Abstract
The development of a next-generation DNA sequencer has provided a method for electrically measuring single molecules. Methods for electrically measuring one molecule are roughly divided into methods for measuring tunneling and ion currents. These methods enable identification of a single molecule of DNA, a RNA nucleotide, or a single protein based on current histograms. However, overlapping of current histograms of molecules with similar properties has been a major barrier to identifying single molecules with high accuracy. This barrier was broken by introducing machine learning. Combining single-molecule electrical measurement and machine learning enables high-precision identification of single molecules. Highly accurate discrimination has been demonstrated for DNA nucleotides, RNA nucleotides, amino acids, sugars, viruses, and bacteria. This combination enables quantitative evaluation of molecular recognition ability. Furthermore, a device structure suitable for high-precision identification has been designed. Combining single-molecule electrical measurement with machine learning enables digital analytical chemistry that can count certain types of molecules. Digital analytical chemistry enables comprehensive analysis of chemical reactions. This new analytical method will lead to the discovery of unknown or missed valuable molecules.
Introduction
Disease prevention is an important global issue. To achieve this, diseases, including infectious diseases, must be quickly, cheaply, and accurately diagnosed at an early stage. Ideally, this should occur before symptoms appear. It is desirable to be able to diagnose cancer when its cells are small. One candidate for achieving this is a method for examining the genome in a single cell. For infectious diseases, it is effective to examine a small number of viruses and bacteria before these multiply in the body. A method of examining a small number, ideally a single DNA molecule, virus, or bacterium, is desirable for early diagnosis. Optical methods such as real-time polymerase chain reaction (RT-PCR) and enzyme-linked immune sorbent assay (ELISA) have been used to test for diseases, including infectious diseases. Since smartphones are an integral component of the world’s infrastructure, an electrical inspection method that can be connected to a smartphone enables in situ diagnosis.1,2 This is because on-site measurements can be transferred to a server and analyzed using a smartphone and the inspection results returned to the hand. The electrical measurement method can be miniaturized, reduced in cost, and highly integrated using semiconductor technology. This measurement method is known as single-molecule electrical measurement.
Single-molecule electrical measurement methods are roughly classified into nanogap3−5 and nanopore methods.3,5−7 These were originally developed for producing next-generation DNA sequencers. The nanogap method detects and discriminates analytes that pass between electrodes with gaps of several nanometers or less using tunneling currents (Figure 1a).4 The tunneling current provides information on the electronic state of the analyte and the analyte–electrode interaction. Particularly, it provides information on the energy levels of the frontier orbitals that determine the current path. The tunneling current–time waveform is characterized by a maximum current (IpT) and a current duration (tdT).4 Two or more analytes are identified by an IpT or tdT histogram or an IpT–tdT heat map. This method has been widely applied to DNA and RNA sequencing, peptide amino acid sequencing, and identification of modified and unmodified amino acid molecules.4
Figure 1.

Single-molecule electrical measurement method. (a) Nanogap method. The current–time waveform corresponding to the single molecule is characterized by a maximum current IpT and a current duration tdT. (b) Nanopore method. The ion current–time waveform is characterized by the maximum current change IpN and the current duration tdN.
The nanopore method is classified into two types, bionanopore3 and solid-state nanopore.6,7 Here, only solid-state nanopores manufactured using semiconductor technology are targeted. A typical solid-state nanopore has a through-hole in SiN with a diameter of several micrometers or less (Figure 1b).6 Both sides of the SiN substrate are filled with an aqueous solution in which the electrolytes are dissolved. When a voltage is applied between the electrodes installed on both sides, an ion current flows through the nanopore. When the sample enters the nanopore, the ion current decreases. The ion current–time waveform is analyzed using two measured quantities: the maximum change in the ion current (IpN) and the duration of the current (tdN). When two or more analytes are measured, they are identified by an IpN or tdN histogram or an IpN–tdN heat map. This method has been widely applied to DNA, RNA, and proteins.3,5−7
However, the nanogap and nanopore methods have a serious problem in common. In the nanogap method, analytes with similar volumes and frontier orbital energy levels have similar IpT and tdT histograms. In the solid-state nanopore method, analytes with similar volumes show similar IpN and tdN histograms. That is, histogram overlap hinders high-precision identification of two or more analytes.
This problem was overcome by analyzing the tunneling current–time and ion current–time waveforms using machine learning.8−16 In the field of single-molecule electrical measurement, support vector machine and random forest are mainly used to handle binary and multiclass classification problems. So far, in the histogram analysis, each waveform has been analyzed using the scalar quantities IpT or IpN. In the machine-learning analysis, the waveform is analyzed with a vector quantity characterizing it. A histogram using IpT shows a one-dimensional data distribution. An analysis using N features shows an N-dimensional data distribution. For example, in a two-dimensional map of a certain IpT–tdT, two analytes are not discriminated (Figure 2a). However, if two appropriate features are used, the data of the two analytes may be classified. Machine learning is similar to learning data and searching for features that classify analytes with high accuracy. The analysis using machine learning quantitatively evaluates false positives and false negatives that are practically important. In discriminating between two analytes, the accuracy, precision, recall, and F-measure are obtained from the confusion matrix (Figure 2b). Each is defined as follows: accuracy = (TP + TN)/(TP + FP + TN + FN); precision = TP/(TP + FP); recall = TP/(TP + FN); F-measure = 2 × recall × precision/(recall + precision), where TP, TN, FP, and FN indicate true positive, true negative, false positive, and false negative, respectively. It should be noted that the evaluation scale differs depending on the application. Different rating scales were used as reviewed below. Analysis by machine learning can identify with high accuracy which sample corresponds to a waveform obtained by measurement. In this mini-review, we introduce the advantages of combining the nanogap method, nanopore method, and machine learning.
Figure 2.
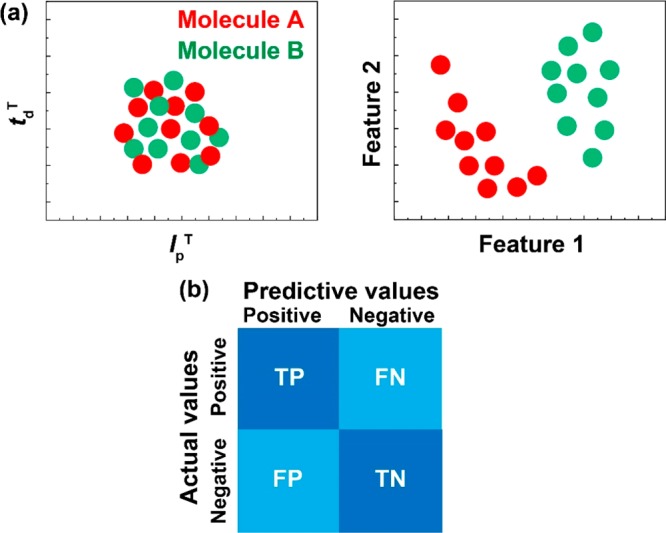
An analysis method using machine learning. (a) In a two-dimensional distribution with feature values IpT and tdT, molecules A and B are not discriminated with high accuracy. The classifier, feature 1, and feature 2 that can be identified with high accuracy are searched by machine learning. (b) A confusion matrix obtained by machine learning. The accuracy is calculated from the confusion matrix. TP, TN, FN, and FP represent true positive, true negative, false negative, and false positive, respectively. These four numbers correspond to the number of data points classified by machine learning.
Combination of Nanogap Methods and Machine Learning
The nanogap method is considered as an ideal principle for next-generation DNA sequencers.3,4 It is classified into two types. One involves modifying an electrode with a molecule that recognizes an analyte. In this method, a single molecule is identified using a tunneling current flowing between the modified electrodes through a single analyte molecule. This current is called the recognition tunneling (RT) current.8−11 The other method involves using a bare electrode.12 These methods enable identification of a single DNA base and amino acid molecules.4 Additionally, the latter approach was used to demonstrate short DNA sequencing and partial peptide sequencing of short peptides.4
However, the proof of concept of these DNA and peptide sequencers was mainly based on the histogram analysis of IpT.4 Many researchers expressed concern that the large overlap of the IpT histograms of the four base molecules and 20 amino acids gave large discrimination errors.4 Histogram-based analysis prompted researchers to stochastically determine which single molecule a tunneling current–time waveform was attributed to. Despite the development of single-molecule measurement, the information obtained from a single molecule could not be handled directly. Machine learning has overcome this dilemma.
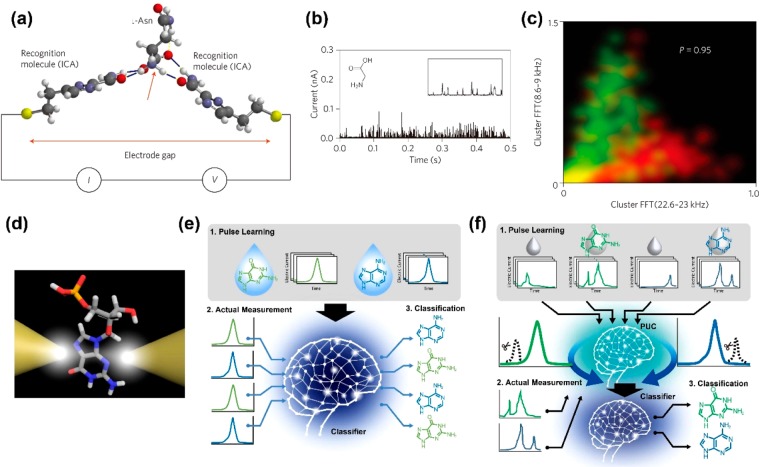
A breakthrough was achieved by single-molecule identification of amino acids using the RT method.8 Pd substrate and Pd electrode surfaces were modified with recognition molecules (Figure 3a). The modifying molecule was 4(5)-(2-mercaptoethyl)-1H-imidazole-2-carboxamide (ICA). Amino acids and modified molecules form hydrogen bonds. When the amino acids were measured, a spike-shaped tunneling current–time waveform was obtained (Figure 3b). Using IpT and tdT histograms, it was not possible to discriminate between two amino acids with high accuracy. Using the results obtained by fast Fourier transform (FFT) of each obtained waveform, 161 features were generated. A support vector machine (SVM)17 was used as a classifier. Analysis using the extracted features and SVM enabled high-precision identification of two amino acids. The discrimination accuracy between leucine (Leu) and N-methylglycine (mGly) was represented by precision = 0.95 (Figure 3c). Analysis using the IpT histogram only gave an accuracy of precision = 0.58. Surprisingly, the optically active forms, d-asparagine (Asn) and l-Asn, were identified with a high accuracy of precision = 0.87. This experiment demonstrated that two amino acid molecules that cannot be identified with high accuracy using IpT and tdT histograms can be identified with high accuracy by analysis using machine learning. Combining the RT method and SVM realized high-precision single-molecule discrimination between DNA9 and RNA10 base molecules. Furthermore, high-accuracy single-molecule identification of sugar was demonstrated.11
Figure 3.
Combination of nanogap method and machine learning. (a)–(c) Combination of recognition tunnel and machine learning. Reproduced with permission from ref (8). Copyright 2014 Springer Nature. (a) Principle diagram of recognition tunnel. (b) Glycine tunneling current vs time waveform. (c) Identification accuracy for Leu and methylated Gly obtained by analysis using a SVM. Red indicates methylated glycine data points, and green indicates Leu data points. The features are obtained by FFT of each pulse. (d)–(f) Principle diagram of a bare nanogap electrode. Reproduced with permission from ref (12). Copyright 2019 American Chemical Society. (e) Machine learning using Rotation Forest. (f) Machine learning using PUC method and Rotation Forest.
Combining the RT method and machine learning enabled quantitative evaluation of the goodness of the combination of the analyte and the recognition molecule.9,10 The stabilization energy due to the interaction between the analyte and the recognition molecule could be obtained from quantum chemical calculations. The strength of the interaction could be obtained by measuring the coupling constant. However, these values are not always an evaluation index of the goodness of the combination of the analyte and the recognition molecule for obtaining high discrimination accuracy. When ICA and three of its derivatives were used as recognition molecules, the DNA nucleoside base molecule identification accuracy was required.9 The average discrimination accuracy for the base molecules ranged from 40.1% ± 35.3% to 73.5% ± 16.2%. Among the four recognition molecules, ICA gave the third most accurate identification. When a recognition molecule having a pyrene structure that interacted with DNA nucleosides and formed π–π interactions was used, higher discrimination accuracy was obtained than with four recognition molecules. Furthermore, the identification accuracy for DNA and RNA nucleosides was required using ICA as a recognition molecule.10 The average discrimination accuracy for RNA nucleosides was 20% higher than that for DNA nucleosides. This difference correlated with the number of hydrogen bonds.
A combination of bare nanogap electrodes and machine learning was applied to single-molecule discrimination of DNA nucleosides (Figure 3d).12 Four nucleoside tunneling current–time waveforms were measured with a 0.54 nm nanogap electrode. Pulses were extracted from each measurement data point. The four nucleoside IpT histograms showed large overlaps. A vector obtained by dividing one waveform corresponding to a single base molecule into 10 in the time direction was used as a feature. A WEKA (Waikato Environment for Knowledge Analysis) workbench18 with many classifiers was used. The classifier with the highest discrimination accuracy, Rotation Forest,19 was selected (Figure 3e). For the two types of discrimination, the mean F-measure >0.70. The average F-measure for the three types of discrimination was 0.70. Using the four types of identification, the F-measures of dAMP, dCMP, dGMP, and dTMP (DNA nucleoside monophosphates) were 0.59, 0.91, 0.55, and 0.86, respectively. A random pick-up gave F-measure = 0.25. Therefore, four nucleosides were identified with high accuracy.
Single-molecule measurement using a tunneling current involves highly sensitive measurement of a minute current. Its high sensitivity enables detection of nonanalyte molecules and environmental signals. All these signals become noise; however, this cannot be avoided. That is, noise data are inevitably included in the data obtained by measuring the analyte. Until now, supervised machine learning has used data obtained by measuring one DNA nucleoside as teacher data.12 The teacher data include a pulse derived from noise. However, data derived from the analyte cannot be identified. Therefore, a solvent containing no sample is measured. All pulses obtained by this measurement are noise and can thus be labeled as noise. The solution containing the analyte is then measured. Since it is not known whether the pulse obtained by this measurement is noise or analyte, it is an unlabeled case. A classifier was developed to extract sample pulses from unlabeled cases using labeled cases (Figure 3f). This is the Positive Unlabeled Classification (PUC) method.20 DNA nucleosides were identified using the PUC method with a 0.54 nm bare nanogap electrode.12 The current vector was used as the feature value, and the WEKA workbench18 was used. The highest F-measure was obtained in Rotation Forest. The F-measures for the four nucleosides, dAMP, dCMP, dGMP, and dCMP, were 0.59, 0.91, 0.55, and 0.86, respectively.
Combining bare nanogap electrodes and machine learning gave a distance between nanogap electrodes that yielded a high F-measure.12 When the distance between the nanogap electrodes was 0.54 nm, the average F-measure of the four DNA nucleosides was 0.77. When the distance between the electrodes was 0.56, 0.58, and 0.60 nm, this changed to 0.57, 0.63, and 0.63. Therefore, the 0.54 nm nanogap electrode had the highest discrimination ability. The electronic coupling between the molecule and the electrode changes with the distance between the electrodes. The interelectrode distance dependency of the F-measure indicates that the waveform corresponding to a single molecule includes information on the electronic coupling between the molecule and the electrode. Physical and chemical interpretation of features and machine learning is difficult. However, external perturbations that change the machine learning results are key to interpreting the features and the reason for the high accuracy.
Combination of Nanopore Methods and Machine Learning
The ion current change provides information on the volume of the analyte passing through the nanopore. This is because the volume that the analyte excludes from the nanopore corresponds to the change in the ion current. When the thickness of the nanopore is less than that of the analyte, this principle provides information on the volume of the analyte, i.e., the volume of the circular slice = the cross-sectional area × the thickness of the nanopore. If the thickness of the nanopore is that of an atomic monolayer, the volume of the analyte can be obtained with atomic resolution. The idea was to develop solid-state nanopores of graphene monolayers7 and MoS2 monolayers21 to develop DNA sequencers. Low-aspect nanopores with nanopore thickness (L)/nanopore diameter (D) < 1 were created.
The flow dynamics of analytes with diameters of several hundred nanometers or more in solid-state nanopores have been analyzed by multiphysics simulations.22,23 The analyte in the nanopore is subjected to the forces of electrophoresis, electroosmotic flow, and fluid resistance. Multiphysics simulation solves the equations describing these forces. This simulation revealed that the ion current–time waveform of the analyte provided information on the volume, structure, and surface charge of the analyte.22,23 Many viruses and bacteria have similar volumes. Thus, it was considered difficult to identify viruses and bacteria with high accuracy using IpN and tdN histograms. The problem with high-accuracy identification is extracting this information from the waveform data. One solution is machine learning.
Combining low-aspect nanopores and machine learning has enabled identification of viruses and bacteria that could not be identified with high accuracy (Figure 4a–c). Influenza A (H1N1), influenza B, and an influenza A subtype (H3N2) were measured using a SiN nanopore with a thickness of 50 nm and a diameter of 300 nm.14 The differences between the three viruses lie in the proteins that they comprise. There were no differences between the IpN histograms of the three viruses. Ten features including IpN and tdN were extracted from the waveform (Figure 4a). Jcurr, td (=tdN), r, θ, and Jtime mainly reflect the translocation time, that is, the surface charge and flow dynamics of the virus. Ip (=IpN), S, and Sr mainly reflect the current change, that is, the volume of the virus, while β reflects its structure. Machine learning was performed using the features created by combining these features and the WEKA workbench.18 Rotation Forest19 gave the highest F-measure. The F-measures for A (H3N2) and B, A (H1N1) and A (H3N2), and A (H1N1) and B were 0.61, 0.68, and 0.72, respectively (Figure 4b). Based on the recall given by each feature as an index, the three influenza viruses showed different flow dynamics (Figure 4c).
Figure 4.
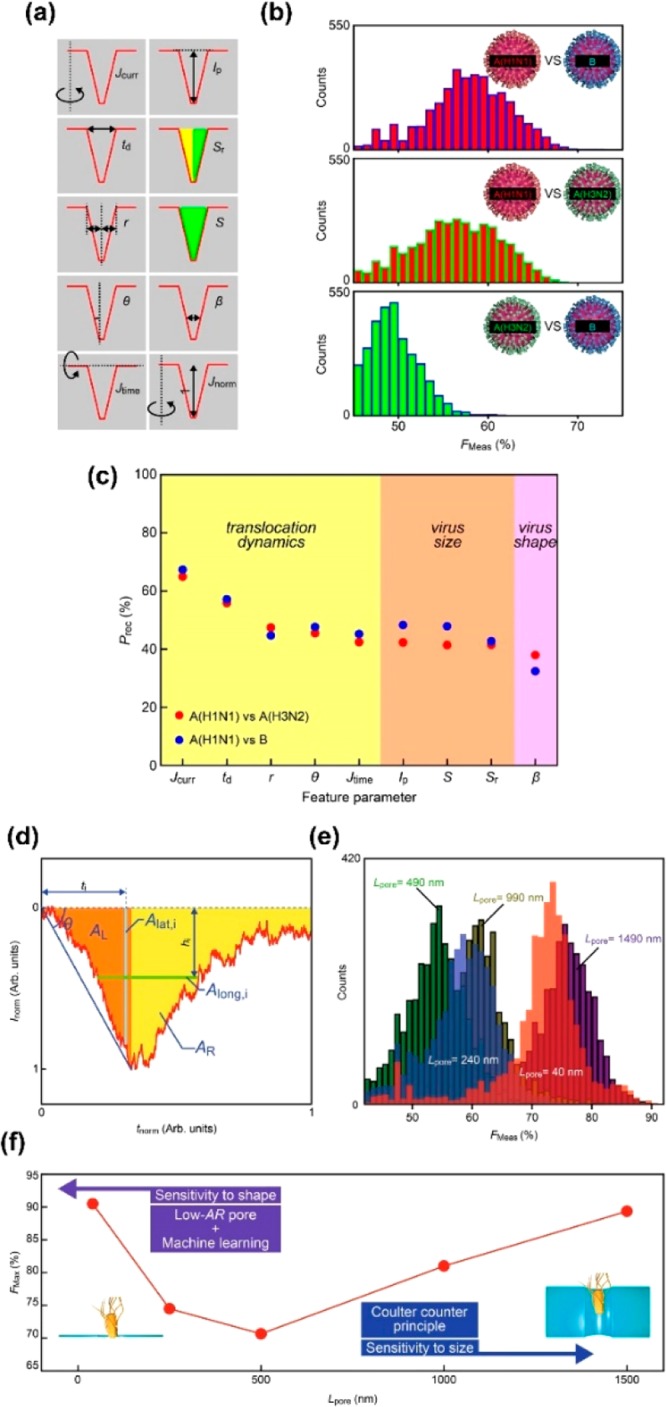
Combination of nanopore method and machine learning. (a)–(c) Identification of influenza virus. Reproduced with permission from ref (14). Copyright 2018 Springer Nature. (a) Definition of feature values used. (b) Identification accuracy for influenza A, influenza B, and influenza A subtype. The accuracy is represented by the F-measure. (c) Correlation between feature and recall. The feature indicating the flow dynamics of the virus gives high identification accuracy. (d)–(f) Discrimination between E. coli and Bacillus subtilis. Reproduced with permission from ref (16). Copyright 2017 Springer Nature. (d) Definition of feature used. (e) Dependence of nanopore thickness on identification accuracy. The identification accuracy is represented by the F-measure. (f) Nanopore thickness dependence of maximum discrimination accuracy.
E. coli and Bacillus subtilis were measured with low-aspect nanopores with L/D = 40/3000 nm (Figure 4d–f).16E. coli and Bacillus subtilis have similar surface potentials. Moreover, although they have the same volume, the curvatures of their structures are different. In the IpN and tdN histograms, the two bacteria could not be discriminated with high accuracy. Multiphysics simulations suggest that the ion current–time waveform differs for different bacterial structures. Therefore, in addition to IpN and tdN, features including θ, reflecting the curvature of the structure, were extracted, and 60 feature vectors were generated (Figure 4d). Machine learning was performed using these features and WEKA workbench18 (Figure 4e). The maximum F-measure was precision >0.85. As with the viruses, analysis of feature values and F-measures revealed that Ip (=IpN), A, Along, and i, reflecting volume and structure, gave high recall. Im and td (=tdN), reflecting surface charge and flow dynamics, gave low recall. Therefore, it is considered that high identification accuracy was obtained by identifying the curvature of the bacterial structure. Developing multiphysics simulation enabled the interpretation of features.
Combining solid-state nanopores and machine learning provides quantitative guidance for designing device structures with high discrimination abilities. The F-measure at each aspect ratio was obtained using the same analysis method by measuring the aspect ratio of the solid-state nanopore (Figure 4f).16 When L/D = 40/3000 nm = 0.013, the F-measure was 0.90. As L increased, the F-measure decreased. This is considered to be because the spatial resolution for examining the structures of bacteria decreases as the nanopore thickness increases. In nanopores with a large thickness, the F-measure was predicted to decrease, but the F-measure increased after L = 500 nm. Even for solid-state nanopores with L/D = 1500/3000 nm, the same F-measure as that of solid-state nanopores with L/D = 0.13 was obtained.
The identification accuracy for a single molecule may be as low as 70%. This was the accuracy obtained from one current–time waveform. In solution, the influenza virus A subtype was identified with an F-measure accuracy of 0.72 in one pulse. When 10 or more pulses of influenza virus A subtype were detected, the identification accuracy for the influenza virus A subtype was represented by F-measure >0.90. A method for improving the identification accuracy by using a plurality of pulses in this way is called an assembly method. Therefore, the higher the identification accuracy for a single molecule, the higher the accuracy of 99% can be achieved with a low number of readings.
Outlook
Combining single-molecule electrical measurement and machine learning enabled high-precision identification of analytes that could not be identified with high accuracy by conventional methods. This allowed quantification of the recognition ability of the recognition molecule. Additionally, this combination provided design criteria for the interelectrode distance and L/D suitable for high-precision identification. Combining single-molecule electrical measurement and machine learning enabled both high-precision discrimination and quantitative evaluation of phenomena and structures that were previously difficult to quantitatively evaluate.
Multiphysics simulation enabled investigation of the flow dynamics of single viruses and bacteria. Combining single-molecule electrical measurement and machine learning enabled physical and chemical discoveries. Developing multiphysics simulation enabled physical and chemical interpretation of features.
Combining single-molecule electrical measurement and machine learning enables quantitative evaluation of the number of molecular species. This is because it is established that the number of pulses of a certain molecule = the number of its molecules. Certain types of molecules can be counted digitally. There are no restrictions on the molecular species that machine learning can handle. Therefore, digital analytical chemistry that determines the molecular species and number of molecules in a solution is possible.
Digital analytical chemistry enables early diagnosis of cancer, virus testing before onset, and bacterial testing. Digital analytical chemistry of chemical reactions allows comprehensive analysis of products. Products that have been missed in previous analyses can be discovered. Digital analytical chemistry has the potential to search for “a needle in a haystack”.
The machine learning methods introduced in this min-review contain features that have been defined by analysts. This enables physical or chemical interpretation of the results obtained from machine learning. Despite this, the arbitrariness of the feature remains. Deep learning eliminates the arbitrary bias that is inadvertently introduced by the analyst because the features have been generated by an algorithm. However, phenomenological interpretation of the results obtained is difficult. Furthermore, when compared with the machine learning methods introduced in this mini-review, deep learning requires anywhere from 10- to 100-fold more data points. Therefore, after a better understanding of the results obtained from the machine learning method has been achieved, it is appropriate to apply deep learning during the practical application of the single-molecule measurement method.
Acknowledgments
This research was supported by KAKENHI Grant No. 19H00852 and JST-CREST Grant N. JPMJCR1666, Japan.
Biography
Masateru Taniguchi obtained a Ph.D. in Chemistry from Kyoto University in 2001. He is a Professor of Osaka University. His current interests include single-molecule science and single-molecule technologies.
The author declares no competing financial interest.
References
- Elhoseny M.; Ramirez-Gonzalez G.; Abu-Elnasr O. M.; Shawkat S. A.; Arunkumar N.; Farouk A. Secure medical data transmission model for IoT-based healthcare systems. IEEE Access 2018, 6, 20596–20608. 10.1109/ACCESS.2018.2817615. [DOI] [Google Scholar]
- Rateni G.; Dario P.; Cavallo F. Smartphone-based food diagnostic technologies: A review. Sensors 2017, 17, 1453. 10.3390/s17061453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branton D.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ventra M.; Taniguchi M. Decoding DNA, RNA and peptides with quantum tunnelling. Nat. Nanotechnol. 2016, 11, 117–126. 10.1038/nnano.2015.320. [DOI] [PubMed] [Google Scholar]
- Restrepo-Perez L.; Joo C.; Dekker C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 2018, 13, 786–796. 10.1038/s41565-018-0236-6. [DOI] [PubMed] [Google Scholar]
- Dekker C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- Heerema S. J.; Dekker C. Graphene nanodevices for DNA sequencing. Nat. Nanotechnol. 2016, 11, 127–136. 10.1038/nnano.2015.307. [DOI] [PubMed] [Google Scholar]
- Zhao Y. A.; et al. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat. Nanotechnol. 2014, 9, 466–473. 10.1038/nnano.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S.; et al. Universal readers based on hydrogen bonding or π-π stacking for identification of DNA nucleotides in electron tunnel junctions. ACS Nano 2016, 10, 11304–11316. 10.1021/acsnano.6b06466. [DOI] [PubMed] [Google Scholar]
- Im J.; Sen S.; Lindsay S.; Zhang P. M. Recognition tunneling of canonical and modified RNA nucleotides for their identification with the aid of machine learning. ACS Nano 2018, 12, 7067–7075. 10.1021/acsnano.8b02819. [DOI] [PubMed] [Google Scholar]
- Im J.; Biswas S.; Liu H.; Zhao Y.; Sen S.; Biswas S.; Ashcroft B.; Borges C.; Wang X.; Lindsay S.; Zhang P. Electronic single-molecule identification of carbohydrate isomers by recognition tunnelling. Nat. Commun. 2016, 7, 13868. 10.1038/ncomms13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M.; Ohshiro T.; Komoto Y.; Takaai T.; Yoshida T.; Washio T. High-precision single-molecule identification based on single-molecule information within a noisy matrix. J. Phys. Chem. C 2019, 123, 15867–15873. 10.1021/acs.jpcc.9b03908. [DOI] [Google Scholar]
- Arima A.; et al. Identifying single viruses using biorecognition solid-state nanopores. J. Am. Chem. Soc. 2018, 140, 16834–16841. 10.1021/jacs.8b10854. [DOI] [PubMed] [Google Scholar]
- Arima A.; et al. Selective detections of single-viruses using solid-state nanopores. Sci. Rep. 2018, 8, 16305. 10.1038/s41598-018-34665-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui M.; et al. Identification of individual bacterial cells through the intermolecular interactions with peptide-functionalized solid-state pores. Anal. Chem. 2018, 90, 1511–1515. 10.1021/acs.analchem.7b04950. [DOI] [PubMed] [Google Scholar]
- Tsutsui M.; et al. Discriminating single-bacterial shape using low-aspect-ratio pores. Sci. Rep. 2017, 7, 17371. 10.1038/s41598-017-17443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jethava V.; Martinsson A.; Bhattacharyya C.; Dubhashi D. The Lovasz θ function, SVMs and finding large dense subgraphs. Neural Inf. Proc. Syst. 2012, 1169–1177. [Google Scholar]
- Holmes G.; Donkin A.; Witten I. H. In WEKA: a Machine Learning Workbench, Proc. ANZIIS ’94 - Australian New Zealnd Intelligent Information Systems Conference; 1994; pp 357–361.
- Rodriguez J.J.; Kuncheva L.I.; Alonso C.J. Rotation forest: A new classifier ensemble method. IEEE T. Pattern Anal. 2006, 28, 1619–1630. 10.1109/TPAMI.2006.211. [DOI] [PubMed] [Google Scholar]
- Elkan C.; Noto K.. Learning classifiers from only positive and unlabeled data. In Proc. The 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; ACM: Las Vegas, Nevada, USA, 2008; pp 213–220.
- Feng J. D.; Liu K.; Bulushev R. D.; Khlybov S.; Dumcenco D.; Kis A.; Radenovic A. Identification of single nucleotides in MoS2 nanopores. Nat. Nanotechnol. 2015, 10, 1070–1076. 10.1038/nnano.2015.219. [DOI] [PubMed] [Google Scholar]
- He Y. H.; Tsutsui M.; Fan C.; Taniguchi M.; Kawai T. Gate manipulation of DNA capture into nanopores. ACS Nano 2011, 5, 8391–8397. 10.1021/nn203186c. [DOI] [PubMed] [Google Scholar]
- He Y. H.; Tsutsui M.; Fan C.; Taniguchi M.; Kawai T. Controlling DNA translocation through gate modulation of nanopore wall surface charges. ACS Nano 2011, 5, 5509–5518. 10.1021/nn201883b. [DOI] [PubMed] [Google Scholar]