Abstract
We examined the impact of gut inflammation on the expression of cytochrome P450 (P450) and other biotransformation genes in male mice using a dextran sulfate sodium (DSS)-induced colitis model. Several P450 isoforms, including CYP1A, CYP2B, CYP2C, and CYP3A, were down-regulated, accompanied by decreases in microsomal metabolism of diclofenac and nifedipine, in the liver and small intestine. The impact of the colitis on in vivo clearance of oral drugs varied for four different drugs tested: a small decrease for nifedipine, a relatively large decrease for lovastatin, but no change for pravastatin, and a large decrease in the absorption of cyclosporine A. To further assess the scope of influence of gut inflammation on gene expression, we performed genome-wide expression analysis using RNA-seq, which showed down-regulation of many CYPs, non-CYP phase-I enzymes, phase-II enzymes and transporters, and up-regulation of many other members of these gene families, in both liver and intestine of adult C57BL/6 mice, by DSS-induced colitis. Overall, our results indicate that gut inflammation suppresses the expression of many P450s and other biotransformation genes in the intestine and liver, and alters the pharmacokinetics for some but not all drugs, potentially affecting therapeutic efficacy or causing adverse effects in a drug-specific fashion.
Keywords: Cytochrome P450, CYP, Colitis, Intestine, Inflammatory bowel disease, Drug metabolism, Pharmacokinetics, Gene expression
Graphical abstract
In a mouse model of dextran-sulfate-sodium (DSS)-induced colitis, the expression of many P450s and other biotransformation genes is changed in the intestine and liver, leading to alterations in the pharmacokinetics for some but not all drugs, potentially affecting therapeutic efficacy or causing adverse effects in a drug-specific fashion.
1. Introduction
The cytochrome P450 (P450) superfamily of monooxygenases controls the homeostasis of many endogenous compounds and dictates the biotransformation and disposition of innumerable drugs and other xenobiotics1,2. The intestine is a major site of P450 expression3, 4, 5, 6. Members of the CYP1, CYP2, CYP3, and CYP4 gene families are the main contributors to the metabolism of therapeutic drugs1,2,4,7,8. For many orally administered drugs, the intestinal P450-mediated metabolism can largely affect their systemic bioavailability.
The expression and activity of drug-metabolizing P450s can be affected by many factors, including genetic polymorphisms, epigenetic modifiers, and non-genetic factors such as age, drug intake, life style, and disease states, leading to changes in first-pass metabolism of drugs and their therapeutic efficacy9. It has been reported that circulating pro-inflammatory cytokines mediate the downregulation of P450s in infection, inflammation, and cancer10, 11, 12, potentially leading to disease–drug interactions. Specific diseases that may alter P450 expression include steatohepatitis13, diabetes14, and inflammatory bowel diseases (IBDs), which are the subject of this study.
IBDs, which consist of ulcerative colitis and Crohn's disease, are chronic diseases with acute phase flare-ups. Though the etiology is not fully understood, IBDs are believed to be multifactorial diseases resulting from abnormal immunological responses to certain environmental triggers in genetically predisposed individuals15. Previous clinical reports and animal studies indicated that the systemic or local tissue concentrations of certain therapeutic drugs, such as cyclosporine A (CsA), 5-aminosalicylic acid, and metronidazole, were different in subjects with colitis, compared to those without colitis, and the efficacy of anti-IBD drugs also varied among patients16, 17, 18, 19. However, the status of drug-metabolism enzymes, a major factor in disease–drug interactions, has not been fully examined in IBD patients.
The dextran sulfate sodium (DSS)-induced colitis models in mice and rats have been widely used as animal models of IBD, due to their similarities to human ulcerative colitis in etiology, pathology, pathogenesis and therapeutic responses20. DSS, a synthetic anionic polymer, is administered to experimental animals through drinking water; it is minimally absorbed when passing through the gastrointestinal tract, leading to accumulation in the colon, where it impairs the tight junctions between colonocytes and initiates colitis20,21. DSS-induced damage, which involves the mucosa, submucosa and muscularis mucosa of the colon, is characterized by ulcers, mucosal edema, goblet cell loss, crypt distortion, abscesses, infiltration of neutrophils, macrophages, plasma cells and lymphocytes, and increased production of pro-inflammatory cytokines, such as TNF-α20. Previous studies demonstrated that several hepatic P450 enzymes were downregulated in a DSS-induced colitis mouse model22, 23, 24. However, the impact of DSS-induced colitis on the expression of intestinal P450s is less well defined, and its effects on the pharmacokinetics of oral drugs are unclear.
In the present study, we have examined the expression and function of some of the major intestinal drug-metabolizing P450 enzymes, including CYP1A, CYP2B, CYP2C, and CYP3A, in the DSS-induced colitis mouse model. We showed that the in vivo impact of DSS-induced colitis on the pharmacokinetics of oral drugs may be different for different drugs. We further demonstrated that the colitis condition affected the expression of a wide range of other genes important for drug disposition in the liver and intestine, including many CYPs, other (non-CYP) phase I biotransformation enzymes, phase II biotransformation enzymes, and drug transporters.
2. Materials and methods
2.1. Chemicals and reagents
DSS was purchased from Affymetrix (Cleveland, OH, USA). Nifedipine (NFP, purity≥98%), oxidized NFP (NFPO, purity≥95%), nitrendipine (purity≥95%), diclofenac (DCF) sodium salt, dimethyl sulfoxide (DMSO), Tween 80, β-nicotinamide adenine dinucleotide phosphate, and reduced tetra (cyclohexyl ammonium) salt (NADPH) (purity≥97%) were from Sigma–Aldrich (St. Louis, MO, USA). CsA (purity≥99%), cyclosporine D (purity≥95%), 4′-OH-DCF-13C6, 4′-OH-DCF, 5-OH-DCF, lovastatin hydroxy acid (LVA, purity≥95%), and simvastatin hydroxy acid sodium salt (SVA, purity≥95%) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Lovastatin (LVS, purity≥98%) and pravastatin sodium salt (PVS, purity≥98%) were purchased from Cayman Chemical (Ann Arbor, MI, USA). The solvents (acetonitrile, methanol and water) for high-performance liquid chromatography (HPLC) were from Thermo Fisher Scientific (Fisher Scientific, Houston, TX, USA).
2.2. Animals and treatments
Male C57BL/6 mice and intestinal epithelium-Cpr-null (IECN) mice25 (8–12-week old) were studied. Mice were housed under 12-h dark–light cycle and provided food and water ad libitum. To induce experimental colitis, mice were given 2.5% DSS (w/v) in drinking water for 7 days; mice in the control group were given drinking water alone. Body weight, liquid intake, physical activity, and occurrence of rectal bleeding were monitored daily. Colon length (from the end of cecum to the anus) was measured on Day 7 of DSS treatment, when mice were sacrificed. For determination of in vivo drug clearance, mice from DSS-treated and control groups were treated once, via oral gavage, with 10 mg/kg NFP [prepared in 10% ethanol, 10% Tween 80, and 80% phosphate-buffered saline (PBS)]25, 25 mg/kg LVS (prepared in 10% DMSO, 20% Tween 80, and 70% PBS)26, 50 mg/kg PVS sodium salt (prepared in PBS), or 10 mg/kg CsA (prepared in 10% ethanol, 10% Tween 80, and 80% PBS)27, on Day 7. All procedures involving animals were approved by the Institutional Animal Care and Use Committees of the Wadsworth Center and the University of Arizona.
2.3. Preparation of microsomes
Intestinal microsomes were prepared, as described25, from isolated intestinal epithelial enterocytes combined from 2 to 3 mice. Liver from individual mouse was used for the preparation of hepatic microsomes, using published procedures28. Protein concentration was determined with the BCA protein assay, using bovine serum albumin as a standard.
2.4. Immunoblot analysis
Intestinal microsomal proteins (15 μg) were separated on 10% NuPAGE Bis-Tris gels (Life Technologies, Grand Island, NY, USA), and then transferred to nitrocellulose membranes, which were incubated in 5% non-fat milk before the addition of primary antibodies. For the immunodetection, rabbit polyclonal anti-CYP1A (Millipore, San Diego, CA, USA), rabbit polyclonal anti-CYP2B (BD Gentest, Woburn, MA, USA)25, goat polyclonal anti-CYP2C (BD Gentest)25, and rabbit polyclonal anti-CYP3A (Abcam, Cambridge, MA, USA)29 were used. Rabbit anti-calnexin30 (Abcam) was used for the detection of calnexin, as a loading control. Peroxidase-conjugated goat anti-rabbit IgG or rabbit anti-goat IgG (Sigma–Aldrich) was used as the secondary antibody. Immunoreactive bands were visualized with the ECL Chemiluminescence kit (GE Healthcare, Piscataway, NJ, USA), and quantified using a ChemiDoc XRS1 System (Bio-Rad, Hercules, CA, USA).
2.5. In vitro assays of diclofenac and nifedipine metabolism
DCF metabolism was assayed as described31. Hepatic or intestinal microsomes (0.1 mg) were incubated with 100 μmol/L DCF in a reaction mixture containing 0.1 mol/L potassium phosphate buffer (pH 7.4), 1.0 mmol/L NADPH, and 3 mmol/L MgCl2, in a final volume of 200 μL. The reaction was initiated by adding NADPH into the reaction mixture, followed by a 30-min incubation at 37 °C. For the control groups, NADPH was omitted. The reaction was terminated by adding 400 μL of ice-cold acetonitrile. The internal standard 4′-OH-DCF-13C6 (10 μL at 4 ng/μL) was added for monitoring extraction efficiency for 4′-OH-DCF and 5-OH-DCF. The mixture was vortexed for 1 min, before centrifugation at 1500×g for 10 min; the supernatant was collected and spun again for another 10 min. The supernatant was subjected to solid-phase extraction (SPE) using ISOLUTE C18 cartridges (Biotage, Charlottesville, VA, USA) before LC/MS analysis.
NFP metabolism was assayed as described previously25; the conditions and procedures used were the same as described above for the DCF assay, except that NFP was added at 25 μmol/L, and the incubation period was 10 min. Nitrendipine (100 pmol) was added as the internal standard for the detection of the reaction product, NFPO.
Quantitative analysis of 4′-OH-DCF, 5-OH-DCF and NFPO was performed using a LC–MS/MS system consisting of a SCIEX Q-Trap 6500 + mass spectrometer (AB SCIEX, Framingham, MA, USA), an Infinity II Series model 1290 ultra-performance LC system (Agilent, Santa Clara, CA, USA), and an Agilent ZORBAX Eclipse Plus C18 column (2.1 mm × 50 mm, 1.8 μm). The assay conditions were modified from previously published protocols31,32, as described in Supporting Information.
2.6. Pharmacokinetic analysis
Blood samples were collected from the tail vein at 10, 30 min, 1, 2, 4, and 10 h after drug treatment, using heparinized capillary tubes (Thermo Fisher Scientific), and plasma was stored at −80 °C until use. The LC–MS/MS system described above was used for determination of NFP, LVA, PVS, and CsA. The conditions for sample processing and LC–MS analysis were adapted from previously published protocols26,32,33, as described in Supporting Information.
2.7. RNA extraction, RNA-sequencing and quantitative real-time polymerase chain reaction
Total RNA was isolated from intestinal epithelium, colonic epithelium, and liver using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), as described34, and then further purified to remove possible DSS contamination using lithium chloride as a precipitant35. RNA purity and quality were examined with bioanalyzer and concentration of total RNA was determined with Nanodrop (Thermo Fisher Scientific). Total RNA (2 μg) was used for first-strand cDNA synthesis using SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative real-time polymerase chain reaction (RT-PCR) was performed as described36. The primers used are listed in Supporting Information Table S1. Transcripts for the ribosomal phosphoprotein 36B4 was also measured and used for data normalization.
For RNA-seq analysis, RNAs extracted from the liver and proximal small intestinal epithelium were further treated with DNase (Qiagen, Germantown, MD, USA). Samples with RNA integrity (RIN) scores above 7.2 were used for RNA-seq. RNA sequencing was performed by Novogene (Sacramento, CA, USA). Insert size of 250–300 bp was used for cDNA library preparation. Libraries were sequenced on the Illumina platform for 150-bp paired-end reads. Reference genome and annotation files were downloaded from Ensembl, and RNA-seq data were aligned to the reference genome using the Spliced Transcripts Alignment to a Reference (STAR) software37. HTSeq was used to obtain read counts of the mapped genes, and the DESeq2 package was used for differential expression analysis. ClusterProfiler and KEGG database were used for enrichment pathway analysis38,39. P values were adjusted by the Benjamini & Hochberg method40, with P < 0.05 as the threshold for significantly differential expression.
2.8. Data analysis
Pharmacokinetic (PK) parameters were calculated using PK solver (Microsoft, Redmond, WA, USA), by assuming a noncompartmental model. Two-way analysis of variance (ANOVA), followed by a post-hoc test for pairwise comparisons, or Student's t-test, was performed using GraphPad Prism (GraphPad Software, La Jolla, CA, USA). P values < 0.05 was considered statistically significant.
3. Results
3.1. Induction of colitis in mice
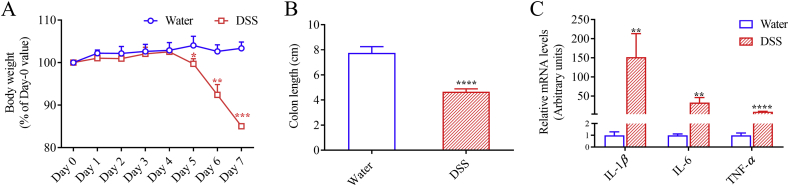
The DSS-induced colitis model displayed the expected signs of colon injury and inflammation. No difference was observed in water intake between DSS group and control group. Symptoms of colitis started to display from Day 5, including body weight loss, rectal bleeding, and diarrhea. In the DSS-treated group, body weight was 85.0% of that on Day 0 after seven-day treatment, which was significantly decreased compared to the control group with body weight at 103% of that on Day 0 (Fig. 1A). Colon was remarkably shortened, by ∼40% (Fig. 1B), in DSS-treated mice compared to control mice. Colon inflammation was further substantiated by significantly elevated mRNA expression levels of IL-1β, IL-6, and TNF-α, by 152-, 33-, and 9-fold, respectively, in the DSS-treated mice, compared to control mice (Fig. 1C).
Figure 1.
Induction of colitis in mice by DSS treatment. Mice (2–3-month old, male) were treated with 2.5% DSS in drinking water or water alone for seven days. (A) Body weight is shown as percentage of initial body weight during treatment with water or 2.5% DSS. (B) Colon length was measured at the end of treatment on Day 7. (C) Expression of IL-1β, IL-6, and TNF-α mRNA was determined by RT-PCR as described in Section Materials and Methods. The relative expression levels were determined by normalizing to 36B4 and are shown in arbitrary units with levels of the control group set to 1. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to control (water alone) group; means ± SD, n = 4, Student's t-test.
3.2. Impact of DSS-induced colitis on intestinal P450 expression
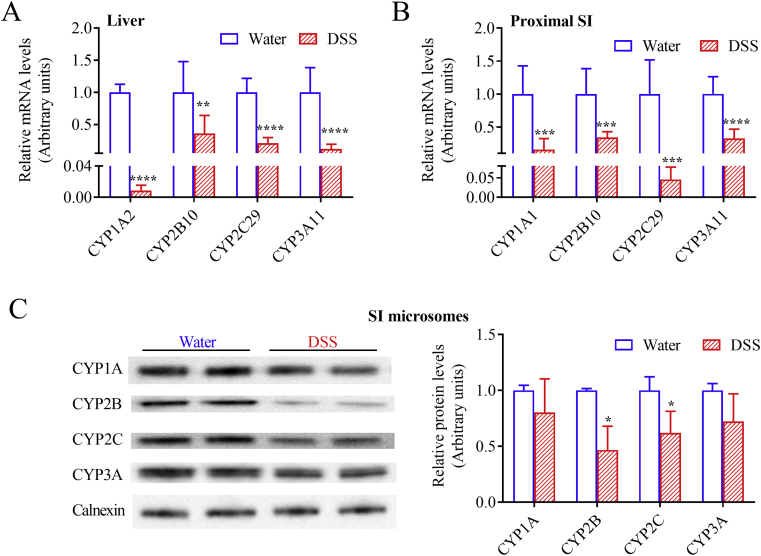
Consistent with previous reports that hepatic P450s were downregulated by DSS treatment22, 23, 24, we confirmed that the DSS-induced colitis was associated with significantly decreased mRNA expression levels of Cyp1a2, Cyp2b10, Cyp2c29, and Cyp3a11 in the liver (Fig. 2A). Similar to what was observed in the liver, intestinal Cyp1a1, Cyp2b10, Cyp2c29, and Cyp3a11 mRNA expression levels, determined for the proximal segment of small intestine, were downregulated after seven days of treatment with DSS, by 3- to 22-fold (Fig. 2B), compared to control mice. Levels of intestinal microsomal CYP2B and CYP2C proteins, determined for the epithelium of the entire small intestine, were also suppressed, by ∼50%, with a trend of decrease also for CYP1A and CYP3A (Fig. 2C).
Figure 2.
Relative expression levels of hepatic and intestinal P450s. Mice (2–3-month old, male) were treated with 2.5% DSS in drinking water or water alone for seven days. (A) and (B) Effects of DSS treatment on P450 mRNA expression in liver (A) and intestine (B). Liver and small intestinal (SI) epithelium were collected on Day 7 for RNA isolation and gene expression analysis. RT-PCR was performed as described in Section Materials and Methods; the relative expression levels were determined by normalizing to 36B4 and are shown in arbitrary units, with levels of the control (water alone) group set to 1. **P < 0.01, ***P < 0.001, ****P < 0.0001 compared with control group; means ± SD, n = 7–8, Student's t-test. (C) Effects of DSS treatment on P450 protein expression in small intestine. SI epithelium were collected on Day 7 for microsome preparation and immunoblot analysis. Microsomal samples, each prepared from pooled SI epithelium from 2 to 3 mice, were analyzed in duplicates (15 μg protein per lane), using anti-CYP1A, anti-CYP2B, anti-CYP2C, anti-CYP3A, and anti-calnexin (loading control) antibodies, as described in Section Materials and Methods. Left, representative immunoblots. Right, relative expression levels were determined by normalizing to calnexin and are shown in arbitrary units, with levels of the control (water alone) group set to 1. Data from three different microsomal samples were used for quantitative analysis. *P < 0.05 compared with control group; means ± SD, n = 3, Student's t-test.
3.3. Impact of DSS-induced colitis on P450-mediated drug metabolism in vitro
DCF was first utilized as a probe substrate for studying CYP2C and CYP3A activities in hepatic and intestinal microsomes prepared from mice that have been treated with DSS in drinking water (or water alone as a control) for 7 days. The rates of formation of 4′-OH-DCF (known metabolite by CYP2C) and 5-OH-DCF (known metabolite by CYP3A) were 23% and 27%, respectively, lower in hepatic microsomes (Fig. 3A), and 58% and 64%, respectively, lower in intestinal microsomes (Fig. 3B), of DSS-treated mice than in control mice. Similarly, the rates of formation of NFPO from NFP (another CYP3A substrate) were 22% lower in hepatic microsomes, and 44% lower in intestinal microsomes, of DSS-treated mice than in control mice (Fig. 3C).
Figure 3.
In vitro metabolism of diclofenac and nifedipine by hepatic and intestinal microsomes. Mice (2–3-month old, male) were treated with 2.5% DSS in drinking water or water alone for seven days. Liver and small intestine were collected on Day 7 for preparation of microsomes. Microsomal proteins (0.1 mg) were incubated with 100 μmol/L DCF (A and B) or 25 μmol/L NFP (C) in reaction mixtures containing 0.1 mol/L potassium phosphate buffer (pH 7.4), 1.0 mmol/L NADPH, and 3.0 mmol/L MgCl2, in a final volume of 200 μL, at 37 °C for 30 min for DCF incubation and 10 min for NFP incubation. Metabolites were determined using LC–MS as described in Section Materials and Methods. Each microsomal sample was prepared from pooled tissues from 2 to 3 mice and three different microsomal samples were used for rate determination. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to control (water) group; means ± SD, n = 4, Student's t-test.
3.4. Impact of DSS-induced colitis on in vivo clearance of drugs
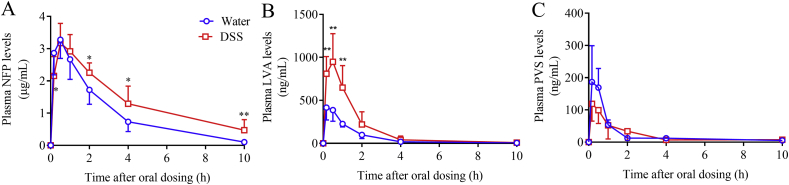
Several probe drugs were used to study the impact of DSS-induced colitis on their in vivo clearance. The plasma levels of NFP were higher in mice exposed to DSS for 7 days than in control mice at 2, 4, and 10 h after a single oral dose of NFP at 10 mg/kg (Fig. 4A). The calculated total NFP exposure (AUC0–t) was significantly higher in mice with colitis than in control mice (Table 1), and the apparent clearance (CL/F) of NFP was slower in DSS-treated mice than in control mice. The plasma levels of LVA, the major circulating form of the pro-drug LVS in mice, were significantly higher in mice with colitis than in control mice after a single oral dose of LVS at 25 mg/kg (Fig. 4B). The calculated values for Cmax and AUC0–t were 2.0- and 2.4-fold higher, respectively, and CL/F was 53% lower, in DSS-treated mice compared to control mice (Table 1). In contrast, no obvious difference in plasma level of PVS was observed between DSS- and water-treated groups after a single oral dose of PVS at 50 mg/kg (Fig. 4C). The calculated PK parameters for PVS did not show significant differences between the two groups of mice (Table 1).
Figure 4.
In vivo clearance of nifedipine, lovastatin, and pravastatin in mice with or without DSS treatment. Plasma levels of NFP (A), LVA (B), and PVS (C) were analyzed at 10 min, 30 min, 1, 2, 4, and 10 h after a single dose of drug administration (at 10, 25, or 50 mg/kg, for NFP, LVS, and PVS, respectively, by oral gavage) in mice (2–3-month-old, male) pretreated with DSS in drinking water or water alone for 7 days. *P < 0.05, **P < 0.01 compared to control (water) group; means ± SD, n = 4–8, Student's t-test.
Table 1.
Pharmacokinetic parameters for orally administered nifedipine, lovastatin, and pravastatin in control and DSS-treated mice.
| Group | Drug | Tmax (h) | Cmax (μg/mL) | t1/2 (h) | AUC0–t (μg/mL·h) | CL/F (L/kg/h) |
|---|---|---|---|---|---|---|
| Water | NFP | 0.52 ± 0.23 | 3.30 ± 0.56 | 2.02 ± 0.55 | 9.89 ± 2.14 | 1.02 ± 0.21 |
| DSS | NFP | 0.52 ± 0.23 | 3.20 ± 0.58 | 3.73 ± 2.13* | 14.0 ± 3.2** | 0.65 ± 0.23** |
| Water | LVS | 0.37 ± 0.18 | 0.43 ± 0.14 | 1.34 ± 0.68 | 0.66 ± 0.09 | 37.7 ± 5.8 |
| DSS | LVS | 0.43 ± 0.15 | 0.99 ± 0.31** | 1.41 ± 0.12 | 1.60 ± 0.75* | 17.8 ± 6.6*** |
| Water | PVS | 0.25 ± 0.17 | 0.22 ± 0.09 | 1.82 ± 1.10 | 0.22 ± 0.06 | 202 ± 53 |
| DSS | PVS | 0.17 ± 0.01 | 0.10 ± 0.04 | 3.32 ± 1.34 | 0.17 ± 0.10 | 290 ± 123 |
Data from Fig. 4 were used for determination of pharmacokinetic parameters. Adult male C57BL/6 mice were treated with DSS in drinking water for 7 days at 2.5% (w/v), or with water alone. On Day 7, mice were given NFP, LVS, and PVS at 10, 25, and 50 mg/kg, respectively, by oral gavage. Values represent means ± SD (n = 5–8). LVS was measured as LVA.
*P < 0.05, **P < 0.01, ***P < 0.001 compared to corresponding water group (Student's t-test).
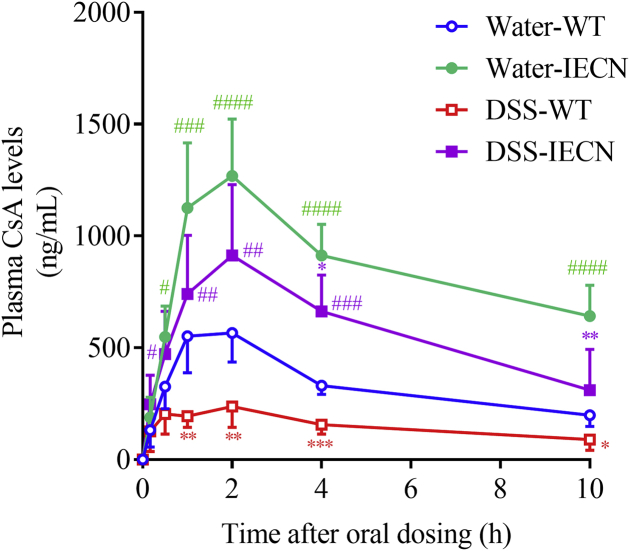
The effects of colitis on drug clearance were also studied for CsA, a substrate for both CYP3A and P-glycoprotein, and a drug with beneficial therapeutic effects in patients with severe ulcerative colitis41,42. Contrary to the observed effects of colitis on the clearance of NFP and LVS, the plasma concentration of CsA was significantly lower in DSS-treated mice than in control mice, after a single oral dose of CsA at 10 mg/kg (Fig. 5). Cmax and AUC0–t values were 60% and 50%, respectively, lower in DSS-treated mice than in water-treated mice (Table 2), despite the decreased P450 expression in the DSS-treated group.
Figure 5.
In vivo clearance of cyclosporine A in WT and IECN mice with or without DSS treatment. Plasma levels of CsA were analyzed at 10, 30 min, 1, 2, 4, and 10 h after a single dose of CsA at 10 mg/kg by oral gavage in WT and IECN mice (2–3-month old, male) pretreated with DSS in drinking water or water alone for 7 days. *P < 0.05, **P < 0.01, ***P < 0.001 compared to corresponding control (water) group; #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001 compared to corresponding WT group; means ± SD, n = 6–10, two-way ANOVA with Tukey's multiple comparison test.
Table 2.
Pharmacokinetic parameters for orally administered cyclosporine A in control and DSS-treated WT and IECN mice.
| Group | Tmax h | Cmax μg/mL | t1/2 h | AUC0–t μg/mL·h | CL/F L/kg/h |
|---|---|---|---|---|---|
| Water-WT | 1.50 ± 0.55 | 0.63 ± 0.12 | 5.95 ± 1.45 | 3.35 ± 0.51 | 2.02 ± 0.40 |
| Water-IECN | 1.70 ± 0.48 | 1.30 ± 0.24#### | 10.2 ± 5.3 | 8.60 ± 1.33#### | 0.60 ± 0.18# |
| DSS-WT | 1.50 ± 0.77 | 0.25 ± 0.09* | 6.10 ± 1.85 | 1.67 ± 0.48* | 4.86 ± 1.70**** |
| DSS-IECN | 1.71 ± 0.49 | 0.94 ± 0.30*,#### | 6.28 ± 3.72 | 5.77 ± 1.53***,#### | 1.31 ± 0.68#### |
Data from Fig. 5 were used for determination of pharmacokinetic parameters. Adult male WT and IECN mice were treated with DSS in drinking water at 2.5% (w/v), or with water alone, for 7 days. On Day 7, mice were given CsA at 10 mg/kg by oral gavage. Values represent means ± SD (n = 6–10).
*P < 0.05, ***P < 0.001, ****P < 0.0001 compared to corresponding water group; #P < 0.05, ####P < 0.0001 compared to corresponding WT group (Two-way ANOVA followed by Tukey's test).
The effects of colitis on CsA clearance were also examined in the intestinal epithelium-Cpr-null (IECN) mice, in which intestinal microsomal P450 activity is abolished due to the ablation of the Cpr gene in enterocytes25. Plasma levels of CsA were much higher in IECN mice than in wild-type (WT) mice, in both DSS-treated or control groups, reflecting the important role of intestinal P450 in CsA clearance (Fig. 5). Nevertheless, colitis caused an (IE-P450-independent) decrease (over the corresponding water control group) in plasma CsA levels (Fig. 5) and calculated Cmax (by 28%) and AUC (by 33%) values (Table 2), in the IECN mice.
3.5. Global gene expression changes in the liver and intestine of mice with DSS-induced colitis
To explore more broadly the impact of DSS-induced colitis on gene expression, RNA sequencing analysis was performed for both liver and proximal intestine, to compare gene expression changes between control mice and mice that have been treated with DSS for 7 days. Approximately 47–54 million (liver) and 46–51 million (intestine) total clean reads were generated for analysis. The uniquely mapping rates were 84.3%–88.5% for liver and 88.8%–90.1% for intestine. Differential expression analysis revealed that, with Padj<0.05 as the criterion for significant changes, 1057 genes were up-regulated and 1280 genes were down-regulated in the liver; whereas, 351 genes were up-regulated and 373 genes were down-regulated in the proximal small intestine (Supporting Information Fig. S1).
Four of the top five down-regulated hepatic genes (based on fold-change), Ces2c (7.3-fold), Cyp2c55 (7.3-fold), Ces2b (7.2-fold) and Cyp2b10 (4.9-fold), encode drug-processing enzymes; the other one was Igfbp3 (6.3-fold), encoding insulin-like growth factor-binding protein 3. Two of the top five down-regulated intestinal genes were also related to drug metabolism, Cyp2c29 (4.9-fold) and Ces2b (2.6-fold); the others were Igfbp3 (4.9-fold), Axin2 (3.4-fold), which is a downstream target of the Wnt/β-catenin pathway43, and Dnase1l3 (2.8-fold), which regulates cytokine secretion44. The colitis-induced decreases in P450 expression that was demonstrated using PCR (Fig. 2) were also confirmed by the RNA-seq analysis (Fig. 6).
Figure 6.
Differential expression of genes between mice with or without colitis analyzed using RNA-seq. Mice (2–3-month old, male) were treated with 2.5% of DSS dissolved in drinking water or water alone for seven days. Hepatic and SI total RNA was used for RNA-sequencing analysis. M-A (log ratio-mean average) plots for hepatic (A) or proximal SI (B) mRNA expression of CYPs, non-CYP phase I enzymes, phase II enzymes, and transporters are shown. Log2 fold-changes (DSS/water) are plotted against averaged read-counts of the two comparison groups. Up to five most highly expressed transcripts (based on counts, in purple box) that are changed, and up to five most increased or decreased transcripts (based on fold change, in orange box), filtered by |log2 (fold change)| >0.585 (1.5-fold) and Padj <0.05, are highlighted by red dots and annotated. CYPs that were found to be changed in mRNA expression using RT-PCR (Fig. 2) are circled in green. Results of global differential gene expression analysis and pathway analysis are shown in Supporting Information Figs. S1 and S2 , respectively.
Fold-change-vs-abundance data (M-A plot) for all detected transcripts in various subgroups are shown graphically in Fig. 6. The differentially expressed genes (with change-fold greater than 1.5, abundance greater than 100 counts, and adjusted P value less than 0.05) that are involved in drug metabolism and transport are selected for presentation in Table 3, with the total number of changed genes in the liver (123) included 25 CYPs (7 up and 18 down), 23 Non-CYP phase I enzymes (1 up and 22 down), 27 phase II enzymes (2 up and 25 down), 9 ATP-binding cassette (ABC) efflux transporters (9 down), and 39 solute carriers (SLC)/solute carrier organic anion (SLCO) uptake transporters (21 up and 18 down). The total number of changed genes in the proximal intestine (26) included 8 CYPs (2 up and 6 down), 6 Non-CYP phase I enzymes (3 up and 3 down), 4 phase II enzymes (2 up and 2 down), 1 ABC efflux transporters (1 up), and 7 SLC/SLCO uptake transporters (4 up and 3 down). The non-CYP phase I enzymes queried included alcohol dehydrogenases (ADHs), aldehyde dehydrogenases (ALDHs), aldo-keto reductases (AKRs), carboxylesterases (CESs), dehydrogenase/reductases (DHRs), dihydropyrimidinase (DPYD), short-chain dehydrogenase/reductases (SDRs), xanthine dehydrogenase (XDH), arylacetamide deacetylase (AADAC), elastases (CELAs), epoxide hydrolases (EPHXs), esterase (ESD), flavin-containing monooxygenases (FMOs), prolidase (PEPD), paraoxonases (PONs), sulfite oxidase (SUOX), carbonyl reductases (CBRs), NAD(P)H: quinone oxidoreductases (NQOs), alternative oxidases (AOXs), monoamine oxidases (MAOs), and P450 oxidoreductase (POR). Phase II enzymes queried included UDP-glucuronosyltransferases (UGTs), glutathione S-transferases (GSTs), microsomal glutathione S-transferases (MGSTs), sulfotransferase (SULTs), methyltransferases (HNMT, COMT, TPMT and AS3MT), acyltransferases (BAAT, GLYAT and NATs), and enzymes for co-substrates of phase II enzymes (GCLC, GCLM, UGDH, UGP2, PAPSSs and MATs). The transporters were divided as ABC efflux transporters and SLC/SLCO uptake transporters.
Table 3.
Summary of differentially expressed genes of CYPs, non-CYP phase I enzymes, phase II enzymes, and transporters.
| Liver |
Proximal small intestine |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Ensembl ID | Fold change (log2) | Adjusted P-value | Gene | Ensembl ID | Fold change (log2) | Adjusted P-value | Gene | Ensembl ID | Fold change (log2) | Adjusted P-value |
| Down-regulated genes | |||||||||||
| Phase I, CYPs | |||||||||||
| Cyp2c44 | ENSMUSG00000025197 | −0.61 | 5.48E-05 | Cyp8b1 | ENSMUSG00000050445 | −1.23 | 1.77E-06 | Cyp3a11 | ENSMUSG00000056035 | −0.77 | 1.36E-02 |
| Cyp2c67 | ENSMUSG00000062624 | −0.63 | 1.12E-03 | Cyp2u1 | ENSMUSG00000027983 | −1.23 | 5.07E-06 | Cyp2b10 | ENSMUSG00000030483 | −0.82 | 2.74E-03 |
| Cyp2d9 | ENSMUSG00000068086 | −0.68 | 3.96E-04 | Cyp4v3 | ENSMUSG00000079057 | −1.27 | 2.73E-16 | Cyp2c55 | ENSMUSG00000025002 | −1.09 | 2.39E-03 |
| Cyp3a59 | ENSMUSG00000061292 | −0.69 | 1.49E-02 | Cyp2f2 | ENSMUSG00000052974 | −1.30 | 1.48E-14 | Cyp2c65 | ENSMUSG00000067231 | −1.12 | 2.91E-10 |
| Cyp1a2 | ENSMUSG00000032310 | −0.75 | 2.12E-03 | Cyp3a11 | ENSMUSG00000056035 | −1.62 | 5.71E-22 | Cyp2c66 | ENSMUSG00000067229 | −1.61 | 2.84E-10 |
| Cyp2c37 | ENSMUSG00000042248 | −0.92 | 1.49E-05 | Cyp4b1 | ENSMUSG00000028713 | −1.63 | 9.87E-10 | Cyp2c29 | ENSMUSG00000003053 | −2.28 | 2.68E-18 |
| Cyp2c38 | ENSMUSG00000032808 | −0.97 | 2.81E-03 | Cyp2a5 | ENSMUSG00000005547 | −2.70 | 4.57E-51 | ||||
| Cyp2c29 | ENSMUSG00000003053 | −0.98 | 2.13E-06 | Cyp4a12b | ENSMUSG00000078597 | −3.05 | 6.28E-32 | ||||
| Cyp3a25 | ENSMUSG00000029630 | −1.06 | 1.94E-09 | Cyp4a12a | ENSMUSG00000066071 | −3.18 | 5.09E-54 | ||||
| Phase I, Non-CYP | |||||||||||
| Aadac | ENSMUSG00000027761 | −1.10 | 3.95E-18 | Ces1f | ENSMUSG00000031725 | −1.21 | 3.78E-09 | Ces2g | ENSMUSG00000031877 | −0.82 | 1.06E-07 |
| Akr1c19 | ENSMUSG00000071551 | −0.77 | 9.31E-03 | Ces2a | ENSMUSG00000055730 | −1.05 | 1.57E-16 | Ces2b | ENSMUSG00000050097 | −1.36 | 5.96E-06 |
| Aldh1b1 | ENSMUSG00000035561 | −0.60 | 2.17E-03 | Ces2c | ENSMUSG00000061825 | −2.87 | 1.48E-11 | Fmo1 | ENSMUSG00000040181 | −1.59 | 3.50E-10 |
| Aldh3a2 | ENSMUSG00000010025 | −2.17 | 3.97E-45 | Ces2e | ENSMUSG00000031886 | −1.44 | 3.67E-22 | ||||
| Aldh5a1 | ENSMUSG00000035936 | −0.69 | 1.96E-05 | Ces3b | ENSMUSG00000062181 | −1.18 | 1.15E-18 | ||||
| Aldh7a1 | ENSMUSG00000053644 | −0.65 | 5.33E-06 | Dpyd | ENSMUSG00000033308 | −0.59 | 1.26E-03 | ||||
| Aldh9a1 | ENSMUSG00000026687 | −0.59 | 7.45E-05 | Ephx1 | ENSMUSG00000038776 | −1.03 | 1.18E-10 | ||||
| Aox1 | ENSMUSG00000063558 | −0.92 | 1.65E-08 | Ephx2 | ENSMUSG00000022040 | −1.20 | 4.39E-22 | ||||
| Aox3 | ENSMUSG00000064294 | −0.91 | 4.28E-10 | Maob | ENSMUSG00000040147 | −0.80 | 3.04E-06 | ||||
| Cela1 | ENSMUSG00000023031 | −1.19 | 2.30E-06 | Nqo2 | ENSMUSG00000046949 | −0.91 | 6.15E-08 | ||||
| Ces1e | ENSMUSG00000061959 | −1.68 | 1.15E-27 | Suox | ENSMUSG00000049858 | −0.86 | 1.34E-10 | ||||
| Phase II | |||||||||||
| Gstk1 | ENSMUSG00000029864 | −0.59 | 5.30E-05 | Ugt2b36 | ENSMUSG00000070704 | −0.87 | 6.51E-08 | Gstm4 | ENSMUSG00000027890 | −0.95 | 6.82E-05 |
| Gstp1 | ENSMUSG00000060803 | −0.92 | 2.42E-04 | Ugt1a5 | ENSMUSG00000089943 | −0.93 | 1.58E-04 | Gstm3 | ENSMUSG00000004038 | −1.19 | 4.98E-05 |
| Gstm4 | ENSMUSG00000027890 | −0.93 | 1.88E-07 | Ugt2a3 | ENSMUSG00000035780 | −0.96 | 3.08E-09 | ||||
| Gstm2 | ENSMUSG00000040562 | −1.14 | 2.21E-06 | Ugt2b35 | ENSMUSG00000035811 | −1.06 | 1.12E-10 | ||||
| Gstm1 | ENSMUSG00000058135 | −1.14 | 2.24E-17 | Ugt2b5 | ENSMUSG00000054630 | −1.33 | 1.25E-20 | ||||
| Gstt1 | ENSMUSG00000001663 | −1.21 | 8.14E-13 | Ugt1a9 | ENSMUSG00000090175 | −1.61 | 1.29E-17 | ||||
| Gstm6 | ENSMUSG00000068762 | −1.32 | 2.20E-19 | Ugt2b1 | ENSMUSG00000035836 | −2.00 | 1.60E-32 | ||||
| Gstm3 | ENSMUSG00000004038 | −1.74 | 6.21E-16 | Ugt2b38 | ENSMUSG00000061906 | −2.83 | 3.65E-21 | ||||
| Gstt3 | ENSMUSG00000001665 | −1.88 | 9.72E-12 | Sult1b1 | ENSMUSG00000029269 | −0.70 | 6.82E-03 | ||||
| Gstt2 | ENSMUSG00000033318 | −1.89 | 1.66E-17 | Gclc | ENSMUSG00000032350 | −0.82 | 1.29E-08 | ||||
| Gsta4 | ENSMUSG00000032348 | −2.16 | 1.91E-36 | Papss2 | ENSMUSG00000024899 | −1.04 | 1.53E-05 | ||||
| Gsta2 | ENSMUSG00000057933 | −3.45 | 4.19E-45 | Ugp2 | ENSMUSG00000001891 | −1.42 | 5.63E-20 | ||||
| Ugt3a1 | ENSMUSG00000072664 | −0.71 | 3.56E-06 | ||||||||
| ABC Transporters | |||||||||||
| Abca2 | ENSMUSG00000026944 | −0.75 | 3.40E-06 | Abcc9 | ENSMUSG00000030249 | −1.02 | 7.68E-07 | ||||
| Abcb11 | ENSMUSG00000027048 | −0.85 | 8.91E-11 | Abcg5 | ENSMUSG00000040505 | −1.04 | 3.88E-10 | ||||
| Abca8a | ENSMUSG00000041828 | −0.97 | 6.72E-06 | Abcc3 | ENSMUSG00000020865 | −1.19 | 6.18E-09 | ||||
| Abcb4 | ENSMUSG00000042476 | −0.98 | 2.28E-10 | Abcd3 | ENSMUSG00000028127 | −1.25 | 1.15E-14 | ||||
| Abcg8 | ENSMUSG00000024254 | −1.02 | 5.16E-09 | ||||||||
| SLC and SLCO transporters | |||||||||||
| Slc1a2 | ENSMUSG00000005089 | −0.59 | 2.89E-04 | Slc25a22 | ENSMUSG00000019082 | −0.85 | 3.27E-07 | Slc9a2 | ENSMUSG00000026062 | −0.64 | 3.44E-02 |
| Slc6a6 | ENSMUSG00000030096 | −0.59 | 3.45E-03 | Slc35g1 | ENSMUSG00000044026 | −0.90 | 5.12E-05 | Slc5a4a | ENSMUSG00000020229 | −1.12 | 6.46E-09 |
| Slc22a30 | ENSMUSG00000052562 | −0.64 | 1.33E-04 | Slc7a2 | ENSMUSG00000031596 | −0.96 | 2.98E-05 | Slc12a2 | ENSMUSG00000024597 | −1.16 | 4.27E-05 |
| Slc25a10 | ENSMUSG00000025792 | −0.65 | 5.07E-05 | Slc22a28 | ENSMUSG00000063590 | −0.98 | 2.20E-04 | ||||
| Slc25a15 | ENSMUSG00000031482 | −0.67 | 2.80E-06 | Slc17a8 | ENSMUSG00000019935 | −1.10 | 5.62E-04 | ||||
| Slc23a2 | ENSMUSG00000027340 | −0.69 | 2.01E-02 | Slc25a30 | ENSMUSG00000022003 | −1.28 | 1.83E-02 | ||||
| Slc9a3r2 | ENSMUSG00000002504 | −0.74 | 5.31E-03 | Slc43a3 | ENSMUSG00000027074 | −1.33 | 1.50E-10 | ||||
| Slc30a10 | ENSMUSG00000026614 | −0.75 | 7.29E-03 | Slc2a2 | ENSMUSG00000027690 | −1.42 | 4.73E-17 | ||||
| Slc10a1 | ENSMUSG00000021135 | −0.77 | 1.85E-06 | Slc17a4 | ENSMUSG00000021336 | −1.68 | 1.29E-17 | ||||
| Up-regulated genes | |||||||||||
| Phase I, CYPs | |||||||||||
| Cyp2d40 | ENSMUSG00000068083 | 0.66 | 3.48E-04 | Cyp51 | ENSMUSG00000001467 | 0.81 | 1.12E-07 | Cyp2j6 | ENSMUSG00000052914 | 0.83 | 5.46E-03 |
| Cyp39a1 | ENSMUSG00000023963 | 0.68 | 1.77E-02 | Cyp4a31 | ENSMUSG00000028712 | 1.39 | 7.22E-05 | Cyp2j9 | ENSMUSG00000015224 | 0.96 | 4.17E-03 |
| Cyp2c70 | ENSMUSG00000060613 | 0.69 | 2.21E-05 | Cyp3a13 | ENSMUSG00000029727 | 1.54 | 3.07E-28 | ||||
| Cyp17a1 | ENSMUSG00000003555 | 0.75 | 3.29E-02 | ||||||||
| Phase I, Non-CYP | |||||||||||
| Por | ENSMUSG00000005514 | 0.71 | 2.95E-03 | Aldh3b1 | ENSMUSG00000024885 | 0.60 | 7.08E-05 | ||||
| Adh1 | ENSMUSG00000074207 | 0.65 | 9.44E-03 | ||||||||
| Akr1b7 | ENSMUSG00000052131 | 0.67 | 3.96E-02 | ||||||||
| Phase II | |||||||||||
| As3mt | ENSMUSG00000003559 | 0.80 | 1.41E-06 | Papss2 | ENSMUSG00000024899 | 0.70 | 3.77E-02 | ||||
| Papss1 | ENSMUSG00000028032 | 0.66 | 1.38E-02 | Mgst1 | ENSMUSG00000008540 | 0.87 | 5.36E-03 | ||||
| ABC Transporters | |||||||||||
| Abcc6 | ENSMUSG00000030834 | 0.84 | 1.26E-03 | ||||||||
| SLC and SLCO transporters | |||||||||||
| Slc30a1 | ENSMUSG00000037434 | 0.60 | 7.53E-04 | Slc1a4 | ENSMUSG00000020142 | 1.15 | 1.81E-06 | Slc25a28 | ENSMUSG00000040414 | 0.64 | 6.26E-05 |
| Slc46a3 | ENSMUSG00000029650 | 0.62 | 5.90E-03 | Slc13a3 | ENSMUSG00000018459 | 1.26 | 2.07E-09 | Slc19a2 | ENSMUSG00000040918 | 0.81 | 4.79E-06 |
| Slc43a1 | ENSMUSG00000027075 | 0.62 | 2.39E-02 | Slc11a2 | ENSMUSG00000023030 | 1.29 | 3.85E-11 | Slc35e3 | ENSMUSG00000060181 | 0.90 | 1.27E-03 |
| Slc35e3 | ENSMUSG00000060181 | 0.65 | 3.34E-03 | Slc10a2 | ENSMUSG00000023073 | 1.30 | 7.16E-03 | Slc6a3 | ENSMUSG00000021609 | 1.03 | 6.05E-03 |
| Slc41a1 | ENSMUSG00000013275 | 0.65 | 1.97E-02 | Slc17a9 | ENSMUSG00000023393 | 1.31 | 4.59E-05 | ||||
| Slc35b1 | ENSMUSG00000020873 | 0.66 | 7.66E-04 | Slc39a14 | ENSMUSG00000022094 | 1.45 | 5.91E-13 | ||||
| Slc11a1 | ENSMUSG00000026177 | 0.68 | 3.24E-02 | Slc41a2 | ENSMUSG00000034591 | 2.16 | 2.22E-08 | ||||
| Slc25a19 | ENSMUSG00000020744 | 0.76 | 7.18E-04 | Slc13a5 | ENSMUSG00000020805 | 2.19 | 2.24E-14 | ||||
| Slc39a1 | ENSMUSG00000052310 | 0.87 | 4.11E-08 | Slc3a1 | ENSMUSG00000024131 | 2.50 | 6.29E-42 | ||||
| Slc16a12 | ENSMUSG00000009378 | 0.90 | 2.39E-05 | Slc37a1 | ENSMUSG00000024036 | 3.01 | 8.82E-30 | ||||
| Slc30a5 | ENSMUSG00000021629 | 1.00 | 4.26E-09 | ||||||||
Differential gene expression was analyzed by RNA-seq for hepatic and SI RNA samples from mice with or without colitis, as described in Fig. 6. Genes with normalized readout mean > 100, adjusted P-value <0.05, |log2 fold change| >0.585 are presented in various subgroups of drug-metabolizing enzymes and transporters.
Enrichment analysis (Supporting Information Fig. S2, for top 20 changed pathways) indicated that drug-metabolism-related pathways are among the most significantly changed by colitis, based on adjusted P value. The top five pathways affected by DSS treatment in liver were metabolic pathways (320 genes differentially expressed), metabolism of xenobiotics by P450s (39 genes differentially expressed), complement and coagulation cascades (39 genes differentially expressed), drug metabolism (41 genes differentially expressed), and citrate cycle (21 genes differently expressed). The top five pathways affected by DSS in the small intestine were drug metabolism (16 genes differentially expressed), metabolism of xenobiotic by P450s (14 genes differentially expressed), linoleic acid metabolism (10 genes differentially expressed), complement and coagulation cascades (13 genes differentially expressed), and osteoclast differentiation (16 genes differentially expressed).
4. Discussion
We have shown evidence for down-regulation of several drug-metabolizing CYPs in the small intestine, in addition to the liver, by DSS-induced colitis in the present study. These CYPs, particularly CYP2C and CYP3A, are among the most important drug-metabolizing P450 enzymes, as they collectively metabolize a large fraction of all prescribed drugs45. CYP3A and CYP2C are also the most predominant P450 isoforms in the intestine2,34. The effects of colitis on CYP expression (Fig. 2) were further supported by data on significant decreases in microsomal activities toward probe substrates, NFP (for CYP3A) and DCF (for CYP2C and CYP3A, Fig. 3).
Our data on colitis-induced downregulation of P450 expression confirm the findings of a previous report, where treatment of male C57BL/6 mice with 5% DSS (we used 2.5%) for 7 days caused downregulation of CYP3A in the upper small intestine at mRNA and protein levels23. Two other studies have examined effects of DSS on intestinal P450 expression; their findings appear to differ from ours and those of Kawauchi and co-workers23. In one, treatment of male ICR mice with 3.5% DSS for 10 days did not change microsomal CYP3A level in the small intestine24. In the other, treatment of male Sprague–Dawley rats with 3% DSS for 7 days did not cause a decrease in CYP activity toward NFP in intestinal microsomes46. Several reasons might account for these apparent discrepancies, including possible species or mouse strain differences in the DSS-induced inflammatory response, the different concentrations and sources of DSS used, and the differences in the specific parts of the intestine that were analyzed, which may show differing extent of response to inflammation. Notably, our study provides, for the first time, data on comprehensive analysis of colitis-induced changes in the expression of various drug-metabolism and disposition-related genes in the liver and intestine, including not only CYPs, but also non-CYP phase I enzymes, phase II enzymes, and drug transporters.
Possible mechanisms of the colitis-induced intestinal CYP down-regulation remain to be determined. In that regard, previous work had indicated that, in the liver of DSS-treated mice, an increase in hepatocyte and serum cytokines might have led to an increased NF-κB nuclear translocation, decreased PXR and CAR nuclear translocation and mRNA expression, and consequent downregulation of P450 expression23,24,47. Nevertheless, DSS treatment has not been associated with histological inflammation or increases in cytokine levels in the small intestine. Hence, studies on whether enterocyte cytokine level is elevated, and the NF-κB pathway is activated, are needed to determine whether the same mechanisms occur in the intestine. Additionally, a reduction in FXR activation, which can lead to decreased activation of PXR and downregulated CYP3A expression in the intestine, may occur, due to DSS-induced decrease in bile acid levels23. It may be necessary to examine the time course of changes in the expression of the various regulatory molecules during disease onset and progression, as well as during recovery following termination of the DSS treatment. In that connection, it has been reported that the extent of post-DSS recovery may differ for different hepatic P450 transcripts in mice48 and for hepatic, renal, or intestinal microsomal P450 activities in rats46.
To investigate whether the downregulation of hepatic and intestinal P450s has an impact on in vivo drug clearance, four probe drugs, NFP, LVS, PVS, and CsA, belonging to different Biopharmaceutics Classification System (BCS)/Biopharmaceutical Drug Disposition Classification System (BDDCS) categories, were studied49. NFP is a BCS I/BDDCS I drug, with relatively high solubility, high permeability, high metabolism, but minimal involvement of transporters in absorption25,49. Mice with colitis displayed slower systemic clearance of NFP, compared to control mice [as indicated by greater total exposure (AUC), longer half-life, and slower apparent clearance (CL/F), Fig. 4 and Table 1), which is consistent with downregulation of its major metabolizing enzyme CYP3A in liver and intestine.
LVS, a BCS II/BDDCS II drug, is rapidly hydrolyzed to become LVA, its active form50 and the major circulating form of the drug in mice51. LVS and LVA are both CYP3A substrates52,53, though they are also metabolized to a lesser extent by other enzymes, such as CYP2C and UGTs26,54. Unlike NFP, LVS absorption involves several transporters54, some of which, including MRP2, MDR1, and BSEP, are suppressed in the liver by DSS treatment23. The pharmacokinetics of plasma LVA is impacted by colitis in similar ways as for NFP, with an increase in AUC and a decrease in CL/F, except that, contrary to the result for NFP, the Cmax value for LVA is also increased, whereas plasma half-life was not changed (Fig. 4 and Table 1). The reason for this subtle difference between NFP and LVS/LVA is unclear; but it may be partly related to the effects of colitis on other drug-metabolizing enzymes and drug transporters, as suggested by the subsequent RNA-seq analysis (Table 3) and by previous studies of DSS effects on hepatic transporters23.
PVS, a BCS III/BDDCS III drug, with high solubility and low permeability, undergoes little P450-mediated metabolism. Its disposition involves several transporters, including OATP1B1, OATP2B1, OATP1A2, OAT1, OAT3, and MDR154,55. In DSS-treated mice, there is no evident change in the various PK parameters for PVS, compared to water-treated mice (Fig. 4 and Table 1), which is consistent with the lack of P450 involvement in PVS metabolism, and also suggests that any changes in transporter expression was insufficient to alter the disposition of PVS.
Interestingly, the effect of colitis on the pharmacokinetics of CsA, another BCS II/BDDCS II drug, was unexpected. When taken orally, CsA undergoes first-pass extraction by CYP3A in the intestine and liver56,57. However, colitis mice showed dramatic decreases in plasma CsA levels after a single oral dose, compared to water-alone control mice. The role of intestinal P450-mediated CsA metabolism on CsA bioavailability was confirmed by comparing WT and IECN mice (Fig. 5). Nevertheless, both WT and IECN mice showed colitis-induced decrease in Cmax and AUC, but not in t1/2, which suggests colitis-associated changes in CsA absorption (Fig. 5 and Table 2). In that regard, the gastrointestinal absorption of CsA can be influenced by many factors, including bile acids in the gut, which can assist in the dissolution of CsA and increase its absorption58. Bile acid levels in the small intestinal lumen were reported to be lower (by 2-fold) in DSS-treated mice than in water-treated mice23, which might have at least partially contributed to the apparent, colitis-associated decrease in CsA absorption observed in this study. CsA absorption may also be impeded by diarrhea59, which occurs in DSS-induced colitis mice. Thus, the anticipated effects of a decreased CYP expression on drug disposition is overshadowed by upstream decreases in drug absorption that might have resulted from a multitude of disease-associated changes, including reductions in bile acid levels and other pathological conditions.
Notably, the effect of DSS-induced colitis on CsA level was also examined in the study of Kawauchi and co-workers23. CsA was given orally to control and DSS-treated mice at 1 mg/kg (10 mg/kg was in our study), and whole-blood CsA levels were determined at two time points (30 and 60 min after dosing). Contrary to our finding, they reported higher (by ∼2-fold) blood CsA levels in DSS-treated mice than in control mice. The reason for this discrepancy is unclear, but it might be related to the large difference in the doses administered between the two studies; it is possible that, at 1 mg/kg, intestinal absorption was not limiting, and the effects of a decreased CYP3A expression on CsA disposition can be demonstrated.
The differing outcomes of colitis on the pharmacokinetics of the four drugs tested demonstrate that the effects of colitis on in vivo drug clearance is drug-specific. The reasons for the drug specificity of the in vivo effects of colitis are complicated and will be important to study. It may include the differential roles of the changed P450s in the first-pass extraction of a particular drug; the role of other phase-I biotransformation enzymes and phase-II metabolism; the involvement of enterohepatic recirculation; and the role of specific transporters in the absorption or excretion of a given drug. In that regard, the results of our broad survey, using RNA-seq, indicate widespread effects of colitis on various groups of drug metabolism and disposition genes (Fig. 6 and Table 3) and pathways (Supporting Information Fig. S2). Notably, though more genes are downregulated by colitis, some genes are upregulated, which further complicates the situation. Validation of these changes at the protein level, and a comprehensive knowledge of the activities of each of these changed enzymes or transporters, is necessary to predict the impact of colitis on the pharmacokinetics of a specific drug.
IBD patients often experience co-morbidities, such as cardiovascular diseases and hypercholesterolemia60. Therefore, the altered pharmacokinetic profiles of anti-hypertensive drugs, such as NFP, nimodipine, and verapamil, and anti-hypercholesterolemia drugs, such as LVS and simvastatin, could impact therapeutic efficacy and potentially cause life-threatening adverse effects in these patients. Colitis-induced changes in pharmacokinetics may also directly impact the safety or efficacy of drugs used in the treatment of IBD, such as CsA, an immunosuppressive agent, which, in addition to its common use in organ transplantation and other immune-related conditions, is also used in ulcerative colitis to delay surgery. Thus, it is important to consider the effects of colitis on drug metabolism and disposition when prescribing for IBD patients. In that regard, given potential species differences between mice and humans, similar analyses to what has been done in mice should be conducted for humans, to yield human gene expression data that can be directly used to build a predictive model for colitis effects on drug disposition.
5. Conclusions
In summary, we have examined the impact of gut inflammation on the expression of intestinal P450 and other biotransformation enzymes and drug transporters in male mice using a DSS-induced colitis model. Our results indicate that colitis suppresses the expression of many P450s and other biotransformation genes in the liver and intestine, and alters the pharmacokinetics for some but not all drugs, potentially affecting therapeutic efficacy or cause adverse effects in a drug-specific fashion.
Acknowledgments
We thank Ms. Weizhu Yang for assistance with mouse production. We gratefully acknowledge the use of the Histopathology Core of the Wadsworth Center (Albany, NY, USA). This work was supported in part by the National Institutes of Health (Grants GM082978 and ES006694, USA).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.12.002.
Author contributions
Xiaoyu Fan, Xinxin Ding and Qing-Yu Zhang participated in research design. Xiaoyu Fan and Qing-Yu Zhang conducted experiments. Xiaoyu Fan and Qing-Yu Zhang performed data analysis. Xiaoyu Fan, Xinxin Ding and Qing-Yu Zhang wrote or contributed to the writing of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest regarding the publication of this article.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kaminsky L.S., Zhang Q.Y. The small intestine as a xenobiotic-metabolizing organ. Drug Metab Dispos. 2003;31:1520–1525. doi: 10.1124/dmd.31.12.1520. [DOI] [PubMed] [Google Scholar]
- 2.Paine M.F., Hart H.L., Ludington S.S., Haining R.L., Rettie A.E., Zeldin D.C. The human intestinal cytochrome P450 “pie”. Drug Metab Dispos. 2006;34:880–886. doi: 10.1124/dmd.105.008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezirtzoglou E.E. Intestinal cytochromes P450 regulating the intestinal microbiota and its probiotic profile. Microb Ecol Health Dis. 2012;23 doi: 10.3402/mehd.v23i0.18370. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding X., Kaminsky L.S. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 5.Galetin A., Houston J.B. Intestinal and hepatic metabolic activity of five cytochrome P450 enzymes: impact on prediction of first-pass metabolism. J Pharmacol Exp Ther. 2006;318:1220–1229. doi: 10.1124/jpet.106.106013. [DOI] [PubMed] [Google Scholar]
- 6.Kaminsky L.S., Fasco M.J. Small intestinal cytochromes P450. Crit Rev Toxicol. 1992;21:407–422. doi: 10.3109/10408449209089881. [DOI] [PubMed] [Google Scholar]
- 7.Thelen K., Dressman J.B. Cytochrome P450-mediated metabolism in the human gut wall. J Pharm Pharmacol. 2009;61:541–558. doi: 10.1211/jpp/61.05.0002. [DOI] [PubMed] [Google Scholar]
- 8.Xie F., Ding X., Zhang Q.Y. An update on the role of intestinal cytochrome P450 enzymes in drug disposition. Acta Pharma Sin B. 2016;6:374–383. doi: 10.1016/j.apsb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Aitken A.E., Richardson T.A., Morgan E.T. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol. 2006;46:123–149. doi: 10.1146/annurev.pharmtox.46.120604.141059. [DOI] [PubMed] [Google Scholar]
- 11.Morgan E. Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin Pharmacol Ther. 2009;85:434–438. doi: 10.1038/clpt.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slaviero K.A., Clarke S.J., Rivory L.P. Inflammatory response: an unrecognised source of variability in the pharmacokinetics and pharmacodynamics of cancer chemotherapy. Lancet Oncol. 2003;4:224–232. doi: 10.1016/s1470-2045(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Lechon M.J., Jover R., Donato M.T. Cytochrome P450 and steatosis. Curr Drug Metabol. 2009;10:692–699. doi: 10.2174/138920009789895543. [DOI] [PubMed] [Google Scholar]
- 14.Kim S.K., Novak R.F. The role of intracellular signaling in insulin-mediated regulation of drug metabolizing enzyme gene and protein expression. Pharmacol Ther. 2007;113:88–120. doi: 10.1016/j.pharmthera.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko K.J., Auyeung K.K. Inflammatory bowel disease: etiology, pathogenesis and current therapy. Curr Pharmaceut Des. 2014;20:1082–1096. doi: 10.2174/13816128113199990416. [DOI] [PubMed] [Google Scholar]
- 16.Bergan T., Bjerke P.E., Fausa O. Pharmacokinetics of metronidazole in patients with enteric disease compared to normal volunteers. Chemotherapy. 1981;27:233–238. doi: 10.1159/000237985. [DOI] [PubMed] [Google Scholar]
- 17.Latteri M., Angeloni G., Silveri N.G., Manna R., Gasbarrini G., Navarra P. Pharmacokinetics of cyclosporin microemulsion in patients with inflammatory bowel disease. Clin Pharmacokinet. 2001;40:473–483. doi: 10.2165/00003088-200140060-00006. [DOI] [PubMed] [Google Scholar]
- 18.Ramírez-Alcántara V., Montrose M.H. Acute murine colitis reduces colonic 5-aminosalicylic acid metabolism by regulation of N-acetyltransferase-2. Am J Physiol Gastrointest Liver Physiol. 2014;306:G1002–G1010. doi: 10.1152/ajpgi.00389.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandborn W.J. A critical review of cyclosporine therapy in inflammatory bowel disease. Inflamm Bowel Dis. 1995;1:48–63. [Google Scholar]
- 20.Solomon L., Mansor S., Mallon P., Donnelly E., Hoper M., Loughrey M. The dextran sulphate sodium (DSS) model of colitis: an overview. Comp Clin Pathol. 2010;19:235–239. [Google Scholar]
- 21.Chen G.Y., Shaw M.H., Redondo G., Núñez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaluvadi M.R., Nyagode B.A., Kinloch R.D., Morgan E.T. TLR4-dependent and -independent regulation of hepatic cytochrome P450 in mice with chemically induced inflammatory bowel disease. Biochem Pharmacol. 2009;77:464–471. doi: 10.1016/j.bcp.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawauchi S., Nakamura T., Miki I., Inoue J., Hamaguchi T., Tanahashi T. Downregulation of CYP3A and P-glycoprotein in the secondary inflammatory response of mice with dextran sulfate sodium-induced colitis and its contribution to cyclosporine A blood concentrations. J Pharmacol Sci. 2014;124:180–191. doi: 10.1254/jphs.13141fp. [DOI] [PubMed] [Google Scholar]
- 24.Kusunoki Y., Ikarashi N., Hayakawa Y., Ishii M., Kon R., Ochiai W. Hepatic early inflammation induces downregulation of hepatic cytochrome P450 expression and metabolic activity in the dextran sulfate sodium-induced murine colitis. Eur J Pharm Biopharm. 2014;54:17–27. doi: 10.1016/j.ejps.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q.Y., Fang C., Zhang J., Dunbar D., Kaminsky L., Ding X. An intestinal epithelium-specific cytochrome P450 (P450) reductase-knockout mouse model: direct evidence for a role of intestinal P450s in first-pass clearance of oral nifedipine. Drug Metab Dispos. 2009;37:651–657. doi: 10.1124/dmd.108.025429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y., D'Agostino J., Zhang Q.Y. Role of intestinal cytochrome P450 (P450) in modulating the bioavailability of oral lovastatin: insights from studies on the intestinal epithelium-specific P450 reductase knockout mouse. Drug Metab Dispos. 2011;39:939–943. doi: 10.1124/dmd.110.037861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kung L., Batiuk T.D., Palomo-Pinon S., Noujaim J., Helms L.M., Halloran P.F. Tissue distribution of calcineurin and its sensitivity to inhibition by cyclosporine. Am J Transplant. 2001;1:325–333. doi: 10.1034/j.1600-6143.2001.10407.x. [DOI] [PubMed] [Google Scholar]
- 28.Fasco M.J., Silkworth J., Dunbar D.A., Kaminsky L.S. Rat small intestinal cytochromes P450 probed by warfarin metabolism. Mol Pharmacol. 1993;43:226–233. [PubMed] [Google Scholar]
- 29.Tanino T., Komada A., Ueda K., Bando T., Nojiri Y., Ueda Y. Pharmacokinetics and differential regulation of cytochrome P450 enzymes in type 1 allergic mice. Drug Metab Dispos. 2016;44:1950–1957. doi: 10.1124/dmd.116.072462. [DOI] [PubMed] [Google Scholar]
- 30.Polanco J.C., Scicluna B.J., Hill A.F., Götz J. Extracellular vesicles isolated from the brains of rTg4510 mice seed tau protein aggregation in a threshold-dependent manner. J Biol Chem. 2016;291:12445–12466. doi: 10.1074/jbc.M115.709485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Y., Zhang Q.Y. Role of intestinal cytochrome P450 enzymes in diclofenac-induced toxicity in the small intestine. J Pharmacol Exp Ther. 2012;343:362–370. doi: 10.1124/jpet.112.198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X.D., Li J.L., Lu Y., Chen X., Huang M., Chowbay B. Rapid and simultaneous determination of nifedipine and dehydronifedipine in human plasma by liquid chromatography–tandem mass spectrometry: application to a clinical herb–drug interaction study. J Chromatogr B. 2007;852:534–544. doi: 10.1016/j.jchromb.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 33.Fang Z.G., You B.G., Chen Y.G., Zhang J.K., Liu Y.Q., Zhang X.N. Analysis of cyclosporine A and its metabolites in rat urine and feces by liquid chromatography–tandem mass spectrometry. J Chromatogr B. 2010;878:1153–1162. doi: 10.1016/j.jchromb.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q.Y., Dunbar D., Kaminsky L.S. Characterization of mouse small intestinal cytochrome P450 expression. Drug Metab Dispos. 2003;31:1346–1351. doi: 10.1124/dmd.31.11.1346. [DOI] [PubMed] [Google Scholar]
- 35.Viennois E., Chen F., Laroui H., Baker M.T., Merlin D. Dextran sodium sulfate inhibits the activities of both polymerase and reverse transcriptase: lithium chloride purification, a rapid and efficient technique to purify RNA. BMC Res Notes. 2013;6:360. doi: 10.1186/1756-0500-6-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Agostino J., Ding X., Zhang P., Jia K., Fang C., Zhu Y. Potential biological functions of cytochrome P450 reductase-dependent enzymes in small intestine novel link to expression of major histocompatibility complex class II genes. J Biol Chem. 2012;287:17777–17788. doi: 10.1074/jbc.M112.354274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 41.Lichtiger S., Present D. Preliminary report: cyclosporin in treatment of severe active ulcerative colitis. Lancet. 1990;336:16–19. doi: 10.1016/0140-6736(90)91521-b. [DOI] [PubMed] [Google Scholar]
- 42.Lichtiger S., Present D.H., Kornbluth A., Gelernt I., Bauer J., Galler G. Cyclosporine in severe ulcerative colitis refractory to steroid therapy. N Engl J Med. 1994;330:1841–1845. doi: 10.1056/NEJM199406303302601. [DOI] [PubMed] [Google Scholar]
- 43.Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., Costantini F. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi G., Abbott K.N., Wu W., Salter R.D., Keyel P.A. Dnase1L3 regulates inflammasome-dependent cytokine secretion. Front Immunol. 2017;8:522. doi: 10.3389/fimmu.2017.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rendic S., Guengerich F.P. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol. 2014;28:38–42. doi: 10.1021/tx500444e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu N., Huang Y., Gao X., Li S., Yan Z., Wei B. Effects of dextran sulfate sodium induced experimental colitis on cytochrome P450 activities in rat liver, kidney and intestine. Chem Biol Interact. 2017;271:48–58. doi: 10.1016/j.cbi.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 47.Alex P., Zachos N.C., Nguyen T., Gonzales L., Chen T.E., Conklin L.S. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis. 2009;15:341–352. doi: 10.1002/ibd.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kusunoki Y., Ikarashi N., Matsuda S., Matsukawa Y., Kitaoka S., Kon R. Expression of hepatic cytochrome P450 in a mouse model of ulcerative colitis changes with pathological conditions. J Gastroenterol Hepatol. 2015;30:1618–1626. doi: 10.1111/jgh.12966. [DOI] [PubMed] [Google Scholar]
- 49.Wu C.Y., Benet L.Z. Predicting drug disposition via application of BCS: transport/absorption/elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm Res. 2005;22:11–23. doi: 10.1007/s11095-004-9004-4. [DOI] [PubMed] [Google Scholar]
- 50.Duggan D., Chen I., Bayne W., Halpin R., Duncan C., Schwartz M. The physiological disposition of lovastatin. Drug Metab Dispos. 1989;17:166–173. [PubMed] [Google Scholar]
- 51.Lodge J.W., Fletcher B.L., Brown S.S., Parham A.J., Fernando R.A., Collins B.J. Determination of lovastatin hydroxy acid in female B6C3F1 mouse serum. J Anal Toxicol. 2008;32:248–252. doi: 10.1093/jat/32.3.248. [DOI] [PubMed] [Google Scholar]
- 52.Ishigami M., Honda T., Takasaki W., Ikeda T., Komai T., Ito K. A comparison of the effects of 3-hydroxy-3-methylglutaryl-coenzyme a (HMG-CoA) reductase inhibitors on the CYP3A4-dependent oxidation of mexazolam in vitro. Drug Metab Dispos. 2001;29:282–288. [PubMed] [Google Scholar]
- 53.Neuvonen P.J., Jalava K.M. Itraconazole drastically increases plasma concentrations of lovastatin and lovastatin acid. Clin Pharmacol Ther. 1996;60:54–61. doi: 10.1016/S0009-9236(96)90167-8. [DOI] [PubMed] [Google Scholar]
- 54.Kitzmiller J.P., Mikulik E.B., Dauki A.M., Murkherjee C., Luzum J.A. Pharmacogenomics of statins: understanding susceptibility to adverse effects. Pharmgenomics Pers Med. 2016;9:97–106. doi: 10.2147/PGPM.S86013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benet L.Z., Broccatelli F., Oprea T.I. BDDCS applied to over 900 drugs. AAPS J. 2011;13:519–547. doi: 10.1208/s12248-011-9290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolars J.C., Awni W.M., Merion R.M., Watkins P.B. First-pass metabolism of cyclosporin by the gut. Lancet. 1991;338:1488–1490. doi: 10.1016/0140-6736(91)92302-i. [DOI] [PubMed] [Google Scholar]
- 57.Tjia J.F., Webber I.R., Back D.J. Cyclosporin metabolism by the gastrointestinal mucosa. Br J Clin Pharmacol. 1991;31:344–346. doi: 10.1111/j.1365-2125.1991.tb05540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pavlović N., Goločorbin-Kon S., Ðanić M., Stanimirov B., Al-Salami H., Stankov K. Bile acids and their derivatives as potential modifiers of drug release and pharmacokinetic profiles. Front Pharmacol. 2018;9:1283. doi: 10.3389/fphar.2018.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schiff J., Cole E., Cantarovich M. Therapeutic monitoring of calcineurin inhibitors for the nephrologist. Clin J Am Soc Nephrol. 2007;2:374–384. doi: 10.2215/CJN.03791106. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein C.N., Nabalamba A. Hospitalization-based major comorbidity of inflammatory bowel disease in Canada. Chin J Gastroenterol Hepatol. 2007;21:507–511. doi: 10.1155/2007/924257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.