Abstract
This article comments on:
Afsharyan NP, Sannemann W, Léon J, Ballvora A. 2020. Effect of epistasis and environment on flowering time of barley reveals novel flowering-delaying QTL allele. Journal of Experimental Botany 71, 893–906.
Keywords: Barley, environment effect, epistasis, flowering time, novel QTL, MAGIC population, QTL analysis
Time to heading is an important target in barley breeding programs due to its high correlation with grain yield. Using genetic analysis of an eight-parent barley MAGIC (multi-parent advanced generation inter-cross) population and phenotype data from two semi-controlled and two field environments, Afsharyan et al. (2020) identified a novel flowering-delaying quantitative trait locus (QTL) allele on chromosome 1H. This novel QTL, known as HvHeading, shows strong epistatic interactions with QTL regions overlapping with prominent flowering time genes, including Vrn-H3, Vrn-H1, and Ppd-H1. The results suggest that studying epistasis among QTL loci, and QTL×environments, and epistasis×environments interactions in high-resolution mapping populations (such as MAGIC) is a promising approach for detecting and identifying novel regulators that act in concert with the large effect QTLs to fine-tune the flowering time and yield formation in response to the changing environment.
Barley is an important cereal crop grown worldwide under a wide range of environments. To maximize yield, it is essential to tailor the life cycle of cereals to the agro-environments in which they are grown. Heading date (HD) marks the transition from vegetative to reproductive growth in barley and is a key adaptive trait that ensures that plants set their flowers at an optimum time for pollination and seed development. This developmental switch is regulated by pathways responsive to changes in the photoperiod and temperature as well as environment-independent pathways such as earliness per se (EPS) (Cockram et al., 2007). Nevertheless, HD is a highly complex trait: genetic variation can greatly alter the plant response to photoperiod and temperature (Jung and Müller, 2009). For this reason, it is one of the most important target traits in crop breeding programs due to its high correlation with final grain yield.
In contrast to maize, where flowering time is controlled by numerous small effect additive quantitative trait loci (QTL) (Buckler et al., 2009), HD in barley is controlled by a small number of QTL loci with large effects that overlap with major genes involved in barley flowering time pathways (Box 1). Due to the high heritability of the HD trait, the large effect QTL loci encompassing main flowering time pathway genes have been consistently detected in both genome-wide association studies (Pasam et al., 2012) and multiple QTL mapping studies, using various bi-parental populations (Khahani et al., 2019), as well as multi-parent populations such as nested association mapping (NAM) and multi-parent advanced generation inter-cross (MAGIC) (Maurer et al., 2015, 2016; Sannemann et al., 2015). Furthermore, epistatic interactions among major flowering pathway genes in barley have already been reported (Maurer et al., 2015; Mathew et al., 2018).
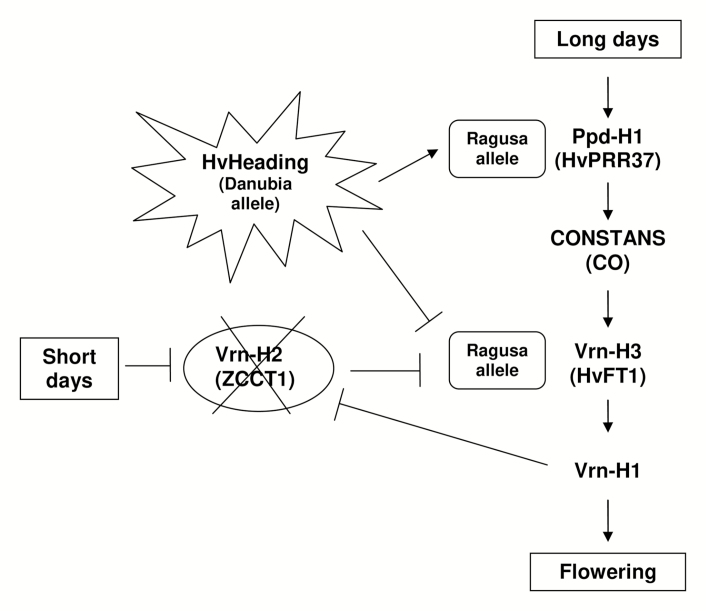
Box 1. Main flowering pathway genes in spring barley showing the interactions of the HvHeading allele from Danubia.
In barley, the daylength-determining light signal is transmitted from a circadian clock oscillator, with Ppd-H1, a PSEUDO-RESPONSE REGULATOR (HvPRR37) gene, in its center (Turner et al., 2005). Under long-day conditions, Ppd-H1 interacts with CONSTANS (CO) and promotes flowering by initiating expression of the Vrn-H3 gene, a homolog of the Arabidopsis thaliana FLOWERING LOCUS T (FT) gene (Yan et al., 2006). The Vrn-H3 gene in turn promotes the expression of the downstream gene Vrn-H1, an APETALA1 family MADS-box transcription factor gene, which induces flowering time by promoting transition from the vegetative to reproductive phase (Hemming et al., 2008). Spring barley lacks the vernalization requirement due to a deletion of Vrn-H2 (von Zitzewitz et al., 2005).
The effects of HvHeading allele originating from the landrace Danubia (Afsharyan et al., 2020), combined with Ppd-H1 and Vrn-H3 alleles from Ragusa, is also depicted. The Danubia allele accelerated HD by 10 d and delayed it by 10.83 d, respectively.
The findings of Afsharyan et al. (2020) add important new information to this body of work by identifying several novel epistasis interactions of major flowering time genes with chromosome segments that have not been previously implicated in flowering time control. Particularly important is a novel flowering-delaying QTL allele, named HvHeading, which is located on chromosome 1H. It originated from the landrace Ackermanns Danubia and explained 6.37% and 7.79% of the proportion of genetic variation in single marker analysis (SMA) and haplotype analysis (HA), respectively (Afsharyan et al., 2020). Analysis of the allelic effect revealed that the presence of this allele delayed HD by 3.5 d as a single QTL in field conditions. HvHeading is also involved in strong epistasis interactions with two other loci, one on chromosome 2H and one on 7H that overlap with the positions of Ppd-H1 and Vrn-H3 (HvFT1), respectively. In field experiments, interaction with the Ppd-H1 locus on chromosome 2H could accelerate HD by up to 10 d, while combinations with alleles of the Vrn-H3 (HvFT1) locus on chromosome 7H could postpone HD by 12.38 d. These strong epistatic interactions were also confirmed by HA, which showed that interaction of the HvHeading haplotype originating from the parent Danubia with Ppd-H1 and Vrn-H3 (HvFT1) haplotypes from Ragusa accelerated and delayed HD by 10 d and 10.83 d, respectively. Significant epistasis interactions of HvHeading with QTL loci that correspond to positions of other important flowering pathway genes, including Vrn-H1, sdw1/denso, HvPRR95, HvPhyC, HvCO8, and HvSS1, were also detected in field conditions. However, in the foil tunnel experiments, where the average temperature was higher and growing degree days (GDD) accumulated faster, HvHeading alone did not show detectable effects on HD (Sannemann et al., 2015) and epistasis influenced it with much lower magnitude, being notably involved in epistatic interaction with Vrn-H3 (HvFT1) alleles (ranging from –1.85 d to +1.50 d) according to SMA. This suggests that both expression and the epistatic effects of the novel chromosome 1H QTL are influenced by environment temperature. This was further confirmed by the strong epistasis×environment interactions of HvHeading with Ppd-H1, Vrn-H3, and Vrn-H1 detected in all four environments, and indicate that HvHeading might be involved in a pathway that regulates flowering time in response to temperature changes. Notably, neither HvHeading nor its epistasis interactions have been detected in previous studies with the same spring barley MAGIC population, using the same genotype data and the phenotype data from 2 years’ foil tunnel experiments (Sannemann et al., 2015; Mathew et al., 2018). Detection of HvHeading epistasis interactions in foil tunnel experiments in the latest study (Afsharyan et al., 2020) suggests that technical improvements in the algorithms used for genotype data preparation and the statistical model for QTL and epistasis detection increased the power to uncover at least the interactions of small effect QTLs with major flowering time genes.
The findings of Afsharyan et al. (2020) paved the way for further development in both theoretical and applied aspects. It could contribute to the identification of novel genes that are involved in the regulation of flowering time pathways in response to temperature changes. Furthermore, the QTL could be integrated into breeding programs to tailor spring barley HD for agro-environment adaptation and climate resilience. However, further research is needed for the advancement in either direction.
Toward identification of the causal gene of HvHeading
The relatively large interval (70.89–71.03 cM by SMA and 60.84–86.47 cM by HA) in which the chromosome 1H QTL is located makes more difficult the task of identifying the causal gene for HvHeading. The in silico analysis performed revealed that the overlapping region between QTL intervals from SMA (7 cM) and HA (30 cM) contained 160 genes with different Gene Ontology (GO) annotations (Afsharyan et al., 2020). The most straightforward approach is to develop near-isogenic lines (NILs) from a bi-parental cross of Danubia×Ragusa, followed by fine-mapping and map-based cloning. However, this approach could be heavily time and resource consuming and might not succeed due to the opposite epistasis effects of the Danubia allele in combination with Ppd-H1 and Vrn-H3 alleles from Ragusa (Box 1). Alternative approaches include targeted NGS sequencing of the HvHeading interval that could provide denser marker coverage and presumably higher mapping resolution. However, there will be no advantage to higher marker density in the case that the QTL resides in a region with reduced recombination, such as those close to the centromere. Therefore, a candidate gene approach could be most appropriate for the identification of the HvHeading causal gene, but its application will require prioritization of the candidate genes as a first step. The only known candidate gene located close to the HvHeading interval at 93.1 cM is Ppd-H2 (HvFT3) which could be readily excluded because no association was detected between this region and HD in spring barley MAGIC double haploid lines (Afsharyan et al., 2020). The prioritization could be based on the GO annotations, sequence information, and expression patterns. A >10-fold reduction in the number of candidate genes for rice QTL loci was achieved by using gene functions predicted on the basis of sequence and expression information (Bargsten et al., 2014). Aligning the HvHeading interval to those reported in other flowering time QTL studies may also help in prioritizing potential candidate genes. A recent gene-set association study (He et al., 2019), using a worldwide barley collection including 952 accessions of cultivated barley, identified single nucleotide polymorphisms (SNPs) in a number of genes showing significant association with flowering time in barley. Two of the reported genes reside within the wider HvHeading interval defined by HA in Afsharyan et al. (2020). One of these genes, HvSTK (HORVU1Hr1G064150), located at 61.47 cM (base pairs 458 871 991–458 877 307), encodes a MADS-box transcription factor, and the second, HvGA20ox2-2 (HORVU1Hr1G070710) at 82.38 cM (base pairs 490 940 150–490 941 780), is a 2-oxoglutarate- and Fe(II)-dependent oxygenase superfamily protein, involved in gibberellin signaling and metabolism. SNPs in 20 MADS box transcription factors, as well as 19 genes related to gibberellin sygnaling and metabolism were significantly associated with flowering time in barley (He et al., 2019). It is known that the early flowering of some barley genotypes is closely linked to gibberellin biosynthesis (Boden et al., 2014). On the other hand, many of the MADS box genes are involved in meristem response and development and are thus related to flowering. Therefore, HvSTK and HvGA20ox2-2 could be considered high priority candidates for HvHeading. Once the candidate genes have been shortlisted, their effects on HD can be identified by utilizing TILLING knockout mutants (Talame et al., 2008) or by CRISPR/Cas gene editing (Kumar and Jain, 2015; Hamada et al., 2018).
Potential breeding applications
Alleles that reasonably delay HD are desirable in breeding programs as they often have pleiotropic effects to increase final grain yield (Jung and Müller, 2009). Once its relationship to grain yield has been established, the flowering delaying HvHeading allele, identified in Danubia (Afsharyan et al., 2020), will have potential for breeding applications to improve adaptation of HD to a wide range of environments. Further research is needed to examine the effect and interactions of this landrace allele with the alleles of major flowering time loci present in elite breeding pools. Identification of the HvHeading underlying gene and subsequent deciphering of the molecular mechanisms of its involvement in flowering time regulation will aid in the employment of various HvHeading alleles to improve barley adaptation to a specific environment.
To conclude, the study of Afsharyan et al. (2020) demonstrates the power of the MAGIC approach to detect novel flowering time regulators involved in epistatic and/or environment interactions. The identification of HvHeading provides us with new opportunities to extend our knowledge of the mechanism of temperature-dependent flowering time control in spring barley. Once the effect HvHeading allele from Danubia on grain yield has been established, it will be a good candidate for breeding programs aiming at fine-tuning HD for the development of climate-resilient, high-yielding barley cultivars.
Acknowledgements
NKC was supported by the Bulgarian Ministry of Education and Science under the National Research Programme ‘Healthy Foods for a Strong Bio-Economy and Quality of Life’ approved by DCM # 577/17.08.2018.
References
- Afsharyan N, Sannemann W, Léon J, Ballvora A. 2020. Effect of epistasis and environment on flowering time of barley reveals novel flowering-delaying QTL allele. Journal of Experimental Botany 71, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargsten JW, Nap JP, Sanchez-Perez GF, van Dijk AD. 2014. Prioritization of candidate genes in QTL regions based on associations between traits and biological processes. BMC Plant Biology 14, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM. 2014. EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. The Plant Cell 26, 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, et al. . 2009. The genetic architecture of maize flowering time. Science 325, 714–718. [DOI] [PubMed] [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA, Greenland AJ. 2007. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. Journal of Experimental Botany 58, 1231–1244. [DOI] [PubMed] [Google Scholar]
- Hamada H, Liu Y, Nagira Y, Miki R, Taoka N, Imai R. 2018. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Scientific Reports 8, 14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T, Hill CB, Angessa TT, Zhang XQ, Chen K, Moody D, Telfer P, Westcott S, Li C. 2019. Gene-set association and epistatic analyses reveal complex gene interaction networks affecting flowering time in a worldwide barley collection. Journal of Experimental Botany 70, 5603–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. 2008. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiology 147, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Müller AE. 2009. Flowering time control and applications in plant breeding. Trends in Plant Science 14, 563–573. [DOI] [PubMed] [Google Scholar]
- Khahani B, Tavakol E, Shariati JV. 2019. Genome-wide meta-analysis on yield and yield-related QTLs in barley (Hordeum vulgare L.). Molecular Breeding 39, 56. [Google Scholar]
- Kumar V, Jain M. 2015. The CRISPR-Cas system for plant genome editing: advances and opportunities. Journal of Experimental Botany 66, 47–57. [DOI] [PubMed] [Google Scholar]
- Mathew B, Léon J, Sannemann W, Sillanpää MJ. 2018. Detection of epistasis for flowering time using bayesian multilocus estimation in a Barley MAGIC population. Genetics 208, 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A, Draba V, Jiang Y, Schnaithmann F, Sharma R, Schumann E, Kilian B, Reif JC, Pillen K. 2015. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A, Draba V, Pillen K. 2016. Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. Journal of Experimental Botany 67, 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, Kilian B, Graner A. 2012. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biology 12, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannemann W, Huang BE, Mathew B, Léon J. 2015. Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Molecular Breeding 35, 86. [Google Scholar]
- Talamè V, Bovina R, Sanguineti MC, Tuberosa R, Lundqvist U, Salvi S. 2008. TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant Biotechnology Journal 6, 477–485. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TH, Hayes PM, Skinner JS. 2005. Molecular and structural characterization of barley vernalization genes. Plant Molecular Biology 59, 449–467. [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. 2006. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences, USA 103, 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]