Abstract
Mutations in genes encoding key players in oncogenic signaling pathways trigger specific downstream gene expression profiles in the respective tumor cell populations. While regulation of genes related to cell growth, survival, and death has been extensively studied, much less is known on the regulation of drug-metabolizing enzymes (DMEs) by oncogenic signaling. Here, a comprehensive review of the available literature is presented summarizing the impact of the most relevant genetic alterations in human and rodent liver tumors on the expression of DMEs with a focus on phases I and II of xenobiotic metabolism. Comparably few data are available with respect to DME regulation by p53-dependent signaling, telomerase expression or altered chromatin remodeling. By contrast, DME regulation by constitutive activation of oncogenic signaling via the RAS/RAF/mitogen-activated protein kinase (MAPK) cascade or via the canonical WNT/β-catenin signaling pathway has been analyzed in greater depth, demonstrating mostly positive-regulatory effects of WNT/β-catenin signaling and negative-regulatory effects of MAPK signaling. Mechanistic studies have revealed molecular interactions between oncogenic signaling and nuclear xeno-sensing receptors which underlie the observed alterations in DME expression in liver tumors. Observations of altered DME expression and inducibility in liver tumors with a specific gene expression profile may impact pharmacological treatment options.
Key words: Xenobiotic metabolism, Hepatocytes, WNT/β-catenin signaling, RAS/MAPK signaling, Gene mutation, Cytochrome P450
Graphical abstract
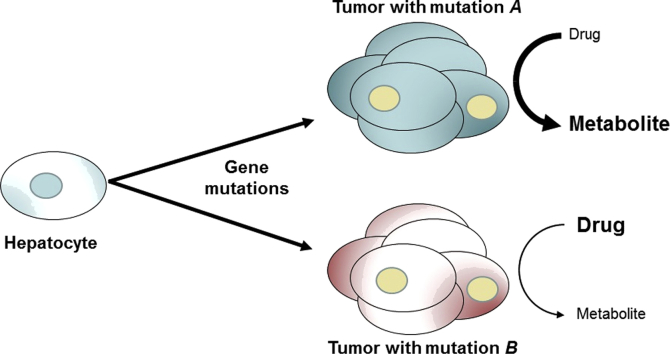
Mutations in key proto-oncogenes or tumor suppressor genes in hepatocellular carcinoma affect the expression of drug-metabolizing enzymes in a specific manner. This review highlights the connection between xenobiotic metabolism and genetic alterations in human or rodent liver tumors.
1. Introduction
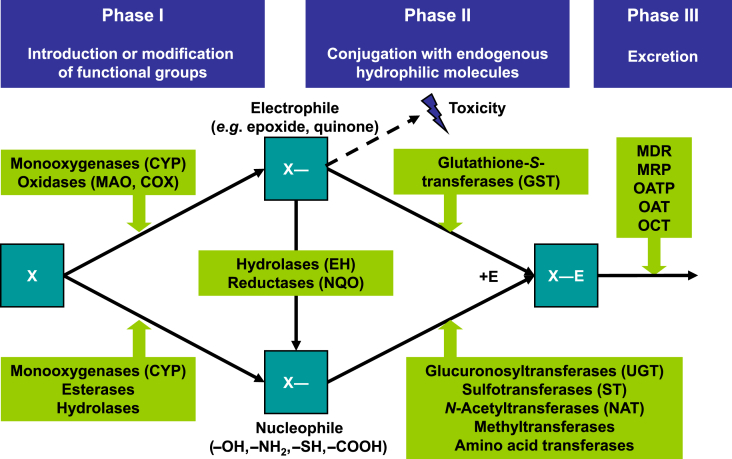
The group of drug-metabolizing enzymes (DMEs) consists of families of enzymes which are involved in the metabolic conversion of both endogenous and exogenous compounds. The latter are often referred to as “xenobiotics” and may comprise drugs, pesticides, herbicides, food additives, and many environmental chemicals of all kind. According to their primary function the underlying proteins are grouped into metabolic enzymes involved in the so-called phase I (target functionalization) and phase II (target conjugation with endogenous molecules) of xenobiotic metabolism, while functionally related transporters of phase 0 and phase III are responsible for the uptake of xenobiotics into cells, or for the active excretion of metabolites out of cells, respectively. For an overview of xenobiotic metabolism see Fig. 1. In mammals, the level and activity of DMEs is highest in the liver; however, many DMEs are also expressed in other organ systems such as for example the gastrointestinal tract. This review will concentrate on the liver and on enzymes of phases I and II.
Figure 1.
Overview of the phases and important enzymes and transporters of xenobiotic metabolism in hepatocytes. Functionalization in phase I is followed by conjugation to endogenous substrates in phase II and excretion in phase III of the biotransformation process. Abbreviations: COX, cyclooxygenase; CYP, cytochrome P450; EH, epoxide hydrolase; GST, glutathione-S-transferase; MAO, monoamine oxidase; MDR, multi-drug resistance protein; MRP, multi-drug resistance-related protein; NAT, N-acetyltransfease; NQO, NAD(P)H-quinone oxidoreductase; OAT, organic anion transporter; OATP, organic anion-transporting peptide; OCT, organic cation transporter; UGT, UDP-glucuronosyl-transferase.
A key group of proteins involved in phase I of xenobiotic metabolism are enzymes belonging to one of the various cytochrome P450 (CYP) subfamilies. In the early 70s of the last century, it was discovered that the content of some CYPs was decreased in experimentally induced hepatomas in rats as compared to normal liver1, 2. Present day omics analysis on global gene expression patterns demonstrates a similar decrease in CYP expression in human hepatocellular carcinoma (HCC); a comprehensive meta-analysis is available by use of the HCCDB database which is available at: www.lifeome.net/database/hccdb/home.html.3 Based on the observation of decreased DME-expression in liver tumors, it was postulated that preneoplastic and neoplastic cells are less sensitive to the toxic action of 2-acetylaminofluorene and other hepatocarcinogens or hepatotoxins which need metabolic activation of the parental compound to toxic intermediates by CYP enzymes1, 4. Based on this observation Farber's group developed the so-called Solt-Farber model which allows for rapid induction of neoplastic nodules in rat liver based on selective pressure given by 2-acetylaminofluorene on preneoplastically transformed hepatocytes produced by single injection of a strong hepatocarcinogen such as N-nitrosodiethylamine5. While potentially toxin-activating enzymes such as CYPs are generally decreased in hyperplastic nodules and hepatomas1, 6, preferentially detoxifying enzymes such as microsomal epoxide hydrolase (mEH), glutathione-S-transferases (GSTs) and UDP-glucuronosyltransferases (UGTs) were found to be increased in premalignant lesions7. This further confers a selective advantage to the preneoplastic and neoplastic liver cells and led to the “selective toxicity resistance” model postulated by Farber8 to be a generalized model for chemical hepatocarcinogenesis.
In a comprehensive immunohistochemical study, Buchmann et al.9 demonstrated that the decreases in phase I enzymes (shown for two phenobarbital (PB)-inducible and two 3-methylcholanthrene-inducible CYPs) along with increases in phase II enzymes (including cytosolic GSTs B and C, and mEH) occurred very early during the carcinogenic process in rat liver, presumably already during its initiation. Since individual lesions showed heterogeneity in DME expression and since some of the DMEs in the preneoplastic lesions were still inducible by PB, it was suggested that the focal enzyme alterations result from genotoxic effects of the carcinogen on “regulatory systems of a higher order” rather than from mutational events in individual genes encoding DMEs10. The nature of these higher order regulatory systems operative in the rat liver lesions was entirely unknown at the time and remained obscure during the following decades. Then, activating mutations in Ctnnb1, encoding β-catenin, were found to be present in about 30% of chemically induced rat liver tumors11 which corresponds in frequency to CTNNB1 mutations found in HCC12. Mutation of Ctnnb1 is associated with constitutive activation of the canonical WNT/β-catenin signaling pathway which, for reasons discussed later, is very unlikely to be responsible for down-regulation of CYP enzymes observed in the rat liver tumors. Mutations in one of the Ras oncogenes, frequently detected in mouse liver tumors, would in principle be much better candidates for reduction of CYP enzymes, but are very rarely present in rat and human liver tumors.
2. Species differences in mutational patterns of liver tumors
The genes most frequently affected by mutation in human12, rat11, 13, 14 and mouse15, 16 primary liver tumors do show some overlap but also divergence, as summarized in Table 1. The reasons for the observed species-specific differences in the mutational patterns of the driver genes affected are not known. However, part of it may be linked to differences in the etiology of the tumors: while rodent liver tumors were mostly experimentally induced by the use of known hepatocarcinogenic chemicals, the occurrence of liver tumors in humans is mostly linked to chronic hepatitis B and C virus infection, alcohol abuse and, to a minor part, to exposure to aflatoxins. In addition, species-specific differences in the biology underlying tumor manifestation and progression are likely to play a role. While primary liver tumors in humans reflect a very heterogeneous group of neoplasms with distinctive clinical and pathologic features17, liver tumors in mice are much more homogeneous in appearance. In the following, we will very briefly discuss effects produced by the mutational changes on cellular signaling pathways before we discuss their consequences for DME expression and drug metabolism in the affected tumor cells.
Table 1.
Genes frequently affected by mutation in human and rodent primary liver tumors.
| Human | Rat | Mouse |
|---|---|---|
| TERT | Nrf2/Keap1 | Hras |
| TP53 | Ctnnb1 | Braf |
| CTNNB1/AXIN1 | Tp53 | Ctnnb1a |
| ARID1A/2 | Egfr |
Ctnnb1 mutations specifically found after tumor promotion with PB-like compounds.
2.1. Human liver tumors
TERT (telomerase reverse-transcriptase, coding for the catalytic subunit of telomerase) promoter mutations are the most frequent genetic alterations found in human primary liver tumors and one of the earliest genomic events in human liver carcinogenesis12, 18. TERT promoter mutations are a common feature of human cancers and are predicted to increase promoter activity and TERT transcription. In fact, in contrast to normal liver, TERT activity is restored in over 90% of human HCCs investigated19. Interestingly, TERT promoter mutations are often found together with mutations in a second gene frequently mutated in human HCC, namely CTNNB118, 20, which encodes the oncoprotein β-catenin, a member of the WNT signaling pathway (see below). The available data suggest that TERT promoter mutations and activation of the WNT/β-catenin pathway cooperate in HCC progression in humans18. Underlying cause may be a recently discovered cross-talk between TERT and the WNT/β-catenin pathway, in which telomerase functions in a “non-canonical” fashion as a cofactor in the β-catenin transcriptional complex, as reviewed by Li and Tergaonkar21, resulting in activation of WNT/β-catenin-dependent transcription. Interestingly, this cofactor-function of TERT is mediated by BRG1, a protein of the SWI/SNF (SWItch/Sucrose Non-Fermentable) complex required for chromatin remodeling. Other SWI/SNF members include ARID1A and ARID2, as discussed below.
TP53 encodes the tumor suppressor protein p53, which has a key function in controlling, amongst others, the induction of senescence and apoptosis; for review see e.g. Hafner et al.22 or Mello and Attardi23. P53 is known to mediate cellular senescence, following e.g. the inappropriate activation of oncogenic signaling pathways, which explains why TP53 is frequently inactivated by mutation in HCC and many other human cancers. Among the various oncogenic pathways that may trigger a p53-senescence-inducing response is the WNT/β-catenin pathway, constitutively activated by mutation of CTNNB124.
CTNNB1 and its rodent ortholog Ctnnb1 encode β-catenin, a central player in the canonical WNT/β-catenin signaling pathway. Cytosolic levels of β-catenin are stringently regulated by a multi-protein complex, which mediates phosphorylation of the protein thus initiating its ubiquitinylation and subsequent degradation by the proteasome; for review see Lustig and Behrens25. Mutation of one of the phosphorylation sites leads to β-catenin accumulation followed by nuclear transfer, where it associates with transcription factors of the T-cell factor (TCF)/lymphoid enhancer factor family and induces target gene transcription. Part of the cytosolic β-catenin degradation complex is AXIN1, the gene of which is also mutated in a certain fraction of human HCC12.
ARID (AT-rich interactive domain-containing protein) 1A and 2 are both members of the ATP-dependent chromatin remodeling SWI/SNF complex, which is required for transcriptional activation of genes normally repressed by chromatin; for review see Savas and Skardasi26. ARID1A (also termed BAF250a) and ARID2 are frequently mutated in diverse human cancers including HCC12. Even though ARID1A and 2 are generally assumed to act as tumor suppressors, their role in HCC development is not entirely clear, since up-regulation of ARID1A expression is observed in a considerable number of HCC27. Deficiency in Arid1a in the respective knockout mouse induces steatohepatitis and HCC28. However, since ARID1A was highly expressed in the primary tumors but was lost in expression in metastatic HCC cases, it may promote carcinogenesis during the early phases of tumor initiation but suppress tumor progression in late-stage HCC29.
It is interesting to note that mutations in one of the oncogenic Ras genes, which are very frequently mutated in mouse liver tumors15, 30, are only very rarely observed in human and rat liver tumors31. The reason for this species difference is not known, but despite the lack of Ras mutations, activation of RAS-downstream mitogen-activated protein kinase (MAPK) signaling is observed in 50%–100% of human HCC and is associated with poor prognosis32.
Another interesting note is that many of the genes recurrently mutated in human HCC encode proteins that have a direct or indirect link to β-catenin, which plays a central role in regulation of DME expression in hepatocytes, as will be discussed in detail later.
2.2. Rat liver tumors
NRF2/KEAP1: Following the observation that in about 6%–8% of cases human HCCs harbor mutations in either the NFE2L2 gene encoding NRF2 [Nuclear factor (erythroid-derived 2)-like 2], or in KEAP1 which encodes the NRF2 inhibitor KEAP1 (Kelch-like ECH-associated protein 1), respectively33, 34, rat liver tumors were also screened for mutations in the two underlying genes13. In this study, preneoplastic lesions as well as tumors of differing stages were induced by a modified Solt-Farber protocol including 2-acetylaminofluorene as selective chemical. More than 70% of preneoplastic lesions and about 60%–80% of HCCs were found to be mutated in Nfe2l2 or, to a lesser extent, in Keap113. The mutations detected in Nfe2l2 or Keap1 all impair the binding between the two proteins and therefore attenuate the inhibitory activity of KEAP1 onto NRF2-mediated signaling. Therefore they should be considered as activating mutations. However, whether NRF2 plays a pro- or anti-tumorigenic role during the early phases of the malignant process is unclear35.
Ctnnb1 mutations are detected in about 20%–30% of chemically induced rat liver tumors11, 13. Since no such mutations were detected in preneoplastic lesions or early HCC, Ctnnb1 mutations are likely associated with a late stage in malignant progression in rat hepatocarcinogenesis13.
Tp53 mutations leading to inactivation of the protein as transcription factor have been described to occur in about 30%–40% of chemically induced rat liver tumors, both in pre-cancerous and cancerous lesions14, 36.
2.3. Mouse liver tumors
Hras mutations leading to the constitutive activation of the HA-RAS/p21 oncoprotein are very frequent in both, spontaneous and chemically-induced mouse liver tumors15, 37, 38. HA-RAS is a monomeric G protein which forwards mitogenic signals received by growth factor receptors to a cascade of downstream kinases, as reviewed by Sun and coworkers39. Interestingly, the frequency of occurrence is dependent on the susceptibility of spontaneous liver tumor development and susceptibility towards chemical induction of liver tumors which differs considerably between different mouse strains: susceptible strains show a high prevalence of Hras mutations in their liver tumors while the prevalence of such mutations is much lower in resistant strains38.
Braf encodes a signaling protein directly downstream of HA-RAS. While Hras is predominantly mutated in mouse liver tumors from susceptible strains, the prevalence of Braf mutations is higher in resistant strains such as C57BL40. Together, mutations in either Hras or Braf are observed in more than 70% of mouse liver tumors40. Very likely, they occur already during initiation of the carcinogenic process41.
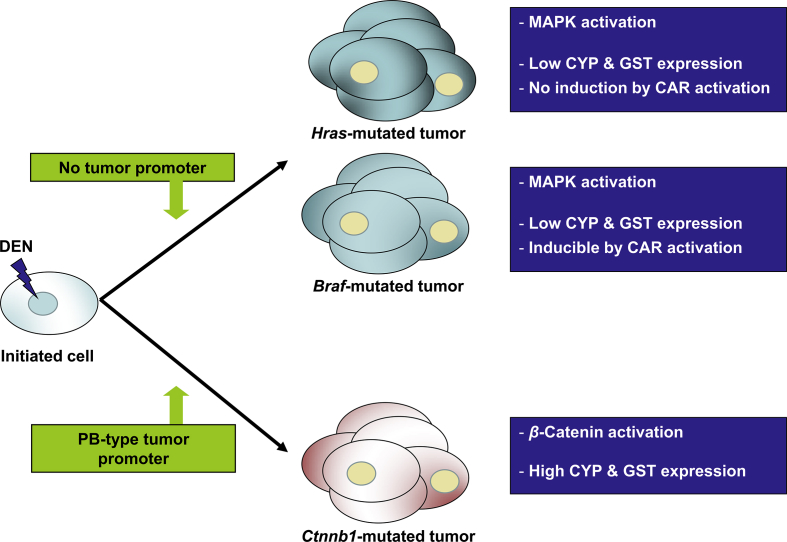
Ctnnb1 mutations are the most prominent type of genetic lesion in mouse liver tumors, occurring in more than 80% of cases. However, this only applies to tumors induced by a regimen including PB or a PB-like agent as tumor promoter16, 42; see also Fig. 2. By contrast, mouse liver tumors chemically induced under a protocol without PB-mediated tumor promotion are often Hras- or Braf-, but not Ctnnb1-mutated16, 30. This finding strongly suggests that in mouse liver PB or similarly acting agents select for hepatocytes mutated in Ctnnb1.
Figure 2.
DME characteristics of chemically induced mouse liver tumors with activating mutations in the Ctnnb1, Hras, or Braf proto-oncogenes. Spontaneous tumors or tumors induced by application of the genotoxic tumor initiator N-nitrosodiethylamine (DEN) mostly leads to tumors with activated MAPK signaling due to mutations in Hras or Braf. By contrast, chronic treatment with the tumor promoter phenobarbital (PB) or similarly acting compounds leads to the outgrowth of liver tumors with activated β-catenin due to activating Ctnnb1 mutations. Tumors with Hras and Braf mutations generally express low basal levels of DMEs (esp. CYPs and GSTs). In contrast to Hras-mutated tumors which are refractory to DME induction by constitutive androstane receptor (CAR) agonists, mouse liver tumors with Braf mutations respond to CAR activation with CYP and GST induction. Hepatomas with activated β-catenin display high constitutive expression of DMEs. For more details, please refer to the main text.
Interestingly, mutations in TP53/Tp53, which are very frequent in both human and rat primary liver tumors, are very rare in mouse liver tumors but occur frequently in cell lines established from the mouse liver tumors43. This evidence suggests that mutational inactivation of the murine p53 tumor suppressor does not confer a selective advantage to the mutated tumor cells. The same seems to be true for the human ortholog TP53 which, when introduced as transgene into mouse hepatocytes in vivo, is also not found inactivated by mutation in liver tumors experimentally induced in the transgenic mice44.
3. Observations on DME expression in liver tumors with specific mutational patterns
3.1. Telomerase-activation and DME expression
To the best of our knowledge, no studies about the regulation of DME expression or activity in liver tumor cells by the promoter-mutated TERT protein have been published so far. Immortalization of human fetal hepatocytes by over-expression of telomerase resulted in some changes to CYP expression, with diminished levels of CYP1A1, CYP2C9, CYP2E1, and CYP3A4, but elevated levels of CYP2B45. In addition, indirect effects through modulation of β-catenin-dependent gene expression programs appears theoretically possible, since TERT affects transcription of WNT/β-catenin target genes through BRG1-mediated interaction with β-catenin/TCF at WNT-responsive gene promoters (for review see Li and Tergaonkar21). Effects of β-catenin on DME expression are discussed below.
3.2. Inactivation of p53 tumor suppressor function and DME expression
To the best of our knowledge, no systematic comparisons of DME expression in TP53/Tp53 wildtype and mutant human or rodent liver tumors are available. However, in vitro evidence suggests a possible role of the p53 tumor suppressor protein in the regulation of some DMEs: a study with human liver tumor cells revealed an induction of various CYPs from families 1–3, including the important isoform CYP3A4, by p5346. Similarly, loss of p53 in mice resulted in decreased metabolism of a CYP3A substrate47.
Mechanistically it appears plausible that interactions of p53 and DMEs are mediated by the transcription factor activity of p53, as has been shown for example for the human CYP2A6 promoter in human liver tumor cells in vitro48. Additional evidence suggests interactions with signaling via nuclear receptors regulating DMEs: inhibition of pregnane-X-receptor (PXR), the prototype CYP3A4-inducing nuclear receptor, by p53 has been reported49. This finding appears to contrast the above observations of p53-dependently increased CYP3A4 expression. An inhibition of the AHR and its target gene CYP1A1 by p53 has also been described, even though not in liver cells50, 51. Thus, more research is needed to clarify the interplay of p53, nuclear receptors and DMEs under varying conditions in different cell types.
3.3. Alterations in chromatin remodeling and DME expression
ARID1A, a member of the SWI/SNF chromatin remodeling complex is often overexpressed in early HCC, while being downregulated in metastatic cancer. In mice, overexpression of ARID1A has been demonstrated to be associated with increased expression of several CYP isoforms including Cyp2e129. Potentially, this increase in CYP expression in the tumor cells may promote the generation of reactive oxygen species mediating liver injury and hepatocarcinogenesis29.
3.4. NRF2-mediated changes in DME expression
NRF2 and KEAP1 are central within a redox-sensitive signaling system that regulates up to 10% of human genes52. Well-known target genes include those encoding reactive oxygen- or electrophiles-inactivating enzymes such as NQO1 [NAD(P)H:quinone oxidoreductase 1], heme oxygenase-1, GSTs, UGTs, and multidrug resistance-associated proteins53. Keap1 knockdown mice showed an increase in NRF2 protein in liver and increases in the expression of NQO1 and GSTs54. Interestingly, there exists an intimate cross-interaction between NRF2- and aryl hydrocarbon receptor (AHR)-dependent signaling pathways (for review see Kohle and Bock55): murine Nfe2l2 is a target gene of the AHR56, while Ahr, on the other hand, is a transcriptional target of NRF257. Therefore, the expression of Ahr and some of its downstream targets, such as Cyp1a1, Cyp1b1 and Gsta1 are higher in expression in Keap1 knockout cells57.
The level of expression of CYPs was not determined in Nfe2l2/Keap1-mutated rat liver tumors, but the NRF2 target genes Nqo1 and Gsta4 were evaluated and found to be increased13. Therefore constitutive activation of NRF2 signaling in Nfe2l2/Keap1-mutated rat liver tumors might potentially explain the observed upregulation of (some) phase II enzymes including GSTs and UGTs. It does not explain, however, why phase I enzymes including CYPs are reduced in expression in these tumors. Rather, one would expect an increase in expression of e.g. CYP1A isoforms, which was not observed in any of in those studies where CYP expression was analyzed. Therefore, other “higher order regulators” must play a role in the regulation of these enzymes in rat liver tumors.
3.5. Activation of the WNT/β-catenin signaling pathway and associated changes in DME expression
In 2005 our group was, to the best of our knowledge, the first to report on the positive regulatory activity of WNT/β-catenin signaling on the expression of CYP enzymes in liver cells58; see also Fig. 2. This conclusion was based on the observation that mouse liver tumors harboring activating mutations in the Ctnnb1 gene, encoding β-catenin, showed higher levels of several CYP isoforms (CYP1A, CYP2B, CYP2C and CYP2E1 proteins), while Ctnnb1 wildtype tumors exhibited decreased levels of these CYP isoforms58. The increase in CYP protein level corresponded to increases in the respective mRNAs indicating that mutation of Ctnnb1 leads to transcriptional activation of a number of CYP isoforms in mouse liver tumors. In the initial studies, the Ctnnb1-mutated tumors in mouse liver were generated by a sequential initiation-promotion regimen: for tumor initiation, mice were first treated with a single dose of N-nitrosodiethylamine, which is converted in hepatocytes to an electrophilic DNA-reactive mutagen; this was then followed by chronic treatment with PB acting as a tumor promoter. PB, however, is also a potent DME inducer. Increased expression of CYPs and other DMEs could therefore, in principle, also result from inducing effects of PB in the Ctnnb1-mutated mouse liver tumors. Later studies, however, confirmed that Ctnnb1-mutated mouse liver tumors show increased DME expression, even after PB had been withdrawn, demonstrating that activated β-catenin by itself is sufficient to drive DME expression in mouse liver (unpublished observation). This was substantiated in a transgenic mouse strain with hepatocyte-specific expression of a point-mutated, constitutively active version of β-catenin, where strongly elevated CYP levels were seen even in periportal hepatocytes, where these enzymes normally are not expressed59.
In wildtype liver, CYPs and other important DMEs are preferentially expressed in perivenous hepatocytes60 which also display physiological activation of the WNT/β-catenin pathway61, 62. It was demonstrated that the preferential perivenous expression of DMEs in mouse liver is regulated by WNT/β-catenin-activating signals derived from the endothelial cells of the central veins63, 64. In line with the aforementioned observations, results from studies conducted by several different groups including ours demonstrated that various CYP isoforms, especially CYP2E1 and CYP1A, are no longer expressed at the mRNA and protein level in livers of mice with conditional hepatocyte-specific knockdown of Ctnnb165, 66. Similarly, down-regulation of a number of GSTs from phase II of xenobiotic metabolism67 and of enzymes engaged in the synthesis of the CYP prosthetic group heme68 were observed in that mouse model. Experiments with xenobiotic inducers of DMEs demonstrated that the knockout of Ctnnb1 resulted in diminished DME induction following exposure to xenobiotics acting via activation of the receptors CAR or AHR66, 69. These results clearly demonstrate that WNT/β-catenin signaling is a key player in regulating CYP expression in Ctnnb1-mutated mouse hepatocytes.
Less is known about human liver tumors. In human hepatoblastoma, a pediatric tumor very frequently mutated in CTNNB1, the human ortholog of mouse Ctnnb1, up-regulation of various CYP isoforms (in particular CYP2E1) was observed in the epithelial parts of the tumors70. By contrast, most CYPs are generally down-regulated in human HCCs71, 72. Of note, in vitro analyses with human HepaRG hepatocarcinoma cells and primary human hepatocytes demonstrated transcriptional regulation of a number of CYPs, especially CYP2E1, by WNT/β-catenin signaling73, 74. In a study from our group, Ctnnb1-mutated and Ctnnb1-wildtype mouse liver tumors were analyzed in parallel with human CTNNB1-mutated or -wildtype HCCs75: glutamine synthetase, a biomarker for increased β-catenin-mediated signaling, and various CYP isoforms were increased in expression in the Ctnnb1-mutated tumors from mice as compared to the surrounding normal liver tissue75. By contrast, while glutamine synthetase was also over-expressed in the CTNNB1-mutated human HCCs, all CYP isoforms investigated were lower in expression in the tumors when compared to normal liver (unpublished observation). However, when CYP expression levels in the CTNNB1-mutated HCCs were compared to those in the corresponding CTNNB1-wildtype tumors, higher expression levels were detected in CTNNB1-mutated tumors (unpublished observation). Interestingly, CTNNB1 mutations were only seen in HCCs associated with hepatitis C virus infection but not in those associated with hepatitis B virus75. This preference of CTNNB1 mutations in HCC with hepatitis C virus association has also been observed by others (e.g. see Pezzuto et al.20 or Tornesello et al.76). This is of interest, since other studies have reported higher expression of CYPs, in particular CYP2E1, in human hepatitis C virus—as compared to hepatitis B virus-associated HCCs77, 78.
3.6. Effects on DME expression upon activation of Ras-downstream signaling
Hras mutations are frequent in spontaneous and chemically-induced mouse liver tumors, particularly in those strains showing a high background prevalence of liver tumor formation38. However, mutations in Hras or one of the other oncogenic Ras genes, Kras or Nras, are only very rarely observed in rat or human liver tumors31. Nonetheless, frequently detected activation of RAS downstream kinases in human HCC32 argues for a relevance of activation of this signaling pathway also in human liver tumors.
With regard to DME expression, the situation in Hras-mutated mouse liver tumors is quite clear: many important enzymes from phase I and II are strongly down-regulated in expression in these tumors at the mRNA and protein levels (Fig. 2). This phenomenon is similarly observed in mouse liver tumors with mutations in Braf, which are indiscernible from their Hras-mutated cousins in terms of global transcriptomic or proteomic expression patterns75, 79, 80. Nonetheless, it has been observed that Hras-mutated mouse liver tumors are refractory to DME induction via activation of the constitutive androstane receptor (CAR), whereas Braf-mutated tumors responded to the presence of the CAR activator PB with pronounced induction of CYPs and GSTs81. Studies with transgenic mice expressing a mutationally activated human HRAS oncogene in some hepatocytes show that the perivenous gene expression profile, including the expression of important DMEs, is abolished in liver cells with activated HA-RAS68, 82.
4. In vitro studies and mechanistic considerations
4.1. WNT/β-catenin signaling
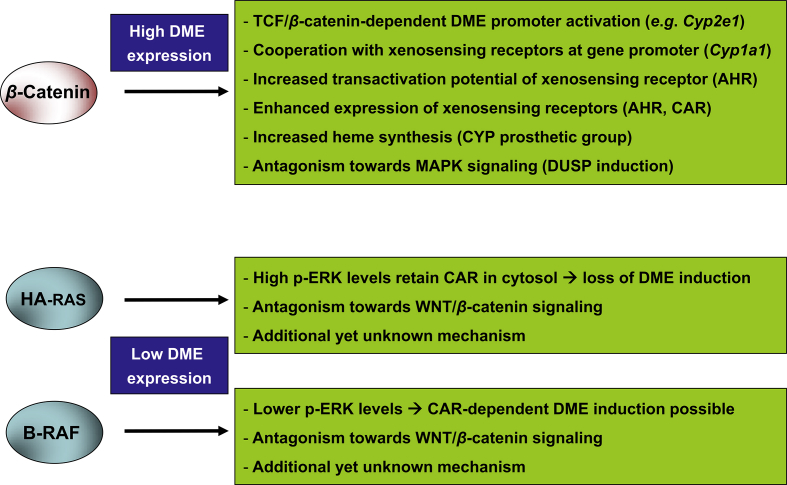
A number of molecular mechanisms have been identified by which signaling through the WNT/β-catenin pathway regulates the transcription of different DMEs (Fig. 3). First, the xeno-sensing receptors CAR and AHR are transcriptionally regulated by the β-catenin pathway66, 69, 83, 84, 85, 86. This way, activated β-catenin may contribute to elevated levels of DME-regulating receptors. Nonetheless, in vitro analyses suggest that AHR up-regulation by β-catenin activation might not be crucial for the observed effects of β-catenin signaling on the expression of the model AHR target gene Cyp1a187. Second, direct transcriptional activation of DMEs by the β-catenin/TCF transcription factor complex has been demonstrated by in vitro gene promoter analyses, for example in case of murine Cyp2e1 and human CYP1A169, 87, 88. Cyp2e1 and Cyp1a2 promoter occupancy by β-catenin/TCF has also been confirmed in mice in vivo86, 88. Third, there is in vitro evidence for a cooperative behavior of β-catenin/TCF and the AHR at the human CYP1A1 promoter, where specific binding sites for these transcription factors are located in close proximity69, 87, 89. Similarly, a cooperation of transcription factor binding sites for hepatocyte nuclear factor 1 alpha (HNF1α) and β-catenin/TCF has been shown for the mouse Cyp2e1 promoter88. Fourth, β-catenin enhances the transcriptional activity of the AHR at its binding sites at the DNA87. The molecular basis of the decreased response of Ctnnb1 knockout hepatocytes to CAR activators66, 85, 90 remains to be studied. In summary, the β-catenin pathway constitutes a master positive regulator of DME expression in hepatocytes, acting through a variety of different molecular mechanisms, especially via a complex interplay with xenobiotic-sensing nuclear receptors; for review see also Braeuning and Schwarz91 or Braeuning92. Moreover, there is evidence that β-catenin activation in mouse liver is able to suppress the DME-repressing signaling program orchestrated by signaling through the MAPK cascade via the induction of dual-specificity phosphatases (DUSPs), negative regulators of the RAS/MAPK pathway93, 94.
Figure 3.
Mechanistic aspects of DME regulation in mouse liver tumors with mutations in either the Ctnnb1, Hras, or Braf oncogene. Signaling through β-catenin affects DME gene expression by different mechanisms involving β-catenin/TCF-dependent promoter activation and various ways of cooperation with nuclear xeno-sensing receptors, e.g., AHR and CAR. Moreover, synthesis of the CYP prosthetic group heme is augmented by β-catenin signaling in hepatocytes. Tumors with mutationally activated HA-RAS harbor high levels of phosphorylated, active extracellular signal-regulated kinase (ERK) 1/2. Phosphorylated ERK retains the constitutive androstane receptor (CAR) in the cytosol to inhibit DME induction by CAR agonists. This phenomenon is not observed in tumors with mutationally activated B-RAF, where ERK phosphorylation is much less pronounced. Antagonistic action of the DME-inhibiting mitogen-activated protein kinase (MAPK) pathway and the DME-inducing β-catenin pathway have been described, for example via the induction of dual-specificity phosphatases (DUSP) by the β-catenin pathway. For more details, please refer to the main text.
4.2. RAS/RAF/MAPK-dependent signaling
Much less is known about the molecular mechanisms by which activation of the MAPK signaling pathway due to mutations in Hras or Braf is able to suppress DME expression (Fig. 3). Observations from mouse liver tumors growing directly next to a branch of the hepatic central vein show that the perivenous gene expression program, including DME expression, which is normally activated in perivenous hepatocytes in close contact to the venous endothelial cells, is not getting activated in Hras-mutated tumor cells82.
This indicates that MAPK-dependent signaling has the ability to block the activation of the β-catenin pathway. Similarly, perivenous-specific gene expression is abolished in hepatocytes from a transgenic mouse model expressing constitutively active HRAS in a fraction of perivenous hepatocytes82. This indicates that MAPK signaling is able to suppress activation of the DME expression-promoting β-catenin signaling pathway94. Based on the antagonistic behavior of both signaling pathways, it has been proposed that gradients of MAPK- and β-catenin-dependent signaling regulate DME expression along the porto–central axis in healthy liver84, 91. Down-regulation of DMEs in liver is also observed under conditions of chronic inflammation95. In this case, suppression of DME expression is mediated by release of inflammatory cytokines which act on receptors located e.g. upstream of the RAS/MAPK pathway. Overexpression of the oncoprotein C-MYC, often seen in human HCC, may also be part of cytokine-mediated DME-repression in liver96, and may also explain down-regulation of DME expression in HCC. Furthermore, interference of interleukins with drug-metabolizing enzymes has been observed, which is based on an inhibition of the retinoid X receptor, the dimerization partner of several nuclear receptors involved in DME regulation97, 98, 99.
A very interesting observation is the fact that Hras- and Braf-mutated mouse liver tumors, even though highly similar with respect to their basal gene expression levels79, 80, behave strikingly different when exposed to PB, a model xenobiotic inducer of CAR-dependent gene expression: transcriptional induction of various CYPs and GSTs is observed in Braf-mutated tumors, but not or only to a very limited degree in their Hras-mutated cousins81. The underlying reason of the latter phenomenon is likely to be the difference in the activating phosphorylation of extracellular signal-regulated kinase (ERK) 1/2, an important kinase within the MAPK cascade: ERK phosphorylation is detected at very high levels in Hras-mutated tumors, whereas the degree of EKR phosphorylation is considerably lower in Braf-mutated tumors81. It has been previously reported that CAR-mediated functions are counteracted by ERK activation, as the phosphorylated kinase retains the receptor in the cytosol thus counteracting its transcriptional activity100. Thus, the different levels of ERK phosphorylation may explain the differences between the two tumors types when exposed to a CAR activator82.
5. Conclusions
Mutations in key proto-oncogenes or tumor suppressor genes trigger specific downstream gene expression profiles in the respective tumor cell populations, with important DMEs being part of the gene batteries regulated by oncogenic signaling. Our synopsis of published literature demonstrates that still a lot of research is needed to fully understand the mechanisms by which oncogenic signaling pathways affect the expression of DMEs in human liver tumors. Most information is available with respect to the oncogenic WNT/β-catenin and MAPK-dependent signaling cascades, which show largely opposing effects on DME expression. Several molecular mechanisms, especially interactions with nuclear xeno-sensing receptors, have been identified by which oncogenic signaling can affect DMEs at the transcriptional level. Many anticancer drugs, including novel targeted therapeutics, are subject to metabolism by DMEs. Thus knowledge on the connection of oncogenic signaling and DME expression may provide information relevant for tumor therapy. For example, it has been demonstrated that the WNT/β-catenin-dependent up-regulation of Cyp2e1 in chemically induced mouse liver adenomas renders these tumors susceptible to selective poisoning with acetaminophen101. Future research will show to which degree oncogene-induced changes in DME expression may be utilized for the optimization of anti-neoplastic therapy.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.06.013.
Author contributions
Both authors, Michael Schwarz and Albert Braeuning, have jointly written the paper based on an intense literature survey conducted by both authors. Both authors have seen and approved the final article.
Conflict of interest
Authors declare absence of conflict of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cameron R., Sweeney G.D., Jones K., Lee G., Farber E. A relative deficiency of cytochrome P-450 and aryl hydrocarbon [benzo(a)pyrene] hydroxylase in hyperplastic nodules induced by 2-acetylaminofluorene in rat liver. Cancer Res. 1976;36:3888–3893. [PubMed] [Google Scholar]
- 2.Hagihara B., Sato N., Fukuhara T., Tsutsumi K., Oyanagui Y. Spectrophotometric analysis of cytochromes in Morris hepatomas. Cancer Res. 1973;33:2947–2953. [PubMed] [Google Scholar]
- 3.Lian Q., Wang S., Zhang G., Wang D., Luo G., Tang J. HCCDB: a database of hepatocellular carcrinoma expression atlas. Genom Proteom Bioinform. 2018;16:269–275. doi: 10.1016/j.gpb.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farber E., Parker S., Gruenstein M. The resistance of putative premalignant liver cell populations, hyperplastic nodules, to the acute cytotoxic effects of some hepatocarcinogens. Cancer Res. 1976;36:3879–3887. [PubMed] [Google Scholar]
- 5.Solt D., Farber E. New principle for the analysis of chemical carcinogenesis. Nature. 1976;263:701–703. [Google Scholar]
- 6.Okita K., Noda K., Fukumoto Y., Takemoto T. Cytochrome P-450 in hyperplastic liver nodules during hepatocarcinogenesis with N-2-fluorenylacetamide in rats. Gan. 1976;67:899–902. [PubMed] [Google Scholar]
- 7.Astrom A., DePierre J.W., Eriksson L. Characterization of drug-metabolizing systems in hyperplastic nodules from the livers of rats receiving 2-acetylaminofluorene in their diet. Carcinogenesis. 1983;4:577–581. doi: 10.1093/carcin/4.5.577. [DOI] [PubMed] [Google Scholar]
- 8.Farber E. The biochemistry of preneoplastic liver: a common metabolic pattern in hepatocyte nodules. Can J Biochem Cell Biol. 1984;62:486–494. doi: 10.1139/o84-066. [DOI] [PubMed] [Google Scholar]
- 9.Buchmann A., Kuhlmann W., Schwarz M., Kunz W., Wolf C.R., Moll E. Regulation and expression of four cytochrome P-450 isoenzymes, NADPH-cytochrome P-450 reductase, the glutathione transferases B and C and microsomal epoxide hydrolase in preneoplastic and neoplastic lesions in rat liver. Carcinogenesis. 1985;6:513–521. doi: 10.1093/carcin/6.4.513. [DOI] [PubMed] [Google Scholar]
- 10.Kunz H.W., Buchmann A., Schwarz M., Schmitt R., Kuhlmann W.D., Wolf C.R. Expression and inducibility of drug-metabolizing enzymes in preneoplastic and neoplastic lesions of rat liver during nitrosamine-induced hepatocarcinogenesis. Arch Toxicol. 1987;60:198–203. doi: 10.1007/BF00296980. [DOI] [PubMed] [Google Scholar]
- 11.Yamada Y., Yoshimi N., Sugie S., Suzui M., Matsunaga K., Kawabata K. Beta-catenin (Ctnnb1) gene mutations in diethylnitrosamine (DEN)-induced liver tumors in male F344 rats. Jpn J Cancer Res. 1999;90:824–828. doi: 10.1111/j.1349-7006.1999.tb00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.S. The mutational landscape of hepatocellular carcinoma. Clin Mol Hepatol. 2015;21:220–229. doi: 10.3350/cmh.2015.21.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zavattari P., Perra A., Menegon S., Kowalik M.A., Petrelli A., Angioni M.M. Nrf2, but not beta-catenin, mutation represents an early event in rat hepatocarcinogenesis. Hepatology. 2015;62:851–862. doi: 10.1002/hep.27790. [DOI] [PubMed] [Google Scholar]
- 14.Lee C.C., Liu J.Y., Lin J.K., Chu J.S., Shew J.Y. p53 point mutation enhanced by hepatic regeneration in aflatoxin B1-induced rat liver tumors and preneoplastic lesions. Cancer Lett. 1998;125:1–7. doi: 10.1016/s0304-3835(97)00415-1. [DOI] [PubMed] [Google Scholar]
- 15.Connor F., Rayner T.F., Aitken S.J., Feig C., Lukk M., Santoyo-Lopez J. Mutational landscape of a chemically-induced mouse model of liver cancer. J Hepatol. 2018;69:840–850. doi: 10.1016/j.jhep.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aydinlik H., Nguyen T.D., Moennikes O., Buchmann A., Schwarz M. Selective pressure during tumor promotion by phenobarbital leads to clonal outgrowth of beta-catenin-mutated mouse liver tumors. Oncogene. 2001;20:7812–7816. doi: 10.1038/sj.onc.1204982. [DOI] [PubMed] [Google Scholar]
- 17.Jiang K., Al-Diffhala S., Cenetno B.A. Primary liver cancers-part 1: histopathology, differential diagnoses, and risk stratification. Cancer Control. 2018;25 doi: 10.1177/1073274817744625. 1073274817744625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nault J.C., Mallet M., Pilati C., Calderaro J., Bioulac-Sage P., Laurent C. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C.M., Hsu C.Y., Eng H.L., Huang W.S., Lu S.N., Changchien C.S. Telomerase activity and telomerase catalytic subunit in hepatocellular carcinoma. Hepatogastroenterol. 2004;51:796–800. [PubMed] [Google Scholar]
- 20.Pezzuto F., Izzo F., Buonaguro L., Annunziata C., Tatangelo F., Botti G. Tumor specific mutations in TERT promoter and CTNNB1 gene in hepatitis B and hepatitis C related hepatocellular carcinoma. Oncotarget. 2016;7:54253–54262. doi: 10.18632/oncotarget.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Tergaonkar V. Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res. 2014;74:1639–1644. doi: 10.1158/0008-5472.CAN-13-3568. [DOI] [PubMed] [Google Scholar]
- 22.Hafner A., Bulyk M.L., Jambhekar A., Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20:199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 23.Mello S.S., Attardi L.D. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damalas A., Kahan S., Shtutman M., Ben-Ze'ev A., Oren M. Deregulated beta-catenin induces a p53- and ARF-dependent growth arrest and cooperates with Ras in transformation. EMBO J. 2001;20:4912–4922. doi: 10.1093/emboj/20.17.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lustig B., Behrens J. The Wnt signaling pathway and its role in tumor development. J Cancer Res Clin Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PubMed] [Google Scholar]
- 26.Savas S., Skardasi G. The SWI/SNF complex subunit genes: their functions, variations, and links to risk and survival outcomes in human cancers. Crit Rev Oncol Hematol. 2018;123:114–131. doi: 10.1016/j.critrevonc.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J., Chen J., Lin H., Jin R., Liu J., Liu X. The clinicopathologic significance of BAF250a (ARID1A) expression in hepatocellular carcinoma. Pathol Oncol Res. 2016;22:453–459. doi: 10.1007/s12253-015-0022-9. [DOI] [PubMed] [Google Scholar]
- 28.Fang J.Z., Li C., Liu X.Y., Hu T.T., Fan Z.S., Han Z.G. Hepatocyte-specific Arid1a deficiency initiates mouse steatohepatitis and hepatocellular carcinoma. PLoS One. 2015;10 doi: 10.1371/journal.pone.0143042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X., Wang S.C., Wei Y., Luo X., Jia Y., Li L. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell. 2017;32:574–589. doi: 10.1016/j.ccell.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaworski M., Buchmann A., Bauer P., Riess O., Schwarz M. B-raf and Ha-ras mutations in chemically induced mouse liver tumors. Oncogene. 2005;24:1290–1295. doi: 10.1038/sj.onc.1208265. [DOI] [PubMed] [Google Scholar]
- 31.Tsuda H., Hirohashi S., Shimosato Y., Ino Y., Yoshida T., Terada M. Low incidence of point mutation of c-Ki-ras and N-ras oncogenes in human hepatocellular carcinoma. Jpn J Cancer Res. 1989;80:196–199. doi: 10.1111/j.1349-7006.1989.tb02290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delire B., Starkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Investig. 2015;45:609–623. doi: 10.1111/eci.12441. [DOI] [PubMed] [Google Scholar]
- 33.Cleary S.P., Jeck W.R., Zhao X., Chen K., Selitsky S.R., Savich G.L. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013;58:1693–1702. doi: 10.1002/hep.26540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I.B. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sporn M.B., Liby K.T. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Y., Deng W., Kawarada Y., Kawagoe M., Ma Y.Z., Li X. Mutation and expression of the p53 gene during chemical hepatocarcinogenesis in F344 rats. Biochim Biophys Acta. 2003;1628:40–49. doi: 10.1016/s0167-4781(03)00113-1. [DOI] [PubMed] [Google Scholar]
- 37.Jackson M.A., Lea I., Rashid A., Peddada S.D., Dunnick J.K. Genetic alterations in cancer knowledge system: analysis of gene mutations in mouse and human liver and lung tumors. Toxicol Sci. 2006;90:400–418. doi: 10.1093/toxsci/kfj101. [DOI] [PubMed] [Google Scholar]
- 38.Buchmann A., Bauer-Hofmann R., Mahr J., Drinkwater N.R., Luz A., Schwarz M. Mutational activation of the c-Ha-ras gene in liver tumors of different rodent strains: correlation with susceptibility to hepatocarcinogenesis. Proc Natl Acad Sci U S A. 1991;88:911–915. doi: 10.1073/pnas.88.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y., Liu W.Z., Liu T., Feng X., Yang N., Zhou H.F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600–604. doi: 10.3109/10799893.2015.1030412. [DOI] [PubMed] [Google Scholar]
- 40.Buchmann A., Karcier Z., Schmid B., Strathmann J., Schwarz M. Differential selection for B-raf and Ha-ras mutated liver tumors in mice with high and low susceptibility to hepatocarcinogenesis. Mutat Res. 2008;638:66–74. doi: 10.1016/j.mrfmmm.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Bauer-Hofmann R., Kress S., Schwarz M. Identification of point mutations at codon 61 of the c-Ha-ras gene by single-strand conformation polymorphism analysis. Biotechniques. 1992;13:192–194. [PubMed] [Google Scholar]
- 42.Strathmann J., Schwarz M., Tharappel J.C., Glauert H.P., Spear B.T., Robertson L.W. PCB 153, a non-dioxin-like tumor promoter, selects for beta-catenin (Catnb)-mutated mouse liver tumors. Toxicol Sci. 2006;93:34–40. doi: 10.1093/toxsci/kfl041. [DOI] [PubMed] [Google Scholar]
- 43.Kress S., Konig J., Schweizer J., Lohrke H., Bauer-Hofmann R., Schwarz M. p53 mutations are absent from carcinogen-induced mouse liver tumors but occur in cell lines established from these tumors. Mol Carcinog. 1992;6:148–158. doi: 10.1002/mc.2940060210. [DOI] [PubMed] [Google Scholar]
- 44.Jaworski M., Hailfinger S., Buchmann A., Hergenhahn M., Hollstein M., Ittrich C. Human p53 knock-in (hupki) mice do not differ in liver tumor response from their counterparts with murine p53. Carcinogenesis. 2005;26:1829–1834. doi: 10.1093/carcin/bgi142. [DOI] [PubMed] [Google Scholar]
- 45.Wege H., Le H.T., Chui M.S., Liu L., Wu J., Giri R. Telomerase reconstitution immortalizes human fetal hepatocytes without disrupting their differentiation potential. Gastroenterology. 2003;124:432–444. doi: 10.1053/gast.2003.50064. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein I., Rivlin N., Shoshana O.Y., Ezra O., Madar S., Goldfinger N. Chemotherapeutic agents induce the expression and activity of their clearing enzyme CYP3A4 by activating p53. Carcinogenesis. 2013;34:190–198. doi: 10.1093/carcin/bgs318. [DOI] [PubMed] [Google Scholar]
- 47.Nantasanti S., Toussaint M.J., Youssef S.A., Tooten P.C., de Bruin A. Rb and p53 liver functions are essential for xenobiotic metabolism and tumor suppression. PLoS One. 2016;11 doi: 10.1371/journal.pone.0150064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu H., Yu T., Arpiainen S., Lang M.A., Hakkola J., Abu-Bakar A. Tumour suppressor protein p53 regulates the stress activated bilirubin oxidase cytochrome P450 2A6. Toxicol Appl Pharmacol. 2015;289:30–39. doi: 10.1016/j.taap.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 49.Elias A., Wu J., Chen T. Tumor suppressor protein p53 negatively regulates human pregnane X receptor activity. Mol Pharmacol. 2013;83:1229–1236. doi: 10.1124/mol.113.085092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wohak L.E., Baranski A.C., Krais A.M., Schmeiser H.H., Phillips D.H., Arlt V.M. The impact of p53 function on the metabolic activation of the carcinogenic air pollutant 3-nitrobenzanthrone and its metabolites 3-aminobenzanthrone and N-hydroxy-3-aminobenzanthrone in human cells. Mutagenesis. 2018;33:311–321. doi: 10.1093/mutage/gey025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wohak L.E., Krais A.M., Kucab J.E., Stertmann J., Ovrebo S., Seidel A. Carcinogenic polycyclic aromatic hydrocarbons induce CYP1A1 in human cells via a p53-dependent mechanism. Arch Toxicol. 2016;90:291–304. doi: 10.1007/s00204-014-1409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi M., Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzym Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Reisman S.A., Yeager R.L., Yamamoto M., Klaassen C.D. Increased Nrf2 activation in livers from Keap1-knockdown mice increases expression of cytoprotective genes that detoxify electrophiles more than those that detoxify reactive oxygen species. Toxicol Sci. 2009;108:35–47. doi: 10.1093/toxsci/kfn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohle C., Bock K.W. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73:1853–1862. doi: 10.1016/j.bcp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 56.Miao W., Hu L., Scrivens P.J., Batist G. Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem. 2005;280:20340–20348. doi: 10.1074/jbc.M412081200. [DOI] [PubMed] [Google Scholar]
- 57.Shin S., Wakabayashi N., Misra V., Biswal S., Lee G.H., Agoston E.S. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loeppen S., Koehle C., Buchmann A., Schwarz M. A beta-catenin-dependent pathway regulates expression of cytochrome P450 isoforms in mouse liver tumors. Carcinogenesis. 2005;26:239–248. doi: 10.1093/carcin/bgh298. [DOI] [PubMed] [Google Scholar]
- 59.Schreiber S., Rignall B., Braeuning A., Marx-Stoelting P., Ott T., Buchmann A. Phenotype of single hepatocytes expressing an activated version of beta-catenin in liver of transgenic mice. J Mol Histol. 2011;42:393–400. doi: 10.1007/s10735-011-9342-6. [DOI] [PubMed] [Google Scholar]
- 60.Oinonen T., Lindros K.O. Zonation of hepatic cytochrome P-450 expression and regulation. Biochem J. 1998;329:17–35. doi: 10.1042/bj3290017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sekine S., Gutierrez P.J., Lan B.Y., Feng S., Hebrok M. Liver-specific loss of beta-catenin results in delayed hepatocyte proliferation after partial hepatectomy. Hepatology. 2007;45:361–368. doi: 10.1002/hep.21523. [DOI] [PubMed] [Google Scholar]
- 62.Benhamouche S., Decaens T., Godard C., Chambrey R., Rickman D.S., Moinard C. Apc tumor suppressor gene is the "zonation-keeper" of mouse liver. Dev Cell. 2006;10:759–770. doi: 10.1016/j.devcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 63.Rocha A.S., Vidal V., Mertz M., Kendall T.J., Charlet A., Okamoto H. The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Rep. 2015;13:1757–1764. doi: 10.1016/j.celrep.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 64.Wang B., Zhao L., Fish M., Logan C.Y., Nusse R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sekine S., Lan B.Y., Bedolli M., Feng S., Hebrok M. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology. 2006;43:817–825. doi: 10.1002/hep.21131. [DOI] [PubMed] [Google Scholar]
- 66.Braeuning A., Sanna R., Huelsken J., Schwarz M. Inducibility of drug-metabolizing enzymes by xenobiotics in mice with liver-specific knockout of Ctnnb1. Drug Metab Dispos. 2009;37:1138–1145. doi: 10.1124/dmd.108.026179. [DOI] [PubMed] [Google Scholar]
- 67.Giera S., Braeuning A., Kohle C., Bursch W., Metzger U., Buchmann A. Wnt/beta-catenin signaling activates and determines hepatic zonal expression of glutathione S-transferases in mouse liver. Toxicol Sci. 2010;115:22–33. doi: 10.1093/toxsci/kfq033. [DOI] [PubMed] [Google Scholar]
- 68.Braeuning A., Schwarz M. Zonation of heme synthesis enzymes in mouse liver and their regulation by beta-catenin and Ha-ras. Biol Chem. 2010;391:1305–1313. doi: 10.1515/BC.2010.115. [DOI] [PubMed] [Google Scholar]
- 69.Braeuning A., Kohle C., Buchmann A., Schwarz M. Coordinate regulation of cytochrome P450 1a1 expression in mouse liver by the aryl hydrocarbon receptor and the beta-catenin pathway. Toxicol Sci. 2011;122:16–25. doi: 10.1093/toxsci/kfr080. [DOI] [PubMed] [Google Scholar]
- 70.Schmidt A., Braeuning A., Ruck P., Seitz G., Armeanu-Ebinger S., Fuchs J. Differential expression of glutamine synthetase and cytochrome P450 isoforms in human hepatoblastoma. Toxicology. 2011;281:7–14. doi: 10.1016/j.tox.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 71.Yan T., Lu L., Xie C., Chen J., Peng X., Zhu L. Severely impaired and dysregulated cytochrome p450 expression and activities in hepatocellular carcinoma: implications for personalized treatment in patients. Mol Cancer Ther. 2015;14:2874–2886. doi: 10.1158/1535-7163.MCT-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kovalszky, Schaff Z., Lapis K., Jeney A. Marker enzymes of rat chemical hepatocarcinogenesis in human liver tumors. Pathol Oncol Res. 1996;2:56–58. doi: 10.1007/BF02893950. [DOI] [PubMed] [Google Scholar]
- 73.Thomas M., Bayha C., Vetter S., Hofmann U., Schwarz M., Zanger U.M. Activating and inhibitory functions of WNT/beta-catenin in the induction of cytochromes P450 by nuclear receptors in HepaRG cells. Mol Pharmacol. 2015;87:1013–1020. doi: 10.1124/mol.114.097402. [DOI] [PubMed] [Google Scholar]
- 74.Gerbal-Chaloin S., Dume A.S., Briolotti P., Klieber S., Raulet E., Duret C. The WNT/beta-catenin pathway is a transcriptional regulator of CYP2E1, CYP1A2 and aryl hydrocarbon receptor gene expression in primary human hepatocytes. Mol Pharmacol. 2014;86:624–634. doi: 10.1124/mol.114.094797. [DOI] [PubMed] [Google Scholar]
- 75.Stahl S., Ittrich C., Marx-Stoelting P., Kohle C., Altug-Teber O., Riess O. Genotype-phenotype relationships in hepatocellular tumors from mice and man. Hepatology. 2005;42:353–361. doi: 10.1002/hep.20768. [DOI] [PubMed] [Google Scholar]
- 76.Tornesello M.L., Buonaguro L., Tatangelo F., Botti G., Izzo F., Buonaguro F.M. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74–83. doi: 10.1016/j.ygeno.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 77.Iiuzaka N., Oka M., Yamada-Okabe H., Mori N., Tamesa T., Okada T. Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method. Cancer Res. 2002;62:3939–3944. [PubMed] [Google Scholar]
- 78.Okabe H., Satoh S., Kato T., Kitahara O., Yanagawa R., Yamaoka Y. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–2137. [PubMed] [Google Scholar]
- 79.Jaworski M., Ittrich C., Hailfinger S., Bonin M., Buchmann A., Schwarz M. Global gene expression in Ha-ras and B-raf mutated mouse liver tumors. Int J Cancer. 2007;121:1382–1385. doi: 10.1002/ijc.22801. [DOI] [PubMed] [Google Scholar]
- 80.Rignall B., Ittrich C., Krause E., Appel K.E., Buchmann A., Schwarz M. Comparative transcriptome and proteome analysis of Ha-ras and B-raf mutated mouse liver tumors. J Proteome Res. 2009;8:3987–3994. doi: 10.1021/pr9002933. [DOI] [PubMed] [Google Scholar]
- 81.Braeuning A., Kollotzek F., Zeller E., Knorpp T., Templin M.F., Schwarz M. Mouse Hepatomas with Ha-ras and B-raf mutations differ in mitogen-activated protein kinase signaling and response to constitutive androstane receptor activation. Drug Metab Dispos. 2018;46:1462–1465. doi: 10.1124/dmd.118.083014. [DOI] [PubMed] [Google Scholar]
- 82.Braeuning A., Menzel M., Kleinschnitz E.M., Harada N., Tamai Y., Kohle C. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by beta-catenin signaling in mouse hepatocytes. FEBS J. 2007;274:4766–4777. doi: 10.1111/j.1742-4658.2007.06002.x. [DOI] [PubMed] [Google Scholar]
- 83.Chesire D.R., Dunn T.A., Ewing C.M., Luo J., Isaacs W.B. Identification of aryl hydrocarbon receptor as a putative Wnt/beta-catenin pathway target gene in prostate cancer cells. Cancer Res. 2004;64:2523–2533. doi: 10.1158/0008-5472.can-03-3309. [DOI] [PubMed] [Google Scholar]
- 84.Hailfinger S., Jaworski M., Braeuning A., Buchmann A., Schwarz M. Zonal gene expression in murine liver: lessons from tumors. Hepatology. 2006;43:407–414. doi: 10.1002/hep.21082. [DOI] [PubMed] [Google Scholar]
- 85.Braeuning A., Heubach Y., Knorpp T., Kowalik M.A., Templin M., Columbano A. Gender-specific interplay of signaling through beta-catenin and CAR in the regulation of xenobiotic-induced hepatocyte proliferation. Toxicol Sci. 2011;123:113–122. doi: 10.1093/toxsci/kfr166. [DOI] [PubMed] [Google Scholar]
- 86.Gougelet A., Torre C., Veber P., Sartor C., Bachelot L., Denechaud P.D. T-cell factor 4 and beta-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–2357. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- 87.Schulthess P., Loffler A., Vetter S., Kreft L., Schwarz M., Braeuning A. Signal integration by the CYP1A1 promoter—a quantitative study. Nucleic Acids Res. 2015;43:5318–5330. doi: 10.1093/nar/gkv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Groll N., Petrikat T., Vetter S., Colnot S., Weiss F., Poetz O. Coordinate regulation of Cyp2e1 by β-catenin- and hepatocyte nuclear factor 1α-dependent signaling. Toxicology. 2016;350–352:40–48. doi: 10.1016/j.tox.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 89.Vaas S., Kreft L., Schwarz M., Braeuning A. Cooperation of structurally different aryl hydrocarbon receptor agonists and beta-catenin in the regulation of CYP1A expression. Toxicology. 2014;325:31–41. doi: 10.1016/j.tox.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 90.Ganzenberg K., Singh Y., Braeuning A. The time point of beta-catenin knockout in hepatocytes determines their response to xenobiotic activation of the constitutive androstane receptor. Toxicology. 2013;308:113–121. doi: 10.1016/j.tox.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 91.Braeuning A., Schwarz M. beta-Catenin as a multilayer modulator of zonal cytochrome P450 expression in mouse liver. Biol Chem. 2010;391:139–148. doi: 10.1515/bc.2010.012. [DOI] [PubMed] [Google Scholar]
- 92.Braeuning A. Interplay of beta-catenin with xenobiotic-sensing receptors and its role in glutathione S-transferase expression. Curr Drug Metabol. 2012;13:203–214. doi: 10.2174/138920012798918381. [DOI] [PubMed] [Google Scholar]
- 93.Zeller E., Mock K., Horn M., Colnot S., Schwarz M., Braeuning A. Dual-specificity phosphatases are targets of the Wnt/beta-catenin pathway and candidate mediators of beta-catenin/Ras signaling interactions. Biol Chem. 2012;393:1183–1191. doi: 10.1515/hsz-2012-0130. [DOI] [PubMed] [Google Scholar]
- 94.Zeller E., Hammer K., Kirschnick M., Braeuning A. Mechanisms of RAS/beta-catenin interactions. Arch Toxicol. 2013;87:611–632. doi: 10.1007/s00204-013-1035-3. [DOI] [PubMed] [Google Scholar]
- 95.Morgan E.T. Regulation of cytochrome p450 by inflammatory mediators: why and how?. Drug Metab Dispos. 2001;29:207–212. [PubMed] [Google Scholar]
- 96.Tinel M., Elkahwaji J., Robin M.A., Fardel N., Descatoire V., Haouzi D. Interleukin-2 overexpresses c-myc and down-regulates cytochrome P-450 in rat hepatocytes. J Pharmacol Exp Ther. 1999;289:649–655. [PubMed] [Google Scholar]
- 97.Klein M., Thomas M., Hofmann U., Seehofer D., Damm G., Zanger U.M. A systematic comparison of the impact of inflammatory signaling on absorption, distribution, metabolism, and excretion gene expression and activity in primary human hepatocytes and HepaRG cells. Drug Metab Dispos. 2015;43:273–283. doi: 10.1124/dmd.114.060962. [DOI] [PubMed] [Google Scholar]
- 98.Tanner N., Kubik L., Luckert C., Thomas M., Hofmann U., Zanger U.M. Regulation of drug metabolism by the interplay of inflammatory signaling, steatosis, and xeno-sensing receptors in HepaRG cells. Drug Metab Dispos. 2018;46:326–335. doi: 10.1124/dmd.117.078675. [DOI] [PubMed] [Google Scholar]
- 99.Keller R., Klein M., Thomas M., Dräger A., Metzger U., Templin M.F. Coordinating role of RXRα in downregulating hepatic detoxification during inflammation revealed by Fuzzy-Logic modeling. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1004431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koike C., Moore R., Negishi M. Extracellular signal-regulated kinase is an endogenous signal retaining the nuclear constitutive active/androstane receptor (CAR) in the cytoplasm of mouse primary hepatocytes. Mol Pharmacol. 2007;71:1217–1221. doi: 10.1124/mol.107.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh Y., Braeuning A., Schmid A., Pichler B.J., Schwarz M. Selective poisoning of Ctnnb1-mutated hepatoma cells in mouse liver tumors by a single application of acetaminophen. Arch Toxicol. 2013;87:1595–1607. doi: 10.1007/s00204-013-1030-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.