Abstract
Background
This study aimed to explore the association of long non‐coding RNA nuclear‐enriched abundant transcript 1 (lncRNA NEAT1) with exacerbation risk, lung function, and inflammatory cytokines in asthma.
Methods
A total of 170 patients with asthma in exacerbation, 170 patients with asthma in remission, and 170 healthy controls (HCs) were enrolled, and their plasma samples were collected. The expressions of lncRNA NEAT1 and microRNA‐124 (miRNA‐124) in plasma were detected by real‐time quantitative polymerase chain reaction; inflammatory cytokines in plasma were measured by the Enzyme‐linked immunosorbent assay (ELISA); and pulmonary ventilation function was detected by examination of forced expiratory volume in 1 second (FEV1) and forced vital capacity (FVC).
Results
LncRNA NEAT1 expression was upregulated in asthma patients in exacerbation compared with HCs and asthma patients in remission, and receiver operating characteristic curve exhibited that it was of good value in distinguishing asthma patients in exacerbation from HCs (AUC: 0.869 (0.830‐0.908)) and asthma patients in remission (AUC: 0.775 (0.724‐0.825)). Furthermore, lncRNA NEAT1 was positively correlated with exacerbation severity, TNF‐α, IL‐1β, and IL‐17, but negatively correlated with IL‐10, FEV1/FVC and FEV1%predicted in asthma patients. Additionally, lncRNA NEAT1 was negatively correlated with miR‐124, and miR‐124 was negatively associated with exacerbation risk, exacerbation severity, and inflammation, but positively associated with lung function in asthma patients.
Conclusion
Circulating lncRNA NEAT1 exhibits potential to be a new biomarker for elevated exacerbation risk and severity of asthma.
Keywords: asthma, exacerbation, lncRNA NEAT1, miR‐124
1. INTRODUCTION
Asthma, a highly prevalent chronic respiratory disease, affects approximately three hundred million people globally.1 Clinically, asthma is characterized by reversible airway obstruction, airway hyper‐responsiveness, and airway inflammation‐related symptoms, such as breathlessness, chest tightness, and dyspnea.2 Asthma exacerbations are acute episodes of asthma symptoms, defined as decreases in expiratory airflows and objective measures of lung function. Once severe exacerbation occurs, patients experience decline of lung function as well as severe airflow obstruction, which contributes to more frequent severe exacerbation in the future.3, 4 Current effective pharmacologic interventions for reducing the asthma symptoms consist of inhaled corticosteroids, long‐acting beta‐agonists, leukotriene modifiers, and anti‐IgE therapy; however, still a large number of asthmatics experience exacerbations despite guideline‐directed treatment.5, 6, 7 Therefore, discovery of potential biomarkers is necessary to identify the risk of asthma exacerbation, which would improve disease control and optimizes the treatment outcomes.
Recently, increasing evidence demonstrates the implication of long non‐coding RNA (lncRNA) in the onset and development of respiratory diseases and inflammation‐related diseases.8, 9 LncRNA nuclear‐enriched abundant transcript 1 (NEAT1) is encoded by the NEAT1 gene and promotes the activation of several inflammasomes in diverse diseases, including sepsis, colitis, and lupus.10, 11, 12 Furthermore, one study elucidates that lncRNA NEAT1 promotes myocardial ischemia‐reperfusion injury via increasing reactive oxygen species (ROS) level, and ROS is an effective predictor for severe exacerbations of asthma; thus, lncRNA NEAT1 is speculated to be involved in the pathology of asthma exacerbation as well.13 Additionally, microRNAs (miRNAs) are a group of small non‐coding RNAs, and existing studies confirm the interactions between lncRNA NEAT1 and miR‐124 through a competing endogenous RNA (ceRNA) regulatory network.14 And mechanically, miR‐124 contributes to development and maintenance of anti‐inflammatory phenotype in lung macrophages of asthma.15 Considering the previous evidence, we hypothesized that lncRNA NEAT1 might play an important role in the development and exacerbation of asthma, while there is still no related research reported currently. Thus, we conducted this study to explore the association of circulating lncRNA NEAT1 expression with exacerbation risk, lung function, and inflammatory cytokines in asthma.
2. MATERIALS AND METHODS
2.1. Participants
From January 2017 to December 2018, 170 patients with asthma in exacerbation, 170 patients with asthma in remission, and 170 healthy controls (HCs) were enrolled at our hospital in this case‐control study. The inclusion criteria for patients with asthma in exacerbation were as follows: (a) confirmed diagnosis of asthma in accordance with the Global Initiative for Asthma (GINA) guideline (2016)16; (b) presenting with an acute worsening in symptoms including breathlessness, chest tightness, dyspnea, and increased cough; and (c) age more than 18 years. And the following patients were excluded: (a) cardiac asthma, bronchogenic carcinoma, endometrial lesions of trachea, or allergic pulmonary infiltration; (b) suffering from autoimmune disorders, hematological diseases, or serious infections; (c) complicated with malignancies or solid tumors; and (d) pregnant or lactating women. The inclusion criteria for patients with asthma in remission were as follows: (a) confirmed diagnosis of asthma in accordance with the Global Initiative for Asthma (GINA) guideline (2016)16; (b) presenting with clinical remission status, which was defined as after treatment or without treatment, the symptoms and signs disappeared, and the pulmonary function recovered to the pre‐acute level and maintained for at least 3 months; and (c) age more than 18 years. And the exclusion criteria for patients with asthma in remission were as same as the patients with asthma in exacerbation, including (a) cardiac asthma, bronchogenic carcinoma, endometrial lesions of trachea, or allergic pulmonary infiltration, (b) suffering from autoimmune disorders, hematological diseases, or serious infections, (c) complicated with malignancies or solid tumors, and (d) pregnant or lactating women. In addition, HCs were recruited from Health Examination Center of our hospital, when they were undergoing health examination at the same period. All of them were presenting with normal lung function and had no history of asthma, allergic diseases, autoimmune disorders, hematological diseases, serious infections, or malignancies.
2.2. Ethics statement
The Institutional Review Board of our hospital approved this study, and all participants provided written informed consents before enrollment.
2.3. Data collection
After collection of written informed consents, basic characteristics of all participants were recorded including age, gender, family history of asthma, and the biochemical index (immune globulin E (IgE)). And the measurement of pulmonary ventilation function was also performed for all participants, which included forced expiratory volume in 1 second (FEV1) and forced vital capacity (FVC), and then, the values of FEV1/FVC and FEV1%predicted were calculated. Besides, the disease severity of the patients with asthma in exacerbation was evaluated according to The Global Strategy for Asthma Management and Prevention (updated 2010) (www. ginasthma.org) (details shown in the Table S1) and documented on the hospital admission.
2.4. Sample collection
On the enrollment, blood samples were collected from all eligible participants using vacuum blood collection tubes containing ethylene diamine tetraacetic acid (EDTA). The collected samples were immediately centrifuged at 1600 g for 15 minutes (4°C) to obtain supernatant; then, the supernatant was centrifuged one more time at 16 000 g for 10 minutes (4°C) to isolate plasma. Finally, the isolated plasma was stored at −80°C until analysis.
2.5. Real‐time quantitative polymerase chain reaction (RT‐qPCR)
The expression of lncRNA NEAT1 and miR‐124 (which was miR‐124‐5p in detail with accession number MIMAT0004591) in the plasma of participants was detected by the RT‐qPCR. The total RNA was extracted from plasma using TRIzol™ Reagent (Thermo Fisher Scientific) and then reversely transcribed to cDNA using PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara). Following that, RT‐qPCR was performed using SYBR® Premix DimerEraser™ (Takara) to quantify lncRNA NEAT1 (GAPDH as internal reference) and miR‐124 expressions (U6 as internal reference). The procedures were carried out as follows: first, 5 minutes at 95 degrees centigrade, 40 cycles of PCR then followed by standard conditions with 15 seconds denaturation at 95 degrees centigrade, and next elongation for 1 minute at 60 degrees centigrade. And the result was calculated using 2−△△Ct method. Primers were as follows: lncRNA NEAT1, forward (5ʹ‐>3ʹ): TGTCCCTCGGCTATGTCAGA, reverse (5ʹ‐>3ʹ): GAGGGGACGTGTTTCCTGAG; miR‐124, forward (5ʹ‐>3ʹ): ACACTCCAGCTGGGCGTGTTCACAGCGGACCT, reverse (5ʹ‐>3ʹ): TGTCGTGGAGTCGGCAATTC; GAPDH, forward (5ʹ‐>3ʹ): GAGTCCACTGGCGTCTTCAC, reverse (5ʹ‐>3ʹ): ATCTTGAGGCTGTTGTCATACTTCT; U6, forward (5ʹ‐>3ʹ): CTCGCTTCGGCAGCACATATACTA, reverse (5ʹ‐>3ʹ): ACGAATTTGCGTGTCATCCTTGC.
2.6. Enzyme‐linked immunosorbent assay
The inflammatory cytokines including tumor necrosis factor alpha (TNF‐α), interleukin‐1 beta (IL‐1β), interleukin‐10 (IL‐10), and interleukin‐17 (IL‐17) in the plasma were determined by Enzyme‐linked immunosorbent assay (ELISA) with ELISA kits, including Human TNF‐α ELISA Kit (Thomas, KHC3014), Human IL‐1β ELISA Kit (Thomas, BMS224HS), Human IL‐10 ELISA Kit (Abcam, ab100549), and Human IL‐17 ELISA Kit (Abcam, ab83707), which were ready‐to‐use, according to the instructions of manufacturer. In brief, firstly, samples or standards were added to the 96‐well plates, followed by the antibody mix. After incubation, the wells were washed to remove unbound material. TMB substrate (tetramethylbenzidine, TMB) was added, generating blue coloration. This reaction was then stopped by addition of Stop Solution completing any color change from blue to yellow. Signal was generated proportionally to the amount of bound analyte (Biotek), and the intensity was measured at 450 nm. Each reaction was run in triplicate by the same operator. Standard curves were prepared before the antibody reaction, and the standard curve range of Human TNF‐α ELISA Kit (Thermo, KHC3014), Human IL‐1β ELISA Kit (Thermo, BMS224HS), Human IL‐10 ELISA Kit (Abcam, ab100549), and Human IL‐17 ELISA Kit (Abcam, ab83707) were 0.5‐32pg/mL, 0.16 ‐10 pg/mL, 2.34‐150 pg/mL, and 3.12‐100 pg/mL, respectively.
2.7. Statistical analysis
Statistical data processing was carried out using SPSS 20.0 software (IBM Corp.), and all figures were made by GraphPad Prism 7.01 software (GraphPad Software Inc). Data were described as the mean and standard deviation (SD), median and interquartile range (IQR), or number (percentage) according to the distribution of data. Difference among three groups was determined by one‐way analysis of variance (ANOVA) and Kruskal‐Wallis test followed by the Dunn's multiple comparison test or chi‐square test. Correlation between variables was assessed by the Spearman's rank correlation test. The receiver operating characteristic (ROC) curve, the area under the curve (AUC), and 95% confidence interval (CI) were used to assess the performance of variables in distinguishing different subjects. All tests were presented as two‐sided, and P value < .05 was considered statistically significant.
3. RESULTS
3.1. Baseline characteristics
The mean age was 37.6 ± 7.9 years of patients with asthma in exacerbation, 36.3 ± 8.0 years of patients with asthma in remission, and 36.4 ± 6.9 years of HCs (Table 1). There were 88 (51.8%) males and 82 (48.2%) females in patients with asthma in exacerbation, 99 (58.2%) males and 71 (41.8%) females in patients with asthma in remission, and 98 (57.6%) males and 72 (42.4%) females in the HCs. No difference of age or gender was observed among patients with asthma in exacerbation, patients with asthma in remission, and HCs (both P > .05). However, family history of asthma, IgE, lung function, and inflammatory cytokines were different among the three groups. The detailed information of participants' characteristics was listed in Table 1.
Table 1.
Characteristics of participants
| Items |
Asthma in exacerbation (N = 170) |
Asthma in remission (N = 170) |
HCs (N = 170) |
P value |
|---|---|---|---|---|
| Age (y), mean ± SD | 37.6 ± 7.9 | 36.3 ± 8.0 | 36.4 ± 6.9 | .220 |
| Gender, No. (%) | ||||
| Male | 88 (51.8) | 99 (58.2) | 98 (57.6) | .414 |
| Female | 82 (48.2) | 71 (41.8) | 72 (42.4) | |
| Family history of asthma, No. (%) | ||||
| No | 120 (70.6) | 135 (79.4) | 146 (85.9) | .003 |
| Yes | 50 (29.4) | 35 (20.6) | 24 (14.1) | |
| Biochemical indexes, median (IQR) | ||||
| IgE (IU/mL) | 225.9 (147.1‐342.9) | 76.5 (52.9‐120.5) | 30.9 (16.4‐47.0) | <.001 |
| Lung function indexes, median (IQR) | ||||
| FEV1/FVC (%) | 66.6 ± 6.6 | 80.1 ± 4.6 | 83.2 ± 4.7 | <.001 |
| FEV1%predicted (%) | 76.0 ± 5.8 | 84.6 ± 5.0 | 100.2 ± 6.9 | <.001 |
| Inflammatory cytokines, median (IQR) | ||||
| TNF‐α (pg/mL) | 50.9 (33.7‐78.4) | 20.46 (10.10‐30.84) | 12.97 (7.29‐18.87) | <.001 |
| IL‐1β (pg/mL) | 3.6 (2.1‐4.8) | 1.3 (0.6‐2.1) | 0.8 (0.5‐1.2) | <.001 |
| IL‐10 (pg/mL) | 7.6 (6.7‐10.6) | 15.1 (10.6‐24.4) | 18.9 (13.3‐29.1) | <.001 |
| IL‐17 (pg/mL) | 42.4 (22.5‐88.3) | 16.0 (5.9‐33.8) | 10.4 (5.2‐16.1) | <.001 |
| Exacerbation severity, No. (%) | ||||
| Mild | 42 (24.7) | – | – | – |
| Moderate | 89 (52.4) | – | – | – |
| Severe | 39 (22.9) | – | – | – |
| Medicine use within 3 mo, No. (%) | – | |||
| ICS | – | 103 (60.6) | – | – |
| ICS + LTRA | – | 38 (22.4) | – | – |
| ICS + β2‐agonists | 60 (35.3) | – | – | – |
| ICS + β2‐agonists + theophylline | 36 (21.2) | – | – | – |
| ICS + β2‐agonists + anticholinergics | 48 (28.2) | – | – | – |
| ICS + β2‐agonists + anticholinergics +anti‐IgE antibody) | 26 (15.3) | – | – | – |
Comparisons were determined by one‐way ANOVA, chi‐square test, or Kruskal‐Wallis H rank sum test.
Abbreviations: FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; HCs, healthy controls; ICS, inhaled corticosteroids; IgE, immune globulin E; IL, interleukin; IQR, interquartile range; LTRA, leukotriene receptor antagonists; SD, standard deviation; TNF, tumor necrosis factor.
3.2. Correlation of lncRNA NEAT1 with exacerbation risk of asthma
The lncRNA NEAT1 relative expression was increased in patients with asthma in exacerbation compared with patients with asthma in remission (P < .001) and HCs (P < .001) (Figure 1A). And the lncRNA NEAT1 relative expression was increased in patients with asthma in remission compared with HCs (P < .001). ROC curve presented that lncRNA NEAT1 relative expression was of good value in distinguishing patients with asthma in exacerbation from HCs (AUC: 0.869 (0.830‐0.908)) (Figure 1B) and patients with asthma in remission (AUC: 0.775 (0.724‐0.825)) (Figure 1C). It could also distinguish patients with asthma in remission from HCs (AUC: 0.631 (0.572‐0.690)) (Figure 1D). These data indicated that lncRNA NEAT1 could predict exacerbation risk of asthma.
Figure 1.
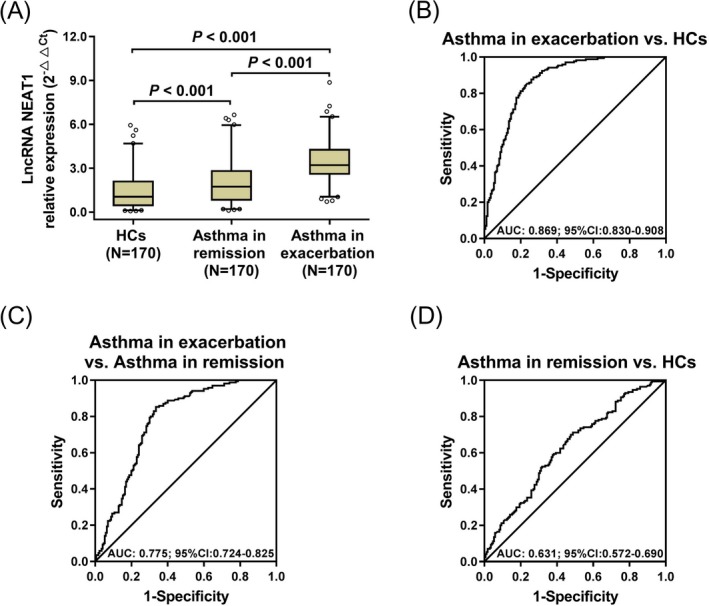
LncRNA NEAT1 relative expression in asthma. The comparison of lncRNA NEAT1 relative expression among HCs, patients with asthma in remission, and patients with asthma in exacerbation (A). The performance of lncRNA NEAT1 in distinguishing asthma in exacerbation from HCs (B), asthma in exacerbation from asthma in remission (C), and asthma in remission from HCs (D). Comparisons among three groups were conducted by Kruskal‐Wallis test followed by the Dunn's multiple comparisons test. And ROC curves were used to assess the ability of lncRNA NEAT1 in distinguishing HCs, patients with asthma in remission, and patients with asthma in exacerbation. P < .05 was considered significant. LncRNA NEAT1, long non‐coding RNA nuclear‐enriched abundant transcript 1; HCs, healthy controls; ROC, receiver operating characteristic
3.3. Correlation of lncRNA NEAT1 with exacerbation severity in asthma patients with exacerbation
The lncRNA NEAT1 relative expression was different among patients with asthma in mild (n = 42), moderate (n = 89), and severe (n = 39) exacerbation severity (P < .001) (Figure 2). The lncRNA NEAT1 relative expression was the highest in patients with severe exacerbation, followed by the patients with moderate exacerbation and the lowest in patients with mild exacerbation. These data suggested that lncRNA NEAT1 was positively correlated with exacerbation severity in patients with asthma in exacerbation.
Figure 2.
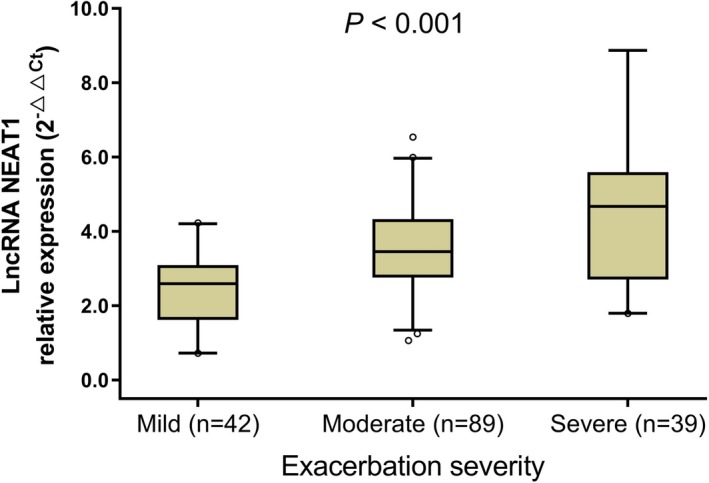
LncRNA NEAT1 relative expression in patients with different exacerbation severity. Correlation of lncRNA NEAT1 relative expression with the asthma exacerbation severity (mild, moderate, severe) was analyzed by Spearman's rank correlation test. P < .05 was considered significant. LncRNA NEAT1, long non‐coding RNA nuclear‐enriched abundant transcript 1
3.4. Correlation of lncRNA NEAT1 with clinical characteristics
In patients with asthma in exacerbation, lncRNA NEAT1 relative expression was positively correlated with TNF‐α (r = .429, P < .001), IL‐1β (r = .342, P < .001), and IL‐17 (r = .416, P < .001), but negatively correlated with FEV1/FVC (r = −.356, P < .001), FEV1%predicted (r = −.273, P < .001), and IL‐10 (r = −.298, P < .001) (Table 2). In patients with asthma in remission, lncRNA NEAT1 relative expression was positively correlated with TNF‐α (r = .253, P = .001), IL‐1β (r = .221, P = .004), and IL‐17 (r = .330, P < .001), but negatively correlated with FEV1/FVC (r = −.170, P = .026) and IL‐10 (r = −.207, P = .007). In HCs, lncRNA NEAT1 was positively correlated with TNF‐α (r = .212, P = .006), IL‐1β (r = .167, P = .029), and IL‐17 (r = .213, P = .005), but negatively correlated with IL‐10 (r = −.269, P < .001). These data suggested that lncRNA NEAT1 positively correlated with poor lung function and higher inflammation, especially in patients with asthma in exacerbation.
Table 2.
Correlation of lncRNA NEAT1 relative expression with clinical features
| Items | LncRNA NEAT1 relative expression | |||||
|---|---|---|---|---|---|---|
| Asthma in exacerbation | Asthma in remission | HCs | ||||
| P value | r | P value | r | P value | r | |
| IgE | .182 | .103 | .465 | .056 | .384 | −.067 |
| FEV1/FVC | <.001 | −.356 | .026 | −.170 | .768 | .023 |
| FEV1%predicted | <.001 | −.273 | .723 | .027 | .616 | .039 |
| TNF‐α | <.001 | .429 | .001 | .253 | .006 | .212 |
| IL‐1β | <.001 | .342 | .004 | .221 | .029 | .167 |
| IL‐10 | <.001 | −.298 | .007 | −.207 | <.001 | −.269 |
| IL‐17 | <.001 | .416 | <.001 | .330 | .005 | .213 |
Correlations were determined by Spearman's rank correlation test.
Abbreviations: FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; HCs, healthy controls; IgE, immune globulin E; IL, interleukin; LncRNA, long non‐coding RNA; r, correlation coefficient; TNF, tumor necrosis factor.
3.5. Correlation of lncRNA NEAT1 and miR‐124
LncRNA NEAT1 relative expression was negatively correlated with miR‐124 relative expression in patients with asthma in exacerbation (r = −.384, P < .001) (Figure 3A), patients with asthma in remission (r = −.256, P = .001) (Figure 3B), and HCs (r = −.295, P < .001) (Figure 3C). This indicated that lncRNA NEAT1 was negatively correlated with miR‐124 especially in patients with asthma in exacerbation. MiR‐124 relative expression was lower in patients with asthma in exacerbation compared with patients with asthma in remission (P < .001) and HCs (P < .001); however, it was similar between patients with asthma in remission and HCs (P = .187) (Figure 3D). ROC curve exhibited that miR‐124 was of good value in distinguishing patients with asthma in exacerbation from HCs (AUC: 0.813 (0.769‐0.857)) (Figure 3E) and patients with asthma in remission (AUC: 0.759 (0.746‐0.843)) (Figure 3F), but it could not differentiate patients with asthma in remission from HCs (AUC: 0.578 (0.516‐0.641) (Figure 3G). These data suggested that lncRNA NEAT1 might contribute to the asthma exacerbation through the interaction with miR‐124.
Figure 3.
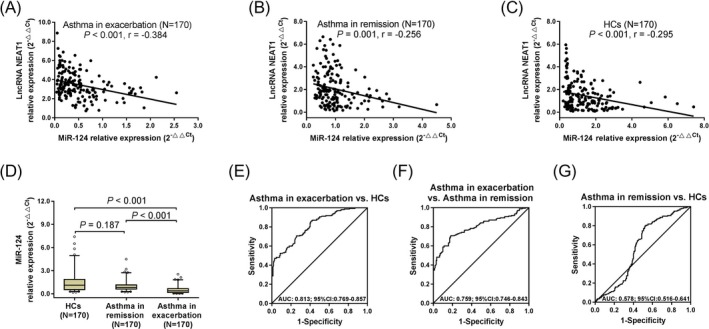
Correlation of lncRNA NEAT1 and miR‐124 in asthma. The correlations of lncRNA NEAT1 and miR‐124 in patients with asthma in exacerbation (A), patients with asthma in remission (B), and HCs (C). The comparisons of miR‐124 relative expression among HCs, patients with asthma in remission, and patients with asthma in exacerbation (D). The performances of miR‐124 in distinguishing patients with asthma in exacerbation from HCs (E), patients with asthma in exacerbation from patients with asthma in remission (F), and patients with asthma in remission from HCs (G). The correlations of lncRNA NEAT1 and miR‐124 were conducted by Spearman's rank correlation test. Comparisons among three groups were conducted by Kruskal‐Wallis test followed by the Dunn's multiple comparisons test. And ROC curves were used to assess the ability of miR‐124 in distinguishing HCs, patients with asthma in remission, and patients with asthma in exacerbation. P < .05 was considered significant. LncRNA NEAT1, long non‐coding RNA nuclear‐enriched abundant transcript 1; miR‐124, microRNA‐124; HCs, healthy controls; ROC, receiver operating characteristic
3.6. Correlation of miR‐124 with clinical characteristics
In patients with asthma in exacerbation, miR‐124 relative expression was positively correlated with FEV1/FVC (r = .503, P < .001), FEV1% predicted (r = .509, P < .001), and IL‐10 (r = .260, P = .001) but negatively correlated with TNF‐α (r = −.387, P < .001), IL‐1β (r = −.348, P < .001), IL‐17 (r = −.244, P = .001), and exacerbation severity (r = −.665, P < .001) (Table 3). In patients with asthma in remission, miR‐124 relative expression was positively correlated with IL‐10 (r = .204, P = .008), but negatively correlated with IgE (r = −.213, P = .005), TNF‐α (r = −.171, P = .026), and IL‐17 (r = −.330, P < .001). And in HCs, miR‐124 relative expression was positively correlated with FEV1/FVC (r = .221, P = .004) and IL‐10 (r = .222, P = .004), but negatively correlated with TNF‐α (r = −.220, P = .004), IL‐1β (r = −.190, P = .013), and IL‐17 (r = −.297, P < .001). Data above suggested that miR‐124 was positively correlated with lung function, but negatively associated with inflammation, especially in patients with asthma in exacerbation. And miR‐124 was negatively associated with exacerbation severity in patients with asthma in exacerbation.
Table 3.
Correlation of miR‐124 relative expression with clinical features
| Items | MiR‐124 relative expression | |||||
|---|---|---|---|---|---|---|
| Asthma in exacerbation | Asthma in remission | HCs | ||||
| P value | r | P value | r | P value | r | |
| IgE | .189 | −.101 | .005 | −.213 | .326 | .076 |
| FEV1/FVC | <.001 | .503 | .579 | −.043 | .004 | .221 |
| FEV1%predicted | <.001 | .509 | .416 | −.063 | .460 | −.057 |
| TNF‐α | <.001 | −.387 | .026 | −.171 | .004 | −.220 |
| IL‐1β | <.001 | −.348 | .126 | −.118 | .013 | −.190 |
| IL‐10 | .001 | .260 | .008 | .204 | .004 | .222 |
| IL‐17 | .001 | −.244 | <.001 | −.330 | <.001 | −.297 |
| Exacerbation severity | <.001 | −.665 | – | – | – | – |
Correlations were determined by Spearman's rank correlation test.
Abbreviations: FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; HCs, healthy controls; IgE, immune globulin E; IL, interleukin; r, correlation coefficient; TNF, tumor necrosis factor.
3.7. Correlation of medications with lncRNA‐NEAT1, miR‐124, and lncRNA NEAT1/miR‐124 axis
In patients with asthma in remission, there was no difference of miR‐124 (P = .216), lncRNA NEAT1 (P = .741), and lncRNA NEAT1/miR‐124 axis (P = .756) expressions among patients without use of medicine and the patients who had medication of ICS and ICS + LTRA (Table S2). As for in patients with asthma in exacerbation, there was also no difference of miR‐124 (P = .964), lncRNA NEAT1 (P = .580), and lncRNA NEAT1/miR‐124 axis (P = .986) expressions among patients with medication of ICS + β2‐agonists, ICS + β2‐agonists + theophylline, ICS + β2‐agonists + anticholinergics, and ICS + β2‐agonists + anticholinergics+anti‐IgE antibody. Data above indicated that medications did not affect lncRNA‐NEAT1, miR‐124, and lncRNA NEAT1/miR‐124 axis.
4. DISCUSSION
In the present study, lncRNA NEAT1 was of good value in differentiating patients with asthma in exacerbation from patients with asthma in remission as well as HCs, especially in predicting asthma exacerbation risk, and it was positively associated with exacerbation severity in asthma patients with exacerbation. Furthermore, it was positively correlated with pro‐inflammatory cytokines, while negatively correlated with anti‐inflammatory cytokine and lung function in asthma patients. Meanwhile, lncRNA NEAT1 was negatively correlated with miR‐124, and miR‐124 was associated with decreased risk of asthma exacerbation, reduced exacerbation severity, improved lung function, and less inflammation.
Recent evidence indicates that lncRNA NEAT1 plays an important role in the transcriptional regulation of cytokines (such as: interleukin‐8 (IL‐8)) in a viral infection, and it stimulates the activation of several inflammasomes (such as NLRP3, NLRC4, and AIM2 inflammasomes), which leads to immune response in immunoinflammatory diseases.10, 17 And the involvement of lncRNA NEAT1 in immune system has been explored in several diseases including sepsis, systemic lupus erythematosus (SLE), mild dengue, and herpes simples.17, 18, 19 For example, lncRNA NEAT1 expression in peripheral blood mononuclear cells is remarkably increased in sepsis patients than that in HCs, and ROC curve illustrates that lncRNA NEAT1 is of good value in predicting sepsis risk with AUC 0.851 (0.812‐0.935).20 In addition, upregulation of lncRNA NEAT1 promotes ROS production and endoplasmic reticulum (ER) stress‐regulated cardiomyocyte apoptosis, aggravating the severity of ischemia‐reperfusion injury.21, 22 Mechanically, increased serum level of ROS is a risk factor for high degree of airway obstruction, worse immune response, and asthma exacerbation, and ER stress induces the abnormal apoptosis as well as dysfunction of airway epithelial, which suggests the promoting role of lncRNA NEAT1 for the higher risk of exacerbation in asthma.22 Therefore, we deduced that lncRNA NEAT1 might be involved in the asthma exacerbation. However, no study to date has been conducted. In our study, lncRNA NEAT1 expression was notably higher in patients with asthma in exacerbation compared with patients with asthma in remission and HCs. ROC curves revealed a good predictive value of lncRNA NEAT1 for asthma exacerbation risk, and it was positively correlated with exacerbation severity. The possible reasons might be that (a) LncRNA NEAT1 might enhance the airway epithelial fibrosis and injury via promoting oxidative stress and ER stress, resulting in airway hyperplasia and airway narrowing in a longer period, thereby increasing the risk of asthma exacerbation.21, 23 (b) Considering lncRNA NEAT1 was of pro‐inflammatory biological property, it might elevate the expression of pro‐inflammatory cytokines and stimulate inflammatory responses, thus increasing susceptibility to asthma exacerbation, which was validated by analyzing association of lncRNA NEAT1 with inflammatory cytokines in our study.10
LncRNA NEAT1 is reported to be a potential contributor to the elevated secretion of diverse cytokines in various diseases.12, 20, 24 For example, lncRNA NEAT1 is upregulated and its overexpression is correlated with the elevated expression of pro‐inflammatory chemokines and cytokines (IL‐1β, IL‐6, CXCL10, etc), increased clinical disease activity, and higher disease severity in patients with SLE.12 However, the association of lncRNA NEAT1 with inflammatory cytokines has not been explored in asthma yet; thus, we conducted related analysis in patients with asthma and observed that lncRNA NEAT1 was positively associated with pro‐inflammatory cytokines while negatively correlated with anti‐inflammatory cytokine and lung function in patients with asthma. The possible reasons might include that (a) expression of lncRNA NEAT1 was activated during the asthma exacerbation, enhancing continuous release of inflammatory factors and cytokines via activating multiple signaling pathways (such as the JNK/ERK MAPK signaling pathways) and inhibiting the anti‐inflammatory miRNAs, accelerating the inflammatory, and leading to the elevated exacerbation severity in patients with asthma. (b) During the acute exacerbation, when the immunity response was abnormally active, and inflammation response was extremely strong, the dysregulation of lncRNA NEAT1 might be aggravated; thus, the association of lncRNA NEAT1 with inflammation and lung function was more obvious during the exacerbation.
The ceRNA hypothesis proposes that numerous lncRNA might serve as molecular sponges for miRNAs to further influence target mRNA expression, which suggests the importance of such interactions in the progression of disease.25 And it is reported that lncRNA NEAT1 facilitates tumorigenesis and development of ovarian cancer cells as well as nasopharyngeal carcinoma via inhibiting miR‐124‐mediated signaling pathway.14 Besides, miR‐124 overexpression results in the downregulation of pro‐inflammatory markers and upregulation of anti‐inflammatory markers in lung residence macrophages, suggesting the anti‐inflammatory role of miR‐124 in asthma exacerbation.15 In order to further understand the possible mechanism of lncRNA NEAT1 in asthma exacerbation, we explored the association of lncRNA NEAT1 with miR‐124 in asthma patients and found that lncRNA NEAT1 expression was negatively associated with miR‐124. Additionally, miR‐124 was negatively correlated with high risk of asthma exacerbation, exacerbation severity, and inflammation, but positively associated with lung function. These data suggested that lncRNA NEAT1 might implicate in the asthma exacerbation through the interaction with miR‐124.
There were some limitations in this study: (a) Patients with cardiac asthma were excluded in our study; therefore, the results might not be suitable for all patients with asthma. (b) The detailed mechanism of lncRNA NEAT1 and miR‐124 in asthma exacerbation was not investigated; thus, further cellular experiments were needed to be conducted in the future. (b) This study was a single‐center, case‐control study, and the patients were from one region; thus, selection bias might exist.
In conclusion, circulating lncRNA NEAT1 exhibits potential to be a new biomarker for elevated exacerbation risk and severity of asthma.
Supporting information
ACKNOWLEDGMENTS
This study was supported by the Clinical Observation of Specific Immunotherapy Combined with Inhalation of Budesonide/Formoterol Aerosol in the Treatment of Respiratory Allergic Diseases (WX17D06).
Li X, Ye S, Lu Y. Long non‐coding RNA NEAT1 overexpression associates with increased exacerbation risk, severity, and inflammation, as well as decreased lung function through the interaction with microRNA‐124 in asthma. J Clin Lab Anal. 2020;34:e23023 10.1002/jcla.23023
Xueying Li and Shenglan Ye contributed equally to this work.
REFERENCES
- 1. Zhang F, Hang J, Zheng B, Su LI, Christiani DC. The changing epidemiology of asthma in Shanghai, China. J Asthma. 2015;52(5):465‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dougherty RH, Fahy JV. Acute exacerbations of asthma: epidemiology, biology and the exacerbation‐prone phenotype. Clin Exp Allergy. 2009;39(2):193‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. National Asthma Education and Prevention Program . Expert panel report 3 (EPR‐3): guidelines for the diagnosis and management of asthma‐summary report 2007. J Allergy Clin Immunol. 2007;120(5 Suppl):S94‐138. [DOI] [PubMed] [Google Scholar]
- 4. Holgate ST. Epithelium dysfunction in asthma. J Allergy Clin Immunol. 2007;120(6):1233‐1244. quiz 1245‐1236. [DOI] [PubMed] [Google Scholar]
- 5. Nannini LJ. Treat to target approach for asthma. J Asthma. 2019;1‐4. 10.1080/02770903.2019.1591443 [DOI] [PubMed] [Google Scholar]
- 6. Kavanagh J, Jackson DJ, Kent BD. Over‐ and under‐diagnosis in asthma. Breathe (Sheff). 2019;15(1):e20‐e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castillo JR, Peters SP, Busse WW. Asthma exacerbations: pathogenesis, prevention, and treatment. J Allergy Clin Immunol Pract. 2017;5(4):918‐927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J, Zhu Y, Wang R. Long noncoding RNAs in respiratory diseases. Histol Histopathol. 2018;33(8):747‐756. [DOI] [PubMed] [Google Scholar]
- 9. Marques‐Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation‐related diseases. FASEB J. 2015;29(9):3595‐3611. [DOI] [PubMed] [Google Scholar]
- 10. Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10(1):1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol. 2019;13(1):46‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang F, Wu L, Qian J, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96‐104. [DOI] [PubMed] [Google Scholar]
- 13. Nakamoto K, Watanabe M, Sada M, et al. Serum reactive oxygen metabolite levels predict severe exacerbations of asthma. PLoS ONE. 2016;11(10):e0164948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng N, Guo Y. Long noncoding RNA NEAT1 promotes nasopharyngeal carcinoma progression through regulation of miR‐124/NF‐kappaB pathway. Onco Targets Ther. 2017;10:5843‐5853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Veremeyko T, Siddiqui S, Sotnikov I, Yung A, Ponomarev ED. IL‐4/IL‐13‐dependent and independent expression of miR‐124 and its contribution to M2 phenotype of monocytic cells in normal conditions and during allergic inflammation. PLoS ONE. 2013;8(12):e81774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Asthma GIf . Global Strategy for Asthma Management and Preventio; 2016.
- 17. Prinz F, Kapeller A, Pichler M, Klec C. The implications of the long non‐coding RNA NEAT1 in non‐cancerous diseases. Int J Mol Sci. 2019;20(3):627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pandey AD, Goswami S, Shukla S, et al. Correlation of altered expression of a long non‐coding RNA, NEAT1, in peripheral blood mononuclear cells with dengue disease progression. J Infect. 2017;75(6):541‐554. [DOI] [PubMed] [Google Scholar]
- 19. Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1‐dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393‐406. [DOI] [PubMed] [Google Scholar]
- 20. Huang S, Qian K, Zhu Y, Huang Z, Luo Q, Qing C. Diagnostic value of the lncRNA NEAT1 in peripheral blood mononuclear cells of patients with sepsis. Dis Markers. 2017;2017:7962836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee HY, Lee GH, Kim HK, Chae HJ. Platycodi Radix and its active compounds ameliorate against house dust mite‐induced allergic airway inflammation and ER stress and ROS by enhancing anti‐oxidation. Food Chem Toxicol. 2019;123:412‐423. [DOI] [PubMed] [Google Scholar]
- 22. Du X‐J, Wei J, Tian D, et al. NEAT1 promotes myocardial ischemia‐reperfusion injury via activating the MAPK signaling pathway. J Cell Physiol. 2019;234(10):18773‐18780. [DOI] [PubMed] [Google Scholar]
- 23. Huang S, Xu Y, Ge X, et al. Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J Cell Physiol. 2019;234(7):11200‐11207. [DOI] [PubMed] [Google Scholar]
- 24. Huang Q, Huang C, Luo Y, He F, Zhang R. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am J Emerg Med. 2018;36(9):1659‐1663. [DOI] [PubMed] [Google Scholar]
- 25. Wang Y, Wang C, Chen C, et al. Long non‐coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation‐resistance through miR‐101‐3p as a competing endogenous RNA mechanism. Oncotarget. 2017;8(41):70156‐70171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


