciferase reporter construct in response to rifampicin, which indicates a possible indirect effect of miR-449a on other factors involved in the PXR-mediated transactivation.
In further experiments, we focused on miR-18a-5p as a potential culprit of PXR negative regulation. MiR-18a-5p is a member of the miR-17/92 cluster dysregulated in cancers and is highly expressed in hepatocellular carcinoma46. Very recently, miR-18a-5p was systematically described as a functional and direct regulator of PXR expression based on luciferase gene reporter studies, loss- and gain-of-function methods, and analysis of PXR-mediated inducibility of CYP3A4 mRNA in LS180 human colorectal adenocarcinoma cells20. In the present study, the functional responsive element for miR-18a-5p within the 3′-UTR of NR1I2 mRNA was confirmed with different luciferase reporter vectors, including the construct with the full-length 3ʹ-UTR (Fig. 5C and Fig. S2A). In contrast to the report by Sharma et al.20, we observed an oscillating pattern of miR-18a-5p regulation in HepG2 cells after treatment with rifampicin (Fig. 5D). During early intervals (6 h), rifampicin significantly induced expression of miR-18a-5p, which was followed by its significant reduction at 12 h and return to a slightly increased level after 24 h. The latter is in line with microarray experiments revealing only weak increases (1.64-fold change) in miR-18a-5 expression after 48 h treatment with rifampicin (10 μmol/L) in pooled human hepatocytes20. In intestinal LS180 cells, different time-dependent profiles of miR-18a-5p expression was reported with decreased expression during short intervals of treatment (3 and 6 h)20. In addition, authors did not observe significant down-regulation of NR1I2 mRNA expression in any interval in LS180 cells after treatment with two PXR ligands. These findings highlight the tissue-specific manner of the regulation and the importance of time profiling of miRNA and mRNA expression with a stress on early periods of treatment. Additionally, HepG2 cells transfected with a pmiRGLO-18a-5p-Compl luciferase reporter construct with the pSG5-PXR expression vector showed downregulation of reporter vector activity after treatment with rifampicin and without treatment, which indicates an impact of overexpressed PXR on miR-18a-5p expression (Fig. 5E). In opposite, the PXR antagonist SPA70 displayed opposite effect that rifampicin in the experiments. Our results suggest that miR-18a-5p is a direct regulator of PXR and that rifampicin-mediated changes in miR-18a-5p expression likely contribute to negative regulation of activated PXR.
Additionally, we studied the role of the 3ʹ-UTR region of NR1I2 mRNA in GR-mediated regulation of PXR expression. In accordance with previous works10,11, we confirmed that activated GR by dexamethasone triggers transcription of NR1I2 gene from its promoter (Fig. 6E and F). Since it is unclear whether GR also induces posttranscriptional changes in NR1I2 mRNA, we performed several experiments using reporter and expression vectors harboring the full-length 3ʹ-UTR region of NR1I2 mRNA. Interestingly, the expression of NR1I2 mRNA was more enhanced in cells that were transfected with the PXR expression construct with the 3ʹ-UTR region than without the 3ʹ-UTR region after treatment with glucocorticoids (Fig. 6A). Moreover, dexamethasone-induced CYP3A4 mRNA expression was significantly higher in the presence of the PXR vector with cloned 3ʹ-UTR than with an expression vector lacking the 3ʹ-UTR (Fig. 6B). We further observed that dexamethasone increased luciferase activity of pmiRGLO-UTR vector, which was reversed by the GR antagonist RU486 in Huh-7 (Fig. 6C) and HepG2 cells (Figs. S3A and B). Based on these results, we postulate that activation of GR leads to increased NR1I2 mRNA via dual mechanisms involving NR1I2 promoter activation and 3ʹ-UTR NR1I2 mRNA stabilization (Fig. 6G). Dexamethasone was used at nanomolar concentrations in our experiments, which is known to selectively activate GR but not PXR10. This experimental precaution limits the downregulation of activated PXR on its 3ʹ-UTR and helps to reveal the specific GR-mediated stabilization of 3ʹ-UTR NR1I2 mRNA.
Next, we proposed that activated GR may increase NR1I2 mRNA expression through changes in miRNA expression levels. In our miRNA expression analysis, we revealed that miR-18a-5p is downregulated by dexamethasone. Dexamethasone-induced downregulation of miR-18a expression has been consistently observed in primary rat thymocytes and cultured leukemic cells47. As a miR-18a-5p responsive element was previously identified within the 3ʹ-UTR of NR1I2 mRNA, we believed that miR-18a-5p may also be a candidate miRNA responsible for GR-mediated posttranscriptional stabilization of NR1I2 mRNA. We observed that dexamethasone increased normalized activation of pmiRGLO-18a-5p-Compl luciferase reporter construct, which was reversed by RU486. Taken together, these data provide insight indicating that dexamethasone treatment downregulates expression of hsa-miR-18a-5p, which may subsequently result in enhanced stability of NR1I2 mRNA.
5. Conclusions
In conclusion, in this study, our data support the role of miRNAs in feed-back and feed-forward regulation and in glucocorticoid-mediated hormonal upregulation of PXR expression. Based on our results, it is tempting to suppose that PXR feed-back regulation could prevent overstimulation of PXR target genes or control augmented and time-limited metabolic elimination of xenobiotic from the body. This could be a protective mechanism that hinders the unfavorable effects of endogenous compounds in the presence of PXR ligands. In this work, we also propose that glucocorticoids increase PXR expression via a dual mechanism involving activation of the NR1I2 gene promoter and by stabilization of the 3ʹ-UTR of NR1I2 mRNA for the first time. The positive effect of glucocorticoids on hepatic DME gene expression is an illustrative example of coordination and hormone-controlled liver response to xenobiotic exposure that occurs via a multifaceted process.
Acknowledgments
This work was supported by grants from the Czech Science Foundation 17-06841S to Petr Pavek and EFSA-CDN (No. CZ.02.1.01/0.0/0.0/16_019/0000841, Czech Republic) co-funded by ERDF to Tomas Smutny.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.09.010.
Author contribution
Tomas Smutny, Stanislav Micuda, Sabine Gerbal-Chaloin and Petr Pavek designed the experiments. Jan Dusek, Lucie Hyrsova, Jana Nekvindova, and Alzbeta Horvatova performed cellular experiments. Stanislav Micuda performed animal studies, and Tomas Smutny, Stanislav Micuda, Sabine Gerbal-Chaloin and Petr Pavek performed statistical analyses.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure S1.
Effect of the 3ʹ-UTR on PXR expression in Huh-7 cells. Huh-7 cells were transiently transfected with either a pSG5-PXR or pSG5-PXR-UTR construct (200 ng) and cotransfected with pSG5-RXRα (200 ng). The next day, cells were cultured in fresh medium for 24 h (A) or for 48 h (B, C), respectively. After treatment, (A, C) total RNA was isolated and analyzed for basal levels of NR1I2 mRNA by RT-qPCR. GAPDH reference gene-normalized data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to control pSG5-PXR-transfected cells (100%). (B) Samples were analyzed for expression of PXR protein by Western blotting. β-Actin was used as a loading control. Representative Western blotting results are shown. *P < 0.05, **P < 0.01 indicate a statistically significant effect of the 3ʹ-UTR region of NR1I2 mRNA.
Figure S2.
miR-18a-5p regulates the 3′-UTR of NR1I2 mRNA in COS-1 and HepG2 cells. (A) COS-1 cells were transiently transfected with either pmiRGLO, pmiRGLO-UTR, pmiRGLO-MRE-1041-1064, pmiRGLO-MRE-1041-1064-REV or pmiRGLO-18a-5p-Compl (50 ng) together with one of the following miRNA inhibitor/mimic reagents, such as negative control A (inhibitor control), hsa-miR-18a-5p inhibitor (50 nmol/L), cel-miR-39-3p (mimic control) or hsa-miR-18a-5p mimic (10 nmol/L) for 48 h. Samples were subsequently analyzed by Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla activities and were further normalized to corresponding firefly luciferase activities obtained from experiments with the empty pmiRGLO vector. The data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to miRNA mimic or inhibitor control (defined as 1). (B) HepG2 cells were transiently transfected with miR-18a-5p inhibitor (50 nmol/L) or mimic (25 nmol/L) reagents for 48 h. Total RNA was isolated and analyzed for levels of miR-18a-5p by RT-qPCR. U6 reference gene-normalized data are shown as the mean ± SD and are expressed as fold change compared to values gained from experiments with mimic or inhibitor controls defined as 1. (C) HepG2 cells were transiently transfected with pmiRGLO, pmiRGLO-UTR, pmiRGLO-18a-5p-Compl, pmiRGLO-MRE-1041-1064, or pmiRGLO-MRE-69-91 (50 ng), respectively. After 48 h, samples were analyzed by Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla activities. The data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to a pmiRGLO empty vector (defined as 100%). **P < 0.01, ***P < 0.001 indicates a statistically significant effect compared to the response of pmiRGLO vector or negative controls.
Figure S3.
Dexamethasone stabilizes the 3′-UTR of NR1I2 mRNA RNA in HepG2 cells. HepG2 cells were transiently transfected with pmiRGLO or pmiRGLO-UTR vector (50 ng) together with pSG5-GRα expression vector (150 ng). The next day, cells were treated with dexamethasone (DEX, 100 nmol/L), a combination of DEX (100 nmol/L)/RU486 (10 μmol/L) or vehicle (DMSO; 0.2%) for 24 h (A) or 48 h (B), respectively. Next, samples were analyzed by Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla activities and were further normalized to the corresponding activity of the pmiRGLO vector. The data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to DMSO-treated controls (defined as 1). *P < 0.05, **P < 0.01 indicates a statistically significant effect compared to vehicle-treated or DEX-treated samples.
Figure S4.
Dexamethasone does not alter the activity of endogenously expressed miR-34a-5p in HepG2 cells. (A) HepG2 cells were transiently transfected with either the pMIR-luc-control or pMIR-luc-34a-5p-Compl (50 ng) construct along with pRL-TK (30 ng) and glucocorticoid receptor expression (pSG5-GRα, 150 ng) vectors. The next day, cells were treated with dexamethasone (DEX, 100 nmol/L) or vehicle (DMSO; 0.1%) for 48 h. After treatment, samples were analyzed by Dual-Luciferase Reporter Assay. Firefly luciferase activities were normalized to Renilla luciferase activities and further normalized to the corresponding activity of the empty pMIR-luc-controls. The data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to DMSO-treated controls (defined as 100%). N.S. indicates a statistically insignificant effect. (B) HepG2 cells were transiently transfected with either pMIR-luc-control or pMIR-luc-34a-5p-Compl (50 ng) along with pRL-TK (30 ng). After 48 h, samples were analyzed by a Dual-Luciferase Reporter Assay System. Firefly luciferase activities were normalized to Renilla luciferase activities. The data are shown as the mean ± SD (n = 3) and are expressed as fold change compared to empty pMIR-luc-control vectors (defined as 1). ***P < 0.001 indicates a statistically significant effect of MRE. (C) HepG2 cells were transiently transfected with either pMIR-luc-control or pMIR-luc-34a-5p-Compl (50 ng) along with pRL-TK (30 ng) and one of the following miRNA inhibitor/mimic reagents, such as negative control A (inhibitor control), hsa-miR-34a-5p inhibitor (50 nmol/L), cel-miR-39-3p (mimic control) or hsa-miR-34a-5p mimic (25 nmol/L) for 48 h. Samples were subsequently analyzed by a Dual-Luciferase Reporter Assay System. Firefly luciferase activities were normalized to Renilla activities and then further normalized to corresponding firefly luciferase activities obtained from experiments with empty pMIR-luc-controls. The data are shown as the mean ± SD (n = 3) and are expressed as relative change compared to miRNA mimic or inhibitor controls (defined as 100%). **P < 0.01, ***P < 0.001 indicate a statistically significant effect compared to negative controls.
References
- 1.Oladimeji P.O., Chen T. PXR: more than just a master xenobiotic receptor. Mol Pharmacol. 2018;93:119–127. doi: 10.1124/mol.117.110155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan J., Xie W. A brief history of the discovery of PXR and CAR as xenobiotic receptors. Acta Pharm Sin B. 2016;6:450–452. doi: 10.1016/j.apsb.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu B., Li S., Dong D. 3D structures and ligand specificities of nuclear xenobiotic receptors CAR, PXR and VDR. Drug Discov Today. 2013;18:574–581. doi: 10.1016/j.drudis.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Hyrsova L., Smutny T., Carazo A., Moravcik S., Mandikova J., Trejtnar F. The pregnane X receptor down-regulates organic cation transporter 1 (SLC22A1) in human hepatocytes by competing for (“squelching”) SRC-1 coactivator. Br J Pharmacol. 2016;173:1703–1715. doi: 10.1111/bph.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavek P. Pregnane X receptor (PXR)-mediated gene repression and cross-talk of PXR with other nuclear receptors via coactivator interactions. Front Pharmacol. 2016;7:456. doi: 10.3389/fphar.2016.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smutny T., Mani S., Pavek P. Post-translational and post-transcriptional modifications of pregnane X receptor (PXR) in regulation of the cytochrome P450 superfamily. Curr Drug Metabol. 2013;14:1059–1069. doi: 10.2174/1389200214666131211153307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson G.G., Phillips A., Aouabdi S., Plant K., Plant N. Transcriptional regulation of the human pregnane-X receptor. Drug Metab Rev. 2006;38:31–49. doi: 10.1080/03602530600569810. [DOI] [PubMed] [Google Scholar]
- 8.Lamba V., Ghodke Y., Guan W., Tracy T.S. Microrna-34a is associated with expression of key hepatic transcription factors and cytochromes P450. Biochem Biophys Res Commun. 2014;445:404–411. doi: 10.1016/j.bbrc.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aouabdi S., Gibson G., Plant N. Transcriptional regulation of the PXR gene: identification and characterization of a functional peroxisome proliferator-activated receptor alpha binding site within the proximal promoter of PXR. Drug Metab Dispos. 2006;34:138–144. doi: 10.1124/dmd.105.006064. [DOI] [PubMed] [Google Scholar]
- 10.Pascussi J.M., Drocourt L., Gerbal-Chaloin S., Fabre J.M., Maurel P., Vilarem M.J. Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Sequential role of glucocorticoid receptor and pregnane X receptor. Eur J Biochem. 2001;268:6346–6358. doi: 10.1046/j.0014-2956.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 11.Pascussi J.M., Drocourt L., Fabre J.M., Maurel P., Vilarem M.J. Dexamethasone induces pregnane X receptor and retinoid X receptor-alpha expression in human hepatocytes: synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol Pharmacol. 2000;58:361–372. doi: 10.1124/mol.58.2.361. [DOI] [PubMed] [Google Scholar]
- 12.Iwazaki N., Kobayashi K., Morimoto K., Hirano M., Kawashima S., Furihata T. Involvement of hepatocyte nuclear factor 4 alpha in transcriptional regulation of the human pregnane X receptor gene in the human liver. Drug Metab Pharmacokinet. 2008;23:59–66. doi: 10.2133/dmpk.23.59. [DOI] [PubMed] [Google Scholar]
- 13.Takagi S., Nakajima M., Mohri T., Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem. 2008;283:9674–9680. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 14.Cai Y., Yu X., Hu S., Yu J. A brief review on the mechanisms of miRNA regulation. Genom Proteom Bioinform. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozomara A., Griffiths-Jones S. Mirbase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano M., Nakajima M. Current knowledge of microrna-mediated regulation of drug metabolism in humans. Expert Opin Drug Metabol Toxicol. 2018;14:493–504. doi: 10.1080/17425255.2018.1472237. [DOI] [PubMed] [Google Scholar]
- 18.Yu A.M., Pang Y.Z. Noncoding micrornas: small RNAs play a big rolein regulation of ADME?. Acta Pharm Sin B. 2009;2:93–101. doi: 10.1016/j.apsb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu A.M., Tian Y., Tu M.J., Ho P.Y., Jilek J.L. Microrna pharmacoepigenetics: posttranscriptional regulation mechanisms behind variable drug disposition and strategy to develop more effective therapy. Drug Metab Dispos. 2016;44:308–319. doi: 10.1124/dmd.115.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma D., Turkistani A.A., Chang W., Hu C., Xu Z., Chang T.K.H. Negative regulation of human pregnane X receptor by microrna-18a-5p: evidence for suppression of microrna-18a-5p expression by rifampin and rilpivirine. Mol Pharmacol. 2017;92:48–56. doi: 10.1124/mol.116.107003. [DOI] [PubMed] [Google Scholar]
- 21.Vachirayonstien T., Yan B. Microrna-30c-1-3p is a silencer of the pregnane X receptor by targeting the 3ʹ-untranslated region and alters the expression of its target gene cytochrome p450 3A4. Biochim Biophys Acta. 2016;1859:1238–1244. doi: 10.1016/j.bbagrm.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramamoorthy A., Li L., Gaedigk A., Bradford L.D., Benson E.A., Flockhart D.A. In silico and in vitro identification of micrornas that regulate hepatic nuclear factor 4alpha expression. Drug Metab Dispos. 2012;40:726–733. doi: 10.1124/dmd.111.040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Zhao J., Wang H., Li X., Liu A., Qin Q. Microrna-140-3p enhances the sensitivity of hepatocellular carcinoma cells to sorafenib by targeting pregnenolone X receptor. OncoTargets Ther. 2018;11:5885–5894. doi: 10.2147/OTT.S179509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagamasbad P., Denver R.J. Mechanisms and significance of nuclear receptor auto- and cross-regulation. Gen Comp Endocrinol. 2011;170:3–17. doi: 10.1016/j.ygcen.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W., Wang Y.M., Chai S.C., Lv L., Zheng J., Wu J. SPA70 is a potent antagonist of human pregnane X receptor. Nat Commun. 2017;8:741. doi: 10.1038/s41467-017-00780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doricakova A., Novotna A., Vrzal R., Pavek P., Dvorak Z. The role of residues t248, y249 and t422 in the function of human pregnane X receptor. Arch Toxicol. 2013;87:291–301. doi: 10.1007/s00204-012-0937-9. [DOI] [PubMed] [Google Scholar]
- 27.Pichard L., Raulet E., Fabre G., Ferrini J.B., Ourlin J.C., Maurel P. Human hepatocyte culture. Methods Mol Biol. 2006;320:283–293. doi: 10.1385/1-59259-998-2:283. [DOI] [PubMed] [Google Scholar]
- 28.Isom H.C., Secott T., Georgoff I., Woodworth C., Mummaw J. Maintenance of differentiated rat hepatocytes in primary culture. Proc Natl Acad Sci USA. 1985;82:3252–3256. doi: 10.1073/pnas.82.10.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smutny T., Bitman M., Urban M., Dubecka M., Vrzal R., Dvorak Z. U0126, a mitogen-activated protein kinase kinase 1 and 2 (MER1 and 2) inhibitor, selectively up-regulates main isoforms of CAP3A subfamily via a pregnane X receptor (PXR) in HerG2 cells. Arch Toxicol. 2014;88:2243–2259. doi: 10.1007/s00204-014-1254-2. [DOI] [PubMed] [Google Scholar]
- 30.Dusek J., Carazo A., Trejtnar F., Hyrsova L., Holas O., Smutny T. Steviol, an aglycone of steviol glycoside sweeteners, interacts with the pregnane X (PXR) and aryl hydrocarbon (AHR) receptors in detoxification regulation. Food Chem Toxicol. 2017;109:130–142. doi: 10.1016/j.fct.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Campos-Melo D., Droppelmann C.A., Volkening K., Strong M.J. Comprehensive luciferase-based reporter gene assay reveals previously masked up-regulatory effects of miRNAs. Int J Mol Sci. 2014;15:15592–15602. doi: 10.3390/ijms150915592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasnicka A., Cermanova J., Hroch M., Dolezelova E., Rozkydalova L., Smutny T. Iron depletion induces hepatic secretion of biliary lipids and glutathione in rats. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:1469–1480. doi: 10.1016/j.bbalip.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Smutny T., Tebbens J.D., Pavek P. Bioinformatic analysis of miRNAs targeting the key nuclear receptors regulating CYP3A4 gene expression: the challenge of the CYP3A4 “missing heritability” enigma. J Appl Biomed. 2015;13:181–188. [Google Scholar]
- 34.Zeng L., Chen Y., Wang Y., Yu L.R., Knox B., Chen J. MicroRNA hsa-miR-370-3p suppresses the expression and induction of CYP2D6 by facilitating mRNA degradation. Biochem Pharmacol. 2017;140:139–149. doi: 10.1016/j.bcp.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey I., Gibson G.G., Plant K., Graham M., Plant N. A PXR-mediated negative feedback loop attenuates the expression of CYP3A in response to the PXR agonist pregnenalone-16alpha-carbonitrile. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S.W., Md H., Cho M., Kim N.H., Choi H.Y., Han J.W. Role of 14-3-3 sigma in over-expression of p-gp by rifampin and paclitaxel stimulation through interaction with PXR. Cell Signal. 2017;31:124–134. doi: 10.1016/j.cellsig.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Berthier A., Oger F., Gheeraert C., Boulahtouf A., Le Guevel R., Balaguer P. The novel antibacterial compound walrycin a induces human PXR transcriptional activity. Toxicol Sci. 2012;127:225–235. doi: 10.1093/toxsci/kfs073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayed-Boussema I., Pascussi J.M., Zaied C., Maurel P., Bacha H., Hassen W. Ochratoxin a induces CYP3A4, 2B6, 3A5, 2C9, 1A1, and CYP1A2 gene expression in primary cultured human hepatocytes: a possible activation of nuclear receptors. Drug Chem Toxicol. 2012;35:71–80. doi: 10.3109/01480545.2011.589438. [DOI] [PubMed] [Google Scholar]
- 39.Maglich J.M., Stoltz C.M., Goodwin B., Hawkins-Brown D., Moore J.T., Kliewer S.A. Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 40.Gerets H.H., Tilmant K., Gerin B., Chanteux H., Depelchin B.O., Dhalluin S. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol Toxicol. 2012;28:69–87. doi: 10.1007/s10565-011-9208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Z.Y., Chen M.J., Zhang Y.T., Wang X.F., Jiang S.S., Wang Y. No correlation of hsa-miR-148a with expression of PXR or CYP3A4 in human livers from Chinese han population. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rieger J.K., Klein K., Winter S., Zanger U.M. Expression variability of absorption, distribution, metabolism, excretion-related micrornas in human liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos. 2013;41:1752–1762. doi: 10.1124/dmd.113.052126. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K., Tatsumi N., Fukami T., Yokoi T., Nakajima M. Integrated analysis of rifampicin-induced microrna and gene expression changes in human hepatocytes. Drug Metab Pharmacokinet. 2014;29:333–340. doi: 10.2133/dmpk.dmpk-13-rg-114. [DOI] [PubMed] [Google Scholar]
- 44.Ramamoorthy A., Liu Y., Philips S., Desta Z., Lin H., Goswami C. Regulation of microRNA expression by rifampin in human hepatocytes. Drug Metab Dispos. 2013;41:1763–1768. doi: 10.1124/dmd.113.052886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Burke P.A. The role of micrornas in hepatocyte nuclear factor-4alpha expression and transactivation. Biochim Biophys Acta. 2013;1829:436–442. doi: 10.1016/j.bbagrm.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mogilyansky E., Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith L.K., Shah R.R., Cidlowski J.A. Glucocorticoids modulate microrna expression and processing during lymphocyte apoptosis. J Biol Chem. 2010;285:36698–36708. doi: 10.1074/jbc.M110.162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.