Influenza A virus (H1N1) is an acute, highly contagious respiratory virus. The use of lactic acid bacteria (LAB) to deliver mucosal vaccines against influenza virus infection is a research hot spot. In this study, two recombinant Lactobacillus plantarum strains expressing hemagglutinin (HA) alone or coexpressing aCD11c-HA to target HA protein to dendritic cells (DCs) by fusion to an anti-CD11c single-chain antibody (aCD11c) were constructed.
KEYWORDS: dendritic cell targeting, Lactobacillus plantarum, cellular immune response
ABSTRACT
Influenza A virus (H1N1) is an acute, highly contagious respiratory virus. The use of lactic acid bacteria (LAB) to deliver mucosal vaccines against influenza virus infection is a research hot spot. In this study, two recombinant Lactobacillus plantarum strains expressing hemagglutinin (HA) alone or coexpressing aCD11c-HA to target HA protein to dendritic cells (DCs) by fusion to an anti-CD11c single-chain antibody (aCD11c) were constructed. The activation of bone marrow dendritic cells (BMDCs) by recombinant strains and the interaction of activated BMDCs and sorted CD4+ or CD8+ T cells were evaluated through flow cytometry in vitro, and cellular supernatants were assessed by using an enzyme-linked immunosorbent assay kit. The results demonstrated that, compared to the HA strain, the aCD11c-HA strain significantly increased the activation of BMDCs and increased the production of CD4+ gamma interferon-positive (IFN-γ+) T cells, CD8+ IFN-γ+ T cells, and IFN-γ in the cell culture supernatant in vitro. Consistent with these results, the aCD11c-HA strain clearly increased the activation and maturation of DCs, the HA-specific responses of CD4+ IFN-γ+ T cells, CD8+ IFN-γ+ T cells, and CD8+ CD107a+ T cells, and the proliferation of T cells in the spleen, finally increasing the levels of specific antibodies and neutralizing antibodies in mice. In addition, the protection of immunized mice was observed after viral infection, as evidenced by improved weight loss, survival, and lung pathology. The adoptive transfer of CD8+ T cells from the aCD11c-HA mice to NOD/Lt-SCID mice resulted in a certain level of protection after influenza virus infection, highlighting the efficacy of the aCD11c targeting strategy.
INTRODUCTION
Influenza A viruses repeatedly cause infectious respiratory tract infections in humans and animals each year, leading to serious illness and even fetal death. The World Health Organization (WHO) predicts that nearly 5 to 10% of young people and 20 to 30% of children become infected with the virus each year. Of these, 3 to 5 million people are seriously threatened by illness, and approximately 290,000 to 650,000 people die annually (1). The swine-origin H1N1 A influenza virus, which was first isolated from humans in North America in 2009, quickly spread around the world and has caused severe economic losses in recent years (2, 3).
Currently, there are inactivated vaccines, attenuated vaccines, subunit vaccines, genetically engineered live vaccines, and liposomal vaccines for influenza virus, whereas most vaccines are administered via the muscle or by subcutaneous injection. Influenza viruses enter the host through the mucosal pathway; therefore, mucosal immunity is crucial, and mucosal delivery vectors have drawn increasing attention recently due to their oral administration route for immunization (4). Influenza virus is an enveloped virus with two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), which are the main antigenic clusters of the virus and the main targets of humoral immunity in humans (5, 6). In antiviral responses, HA is the main target of neutralizing antibodies (7). Therefore, research on mucosal vaccines using HA as a model antigen is crucial for influenza virus.
Lactic acid bacteria (LAB), as a mucosal vaccine vector, can deliver heterogeneous antigens or DNA vaccines to host cells and have become a research hot spot (8, 9). In particular, Lactobacillus plantarum strains can target a series of pathogens, such as Helicobacter felis (10), Mycobacterium tuberculosis (11), and even tumor antigens (12). In addition, recombinant L. plantarum NC8 strains expressing the M2e or HA2 antigen constructed by Yang et al. have a protective effect after H9N2 subtype avian influenza virus (AIV) or H1N1 influenza A virus infection in both mice and chickens (2, 13, 14). Therefore, L. plantarum strains are regarded as a potential oral vaccine delivery vector.
Dendritic cells (DCs), as important antigen-presenting cells, can effectively induce cytotoxic T cell (CTL) responses in antiviral infections. On the surfaces of DCs, there are many receptors, such as DEC205 and CD11c. It has been demonstrated that the host immune response can be significantly improved by the production of desired antigens fused to individual antibodies targeting specific surface markers on DCs. For instance, the expression of a single-chain antibody against DEC205 (scFv-DEC205) in L. plantarum clearly increased the cellular uptake of bacteria, as well as plasmid transfer to DCs (15). A single-chain variable fragment against CD11c (scFv-CD11c) fused to the immunodominant peptide of a retrovirus induces a virus-specific T cell response (16). In addition, we confirmed that the expression of scFv-CD11c in L. plantarum NC8 clearly enhanced the cellular uptake of L. plantarum strains into DCs and improved the delivery efficiency of the plasmid to host cells, demonstrating a prospective strategy for vaccine research (17).
In this study, HA from the H1N1 subtype influenza virus was coexpressed with scFv-CD11c on the surface of L. plantarum, and the immunological effects of DC-targeting strains were evaluated in mice.
RESULTS
Construction of L. plantarum strains expressing HA and aCD11c-HA.
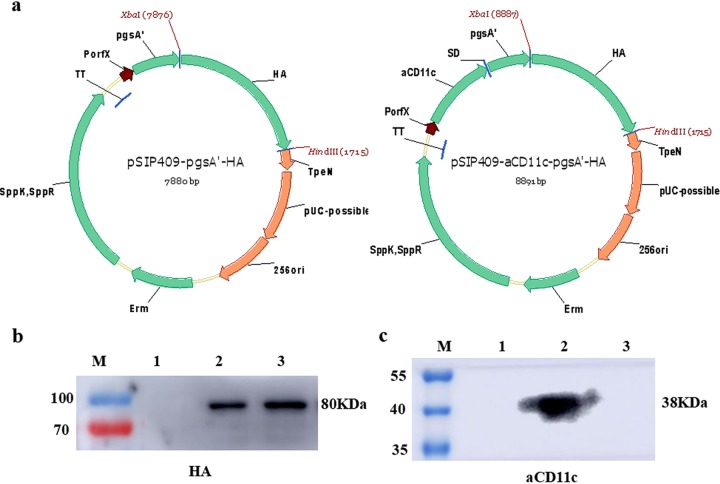
To determine the expression of HA and anti-CD11c single-chain antibody (aCD11c) by the HA and aCD11c-HA strains, cell membrane and cell wall fractions were collected, and Western blotting was performed with HA and His antibody to determine the expression of HA (Fig. 1b) and aCD11c (Fig. 1c), respectively. Using an HA-specific antibody, an 80-kDa band of pgsA′-HA was detected in the aCD11c-HA strain (Fig. 1b, lane 2) and the HA strain (Fig. 1b, lane 3), but no band was detected in the 409p′ strain (Fig. 1b, lane 1). Using anti-His as the primary antibody, a 38-kDa band of aCD11c was detected in the aCD11c-HA strain (Fig. 1c, lane 2), and no bands were detected in the HA strain (Fig. 1c, lane 3) and 409p′ strain (Fig. 1c, lane 1). These results suggest that HA and aCD11c were successfully expressed in the HA strain and aCD11c-HA strain, respectively.
FIG 1.
Structural diagrams of pSIP409-pgsA′-HA and pSIP409-aCD11c-pgsA′-HA plasmids and detection of HA and aCD11c expression. (a) pSIP409-pgsA′-HA and pSIP409-aCD11c-pgsA′-HA were constructed as described in the text. pgsA′, anchoring sequence; HA, heterologous protein (hemagglutinin); aCD11c, scFv-CD11c. The expression of HA (b) and aCD11c (c) in L. plantarum was measured by Western blotting. HA and His were used as primary antibodies in panels b and c, respectively. M, prestained marker; lane 1, 409p′ strain; lane 2, aCD11c-HA strain; lane 3, HA strain.
Activation of BMDCs by the strains expressing HA.
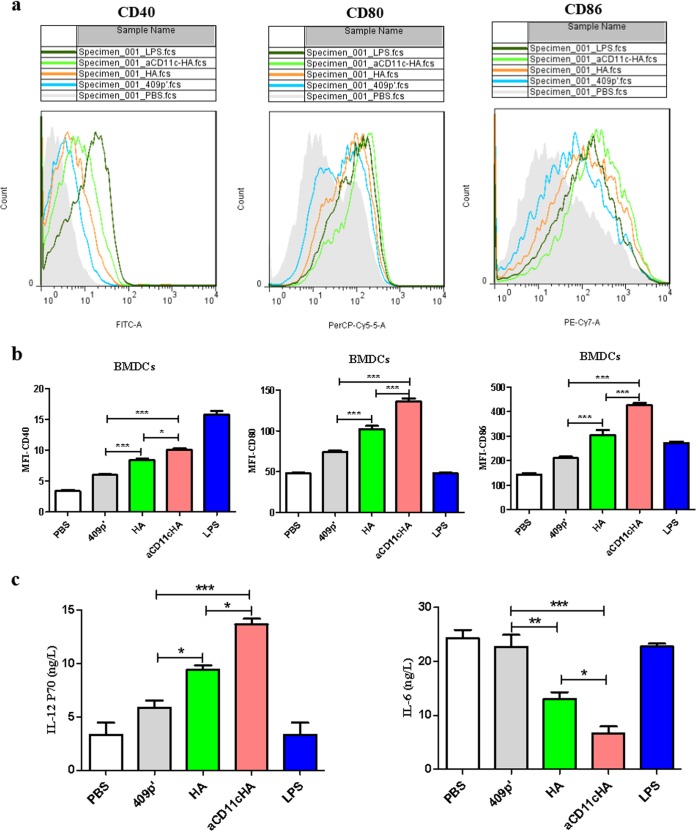
The recombinant strains were cultured with bone marrow dendritic cells (BMDCs) as described in Materials and Methods. The median fluorescence intensities (MFIs) of CD40, CD80, and CD86 surface markers were analyzed at 12 h using flow cytometry (FCM) (Fig. 2a). The results suggested that, compared to the HA strain, the CD11c-HA strain obviously improved the MFIs of CD40 (P < 0.05), CD80 (P < 0.001) and CD86 (P < 0.001; Fig. 2b), indicating that the expression of aCD11c could promote the activation of DCs. We were also interested in the secretion of cytokines caused by the strains; therefore, we collected the culture medium to assess the production of interleukin-6 (IL-6) and IL-12P70. Compared to the HA strain, the aCD11c-HA strain obviously stimulated the production of IL-12P70 and reduced the secretion of IL-6 (P < 0.05; Fig. 2c), indicating that the aCD11c-HA strain may polarize T cells toward the Th1 subtype.
FIG 2.
The aCD11c-HA strain promotes the activation of BMDCs and the secretion of cytokines. (a) The flow peak diagrams from PBS, 409p′, HA, and aCD11c-HA groups are displayed. LPS was used as a positive control. (b) MFIs of CD40, CD80, and CD86 from each group analyzed by FCM. (c) Concentrations of IL-12P70 and IL-6 in cell culture supernatants measured by ELISA. The statistical significance was calculated using one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The strains expressing HA induce T cell activation in vitro.
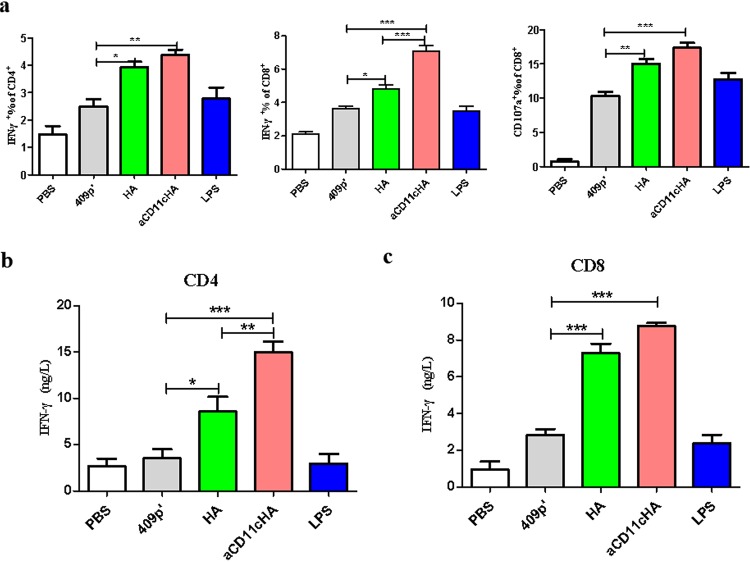
To further detect the influence of BMDCs differentiated by the aCD11c-HA strain on T cells, CD4+ and CD8+ T cells were sorted and then cocultured with the differentiated BMDCs described above. Gamma interferon (IFN-γ)-producing CD4+ T cells and IFN-γ-, CD107a-producing CD8+ T cells were detected through FCM. Furthermore, the cell culture supernatants were collected to detect the secretion of IFN-γ from CD4+ or CD8+ T cells using enzyme-linked immunosorbent assay (ELISA) in vitro. The results demonstrated that, compared to the HA strain, the aCD11c-HA strain obviously stimulated the production of CD8+ IFN-γ+ T cells (P < 0.001; Fig. 3a) and the secretion of IFN-γ from CD4+ T cells (P < 0.01) and CD8+ T cells (P < 0.01; Fig. 3b and c), whereas the production of CD4+ IFN-γ+ T cells was not significantly different between the two strains (Fig. 3a). For CD8+ CD107a+ T cells, the aCD11c-HA (P < 0.001) and HA (P < 0.01) strains, compared to the 409p′ strain, markedly increased the production of CD8+ CD107a+ T cells (Fig. 3a), further indicating that the aCD11c-HA strain stimulates the polarization of T cells toward Th1 subtypes.
FIG 3.
T cell activation in vitro. Sorted CD4+ T cells and CD8+ T cells were cocultured with the differentiated BMDCs for 48 h. (a) IFN-γ-producing CD4+ T cells or IFN-γ-, CD107a-producing CD8+ T cells evaluated by FCM. The cell culture supernatants were used to detect the secretion of IFN-γ from CD4+ (b) and CD8+ T cells (c) by ELISA. The statistical significance was calculated via one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The HA strains promote the activation and maturation of DCs.
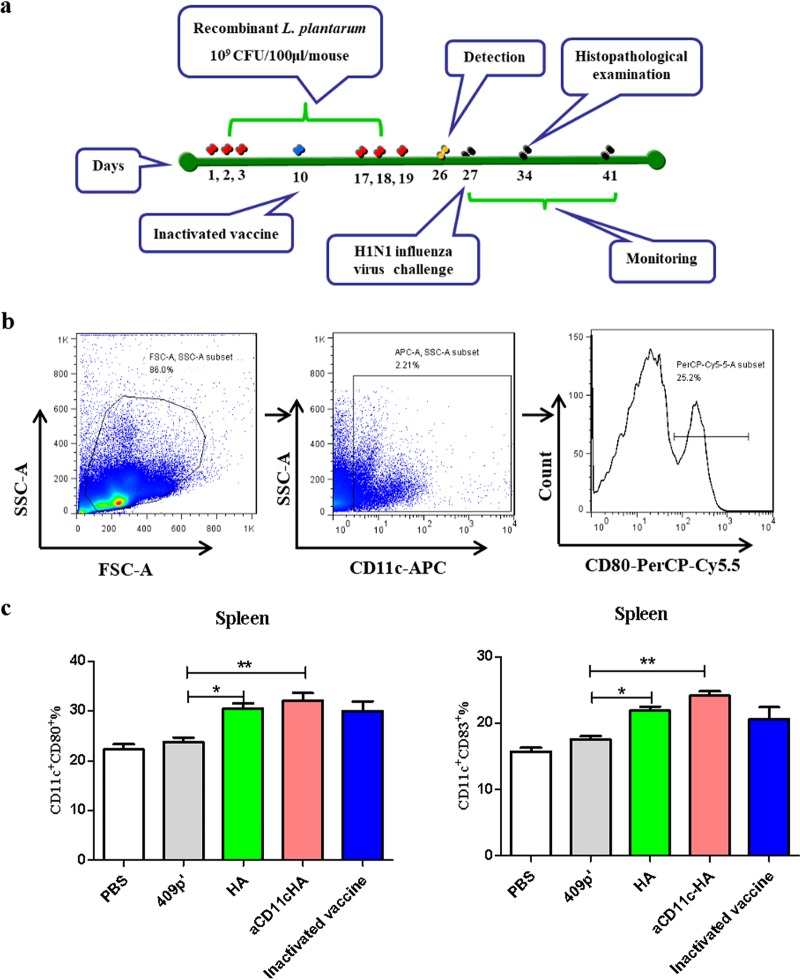
The aCD11c strain promotes the activation and maturation of DCs according to previous research (17). Mice were immunized as outlined in the protocol schematic (Fig. 4a), and the influence of the strains expressing HA on DCs on the model mice was evaluated by FCM (Fig. 4b). The results showed that compared to the 409p′ strain, the aCD11c-HA (P < 0.01) and HA (P < 0.05) strains significantly increased the percentages of CD11c+ CD80+ (Fig. 4c) and CD11c+ CD83+ (Fig. 4c) DCs in the spleen and further stimulated the differentiation and maturation of DCs.
FIG 4.
Immunization protocol and the in vivo differentiation and maturation of DCs. (a) Protocol for mouse immunization. (b and c) Proportions of CD11c-, CD80-, and CD83-expressing splenocytes, as evaluated by FCM. The statistical significance was calculated by using one-way ANOVA (*, P < 0.05; **, P < 0.01).
HA-specific T cells are activated by the HA strains.
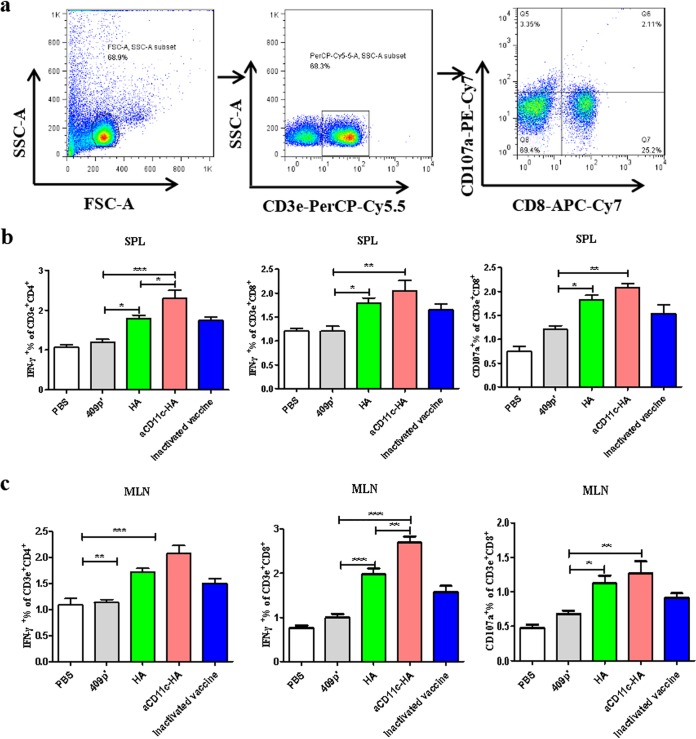
To further assess the HA-specific T cell response induced by the strains, HA protein was used as a stimulant, and specific T cell responses were evaluated using FCM (Fig. 5a). The results suggested that compared to the HA strain, the aCD11c-HA strain clearly improved the response of HA-specific CD4+ IFN-γ+ T cells (P < 0.05) and CD8+ IFN-γ+ T cells (P < 0.05) in the spleen (Fig. 5b), an observation consistent with the in vitro results presented above, indicating that the aCD11c-HA strain polarizes T cells toward Th1 subtypes. Compared to the HA strain, the aCD11c-HA strain clearly enhanced the responses of HA-specific CD8+ IFN-γ+ T cells (P < 0.01) in the mesenteric lymph nodes (MLNs) (Fig. 5c). Furthermore, compared to the 409p′ strain, the aCD11c-HA (P < 0.01) and HA (P < 0.05) strains clearly improved the HA-specific CD8+ CD107a+ T cell response in the spleen and MLN (Fig. 5b and c).
FIG 5.
T cell immune responses in vivo. (a) HA-specific cytokine secretion from splenocytes and MLNs was measured by FCM after oral immunization. The percentages of HA-specific CD4+ IFN-γ+ T cells, CD8+ IFN-γ+ T cells, and CD8+CD107a+ T cells from the spleen (b) and MLN (c) were detected by using FCM. The mean values ± the SEM of three independent experiments are shown. The statistical significance was calculated by using one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The strains expressing HA induce T cell proliferation.
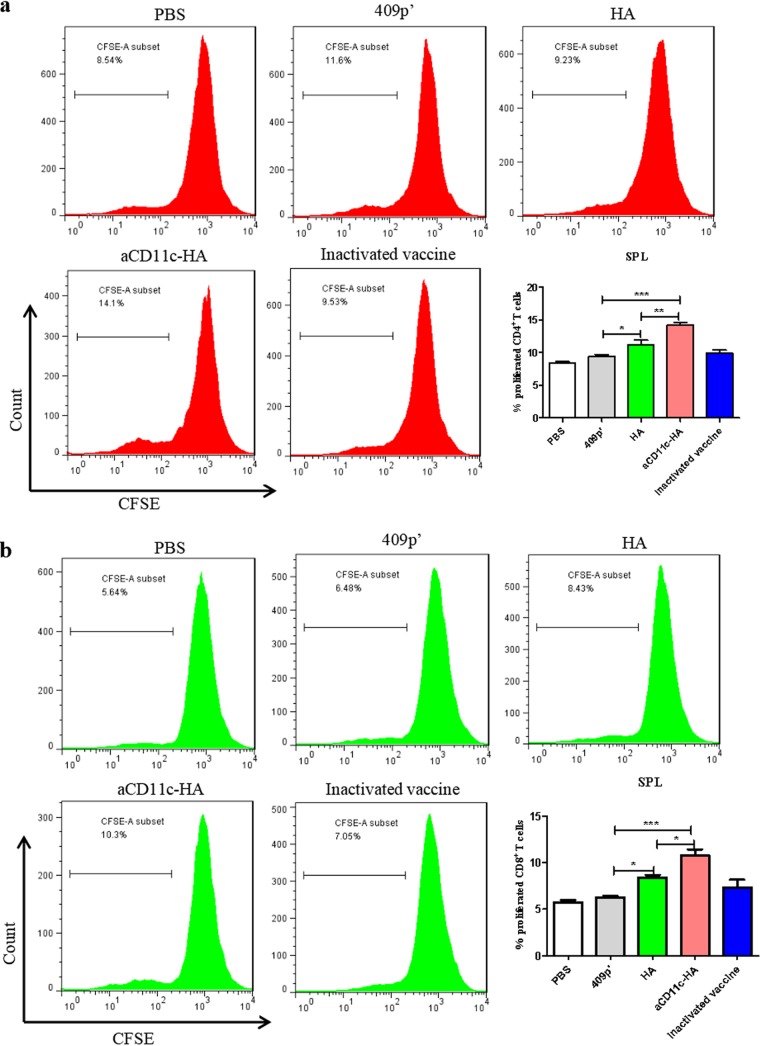
To assess the T cell proliferation induced by HA protein restimulation, the T cells from mice immunized with the strains were analyzed. Splenocytes isolated from different groups of mice were incubated with HA protein in vitro. Compared to the HA strain, the aCD11c-HA strain significantly improved the HA protein-induced CD4+ (P < 0.01; Fig. 6a) and CD8+ (P < 0.05; Fig. 6b) T cell proliferation response in the spleen.
FIG 6.
T cell proliferation assay. HA protein was used to stimulate the proliferation of mouse splenic T cells. The percentages of CD4+ T cell (a) and CD8+ T cell (b) proliferation were evaluated in CFSE (carboxyfluorescein succinimidyl ester)-stained splenocytes by FCM. The statistical significance was calculated by one-way ANOVA (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The HA strains increase the percentages of B220+ IgA+ B cells.
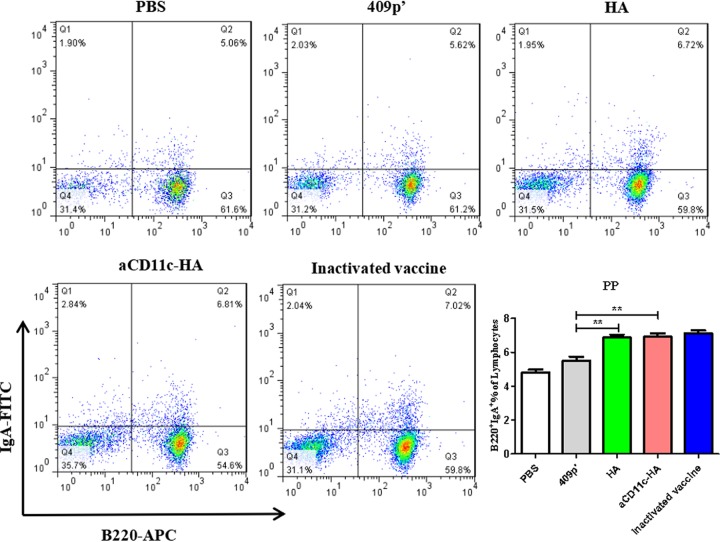
We detected the expression of B220+ IgA+ B cells from Peyer’s patches (PPs) using FCM. Compared to the 409p′ strain, the HA-expressing strain clearly increased the percentage of B220+ IgA+ B cells, whereas the percentage of B220+ IgA+ B cells induced was not significantly different between the aCD11c-HA and HA strains (Fig. 7).
FIG 7.
Percentages of B220+ IgA+ B cells in PPs. The flow scatter diagrams of each group are displayed, and the percentages of B220+ IgA+ B cells in the PPs were evaluated. The statistical significance was calculated using a one-way ANOVA (**, P < 0.01).
HA-expressing strains induce the production of humoral antibodies and neutralizing antibodies.
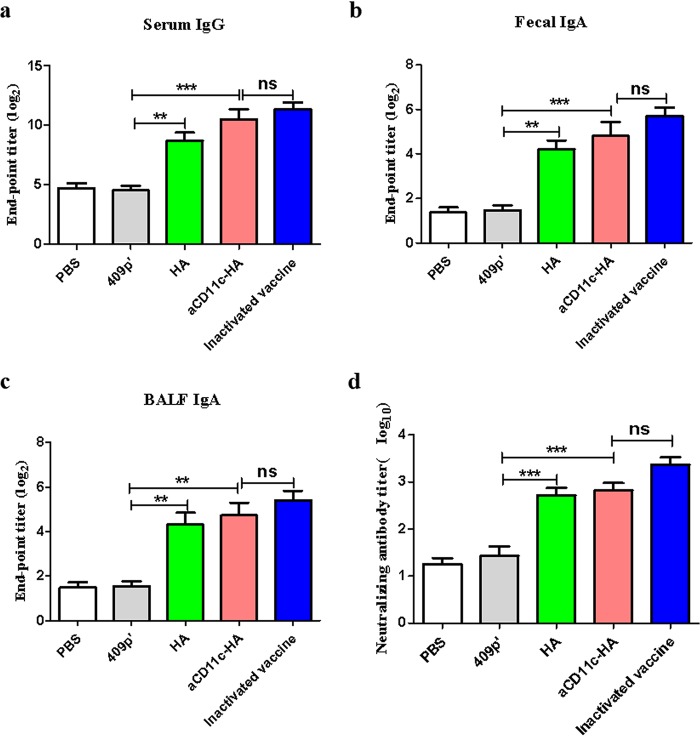
To assess the humoral antibody and neutralizing antibody levels, HA protein was used to coat 96-well plates. Compared with those immunized with 409p′, mice immunized with the aCD11c-HA (P < 0.001) and HA (P < 0.01) strains produced increased levels of HA-specific IgG in the serum (Fig. 8a) and IgA in the feces (Fig. 8b) and bronchoalveolar lavage fluid (BALF) (Fig. 8c). The level of neutralizing antibody was evaluated due to the expression of HA protein. The results showed that the aCD11c-HA group and HA group significantly improved the neutralizing antibody titers (P < 0.001) compared to the 409p′ group (Fig. 8d). No significant differences were shown between the aCD11c-HA group and the inactivated vaccine group in the specific antibody and neutralizing antibody.
FIG 8.
Determination of HA-specific antibodies and neutralizing antibodies. The HA-specific serum IgG titer (a) and the fecal (b) and BALF (c) IgA antibody titers were detected by indirect ELISA. Purified HA protein was used as the plate-coating antigen. Endpoint titers (log2) were calculated in accordance with the dilution of the serum, feces, and BALF by indirect ELISA. (d) Neutralizing antibody levels were detected in 10-day-old embryonated SPF eggs. Neutralizing antibody titers (log10) were calculated by the Reed and Muench method (39). The statistical significance was calculated using one-way ANOVA (**, P < 0.01; ***, P < 0.001).
Protective immunity against influenza virus infection is induced by the strains expressing HA.
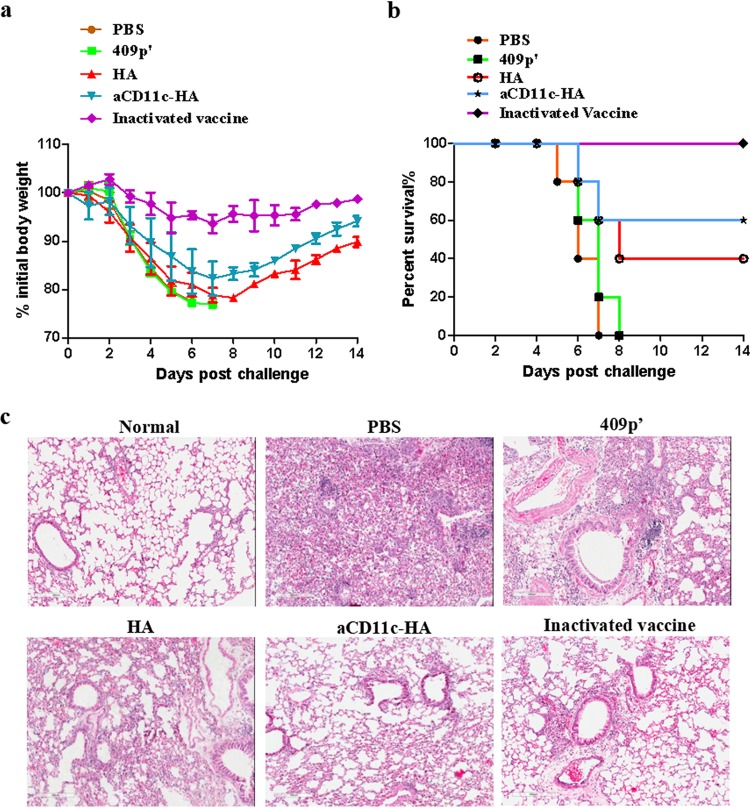
To evaluate the protective effect of the strains expressing HA against influenza virus, mice were orally immunized with the aCD11c-HA, HA, and 409p′ strains and phosphate-buffered saline (PBS) on days 1, 2, 3, 17, 18, and 19. All of the remaining mice (n = 5) were intranasally infected with A/PR/8/34 (H1N1) virus on day 27. The body weights and survival rates were measured for up to 2 weeks (Fig. 4a). The results suggested that, compared to those from the HA and 409p′ groups, the mice from the aCD11c-HA group had decreased weight loss after influenza virus infection (Fig. 9a). Furthermore, after immunization, 60% of the mice in the aCD11c-HA group survived, whereas only 40% of the mice vaccinated with the HA strain survived. Mice from the inactivated vaccine group exhibited 100% protection after H1N1 influenza virus infection (Fig. 9b). Consistent with these results, the lung damage in mice immunized with the aCD11c-HA strain was clearly improved compared to that in influenza virus-infected mice immunized with the HA or 409p′ strain or PBS (Fig. 9c). These data suggest that oral vaccination with the aCD11c-HA strain efficiently produces protective effects against influenza virus challenge.
FIG 9.
Protection against influenza virus challenge. Weight loss (a) and survival (b) were recorded for 2 weeks. The weight loss and mortality data are presented as the means ± the SEM of triplicate tests (n = 5 mice per group). (c) Lung samples from different influenza virus-infected groups were acquired, fixed, and embedded in paraffin. Sections were stained with H&E (magnification, ×200). Scale bar, 200 μm.
Protection against influenza virus by the adoptive transfer of CD8+ T cells.
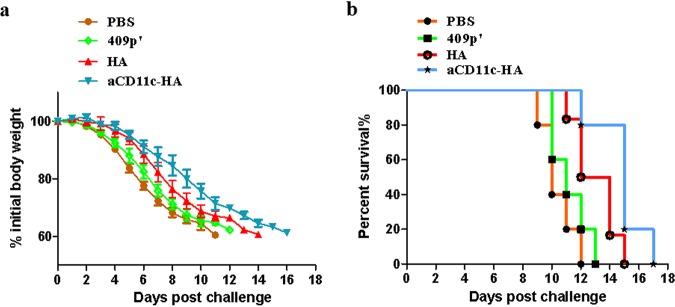
To assess whether the protective effect of aCD11c-HA against influenza virus is related to an increased specific CD8+ T cell response, CD8+ T cells were acquired from the spleens of the aCD11c-HA, HA, 409p′, and PBS groups of mice after immunization at day 26 and then injected into NOD/Lt-SCID mice via the tail vein. The results demonstrated that even though the body weights of all NOD/Lt-SCID mice decreased during the challenge, the CD8+ T cells isolated from aCD11c-HA immunized BALB/c mice alleviated these effects to some extent (Fig. 10a). A similar trend was observed for the survival rates, demonstrating that the CD8+ T cells from the aCD11c-HA group could extend the survival time from 13 to 17 days compared to those from the HA group (Fig. 10b), although no mice survived to the end of the experiment.
FIG 10.
Protective effect of CD8+ T cells transferred from aCD11c-HA mice to NOD/Lt-SCID mice. (a) The weight loss of the mice after H1N1 influenza virus infection was detected every day until day 16. (b) The survival rates of the NOD/Lt-SCID mice were observed after virus infection. The weight loss and mortality data are shown as means ± the SEM of triplicate tests (n = 5 mice per group).
DISCUSSION
Antibody-mediated passive immunization can provide protection against infection by invading pathogens (18). Therefore, it is important to study novel passive immunization approaches to prevent H1N1 influenza A virus infection. For influenza viruses, HA glycoproteins induce neutralizing antibody production and mediate adaptive immune responses (19). Vaccination with a mixture of HAs from different H1N1 influenza A viruses has been shown to be useful for the induction of specific immunity against the viruses represented in the mixture and confer some degree of cross-protection against unrelated influenza virus strains. Cross-reacting antibodies do not neutralize in microneutralization assays in vitro (20). However, antibodies might confer protection through antibody-dependent cell-mediated cytotoxicity (21, 22) or complement-dependent lysis (23).
In contrast, T cell-mediated immune responses, when combined with humoral immune responses, provide more comprehensive protection, which is important for influenza virus vaccine research (24). Cytotoxic CD8+ T cells have been shown to be critical for the protection against influenza virus in humans and animal models (25) and provide cross-reactivity against different influenza virus strains (26). Upon influenza A virus infection, CD8+ T cells mainly rely on the action of CTLs to eliminate the virus. The main antiviral mechanism involves the perforin/granzyme and Fas/Fas ligand pathways (27). The delivery of vaccine antigens to human leukocyte antigen (HLA) class II molecules on antigen-presenting cells has been achieved using a targeting unit such as major histocompatibility complex class II (MHC-II) (20, 28).
Thus, in this study, we mainly evaluated the expression of IFN-γ and perforin in CD8+ T cells. After immunization, the production of IFN-γ in CD8+ T cells was significantly higher in the aCD11c-HA group than the HA group owing to the specific combination of the scFv-CD11c and CD11c receptor in DCs in the mouse spleen, resulting in the increased uptake of HA antigens by DCs and subsequently increased activation and maturation of DCs and further stimulation of T cell differentiation. Notably, similar trends were observed in both in vivo and in vitro studies. The in vitro results showed that, compared to the HA strain, the aCD11c-HA strain significantly promoted the activation of BMDCs. After antigen activation, DCs secrete cytokines such as interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), IL-12P70, and IL-2. The increase in IL-12P70 and the decrease in IL-6 indicated that T cells were polarized toward the Th1 phenotype. Therefore, we evaluated the expression of IL-6 and IL-12P70 in the cell culture supernatants after BMDC stimulation. The results suggested that the aCD11c-HA strain could polarize T cells toward the Th1 direction. To verify this result, the above-activated BMDCs were cocultured with the sorted splenic CD4+ or CD8+ T cells from mice. At a T cell/BMDC ratio of 1:1, the expression of IFN-γ in CD8+ T cells was enhanced to a greater extent by the aCD11c-HA strain than by the HA strain, which is consistent with the experimental results of Ejaz et al. (16). In addition, the concentration of IFN-γ in the cell culture supernatants was also significantly increased, which promoted the Th1-type response. This result is consistent with the DC cytokine secretion results. In addition, an increased production of CD8+ CD107a+ T cells was also observed in the aCD11c-HA group. All these results indicate that the presence of scFv-CD11c significantly increases the host cellular immune response. However, although HA-specific humoral antibodies and neutralizing antibodies were observed in both the HA group and the aCD11c-HA group, no significant difference was observed between the groups, indicating that the presence of scFv-CD11c had fewer effects on the humoral immune response than on the cellular immune response.
To further confirm the role of the CD8+ T cell-mediated cellular immune response in the protection against virus challenge, NOD/Lt-SCID mice were used as a model to receive antigen-specific CD8+ T cells from BALB/c mice. The results showed that the aCD11c-HA-primed CD8+ T cells increased the mouse survival time and had a certain antiviral effect on body weight change. Similar results have been reported previously using a DC-targeting peptide in an influenza virus challenge mouse model (29). Notably, we did not observe complete protection against viral challenge during this study; one of the reasons could be that the challenge dose was slightly higher than expected. The other possible reason could be that both cellular and humoral immune responses are necessary for full protection.
Notably, compared to the 100% protection against virus challenge in the inactivated vaccine group, our recombinant L. plantarum strains produced a relatively low 60% protection rate, indicating that the direct comparison between our recombinant strains with the inactivated vaccine may not be reasonable. In fact, it has been demonstrated previously that L. plantarum-based influenza virus vaccine candidates are less effective than commercial inactivated vaccines. For example, Yang et al. produced an NP-M1 fusion protein in L. plantarum using the DC-targeting peptide DCpep, resulting in 80% protection against the same strain of H9N2 AIV (29). In addition, the production of 3M2e-Fc by L. plantarum NC8 strain has been shown to provide 60% protection against an A/PR/8/34 (H1N1) influenza virus challenge (2). All of these results indicate that there is still much room to improve the immune responses induced by recombinant L. plantarum strains. However, the properties of L. plantarum strains make their use a very promising approach for further vaccine studies. For example, the strains offer attractive advantages, including low cost, simple use, noninvasive administration by the oral or intranasal route, high safety levels, and genetic manipulability (9). In addition, the strains usually elicit minimal immune responses against themselves and stimulate high levels of humoral immune responses against foreign antigens (30). In particular, the novel DC-targeting strategy based on surface-displayed scFv-CD11c provides a novel platform to deliver any possible antigens from any potential pathogens, significantly enlarging its applicability in the oral vaccine field.
In conclusion, L. plantarum strains coexpressing influenza virus aCD11c and HA proteins were constructed and then used to immunize BALB/c mice. The presence of scFv-CD11c significantly enhanced the humoral immune response and promoted the production of IFN-γ and perforin in CD8+ T cells, as shown by FCM. Then, flow sorting and adoptive transfer to NOD/Lt-SCID mice further revealed the molecular mechanism of the immune response induced by the DCs targeting the L. plantarum strain, laying the foundation for the development of a new LAB-based vaccine.
MATERIALS AND METHODS
Construction of the strains expressing HA and aCD11c-HA.
The strains, plasmids and primers used in this study are shown in Table 1. L. plantarum strain NC8 was generously contributed by A. Kolandaswamy (Madurai Kamaraj University, Madurai, India). The NC8-pSIP409-pgsA′ and NC8-pSIP409-aCD11c strains and corresponding plasmids were constructed by Cai et al. (31) and Liu et al. (17), respectively. Luria-Bertani (LB) medium and MRS medium were used for the culture of Escherichia coli and L. plantarum strains, respectively. The concentration of erythromycin (Sigma-Aldrich) was 200 μg/ml in LB medium and 10 μg/ml in MRS medium.
TABLE 1.
Bacterial strains, plasmids, and primers used in this studya
| Plasmid, strain, or primer | Description or sequence | Source or reference |
|---|---|---|
| Plasmids | ||
| pSIP409-pgsA′ | p256rep/pUC (pGEM) PsppIP sppR sppK pgsA′ Emr | 31 |
| pSIP409-aCD11c | PsppIP sppR sppK aCD11c Emr | 17 |
| pSIP409-pgsA′-HA | PsppIP sppR sppK pgsA′ HA Emr | This study |
| pSIP409-aCD11c pgsA′-HA | PsppIP sppR sppK aCD11c pgsA′ HA Emr | This study |
| Strains | ||
| E. coli TOP10 | Host strain | TaKaRa |
| L. plantarum | ||
| NC8 (CCUG 61730) | Host strain, plasmid-free, silage isolate | 38 |
| NC8-pSIP409-pgsA′ | NC8 with pSIP409-pgsA′ | 31 |
| NC8-pSIP409-aCD11c | NC8 with pSIP409-aCD11c | 17 |
| NC8-pSIP409-pgsA′-HA | NC8 with pSIP409-pgsA′-HA | This study |
| NC8-pSIP409-aCD11c-pgsA′-HA | NC8 with pSIP409-aCD11c-pgsA′-HA | This study |
| Primers | ||
| aCD11c-F | CTATTACAAGGAGATTTTAGCCATGAAAAAATTGGTTTCA | This study |
| aCD11c-R | TTAATTCTTTCTTGCCCATATGGAGTGCCTCCTTATAATTTAAGCAGCCCGCTTAATTTC | This study |
| V-F | ATGGGCAAGAAAGAATTAAG | This study |
| V-R | GGCTAAAATCTCCTTGTAATAG | This study |
pgsA′ indicates an anchoring sequence for L. plantarum, a short version of poly-γ-glutamic acid synthetase A (pgsA). Em, erythromycin.
The HA sequences were optimized for L. plantarum and synthesized by Genewiz (Suzhou, China). In detail, the HA gene and pSIP409-pgsA′ vector were digested with XbaI and HindIII, respectively, and the recombinant plasmid, named pSIP409-pgsA′-HA, was acquired by ligation using T4 ligase (NEB). The pSIP409-aCD11c plasmid was used as a template to amplify the aCD11c fragment using primers aCD11c-F and aCD11c-R (Table 1), and then pSIP409-pgsA′-HA was used as a template to acquire the linearized pSIP409-pgsA′-HA vector using the primers V-F and V-R (Table 1). The linearized pSIP409-pgsA′-HA and aCD11c were ligated to produce the recombinant pSIP409-aCD11c-pgsA′-HA plasmid using seamless cloning technology (Clone Smarter). These plasmids were electroporated into the L. plantarum NC8 strain, producing NC8-pSIP409-pgsA′-HA and NC8-pSIP409-aCD11c-pgsA′-HA (Fig. 1a).
Synthesis of HA and aCD11c-HA.
For convenience, the recombinant strains NC8-pSIP409-pgsA′-HA and NC8-pSIP409-aCD11c-pgsA′-HA were named HA and aCD11c-HA, respectively. The empty strain named 409p′ was used as a negative control. The recombinant strains were established and cultivated, and then protein samples were collected by extracting cell membrane and cell wall fractions, as previously described (17, 31). Western blotting was performed using an anti-rabbit H1N1 HA monoclonal antibody (GenScript, China) and a rabbit anti-His monoclonal antibody (Abbkine) as the primary antibodies, followed by goat anti-rabbit IgG-horseradish peroxidase (HRP) as the secondary antibody (Abbkine). A Chemiluminescence Plus detection kit (Thermo Scientific) was used to detect the expression of HA and aCD11c.
Activation of bone marrow dendritic cells.
Bone marrow precursors were acquired from C57BL/6 mice, and granulocyte-macrophage colony-stimulating factor and IL-4 (PeproTech) were added to prepare BMDCs, as described previously (32). Cells were then cultured in 24-well plates with 2 × 105 cells per well at day 8. The 409p′, HA, and aCD11c-HA strains were cocultured with BMDCs on the basis of 10 bacteria per BMDC for 12 h according to previous protocols (33). The cell culture supernatants were collected, and cytokines, including IL-12P70 and IL-6, were detected using an ELISA kit (Liuhebio, China) according to the manufacturer’s instructions. The cells were then collected, and antibodies, including 10 μl of hamster anti-mouse CD11c-allophycocyanin (APC), 10 μl of hamster anti-mouse CD40-fluorescein isothiocyanate (FITC), 10 μl of hamster anti-mouse CD80-peridinin chlorophyll protein (PerCP)-Cy5.5, and 10 μl of hamster anti-mouse CD86-phycoerythrin (PE)-Cy7 (BD Pharmingen), were used for incubation for 20 min at room temperature in darkness. Subsequently, the cells were resuspended and washed three times with PBS. The samples were detected by FCM using a BD Pharmingen FCM LSR Fortessa flow cytometer. The data were analyzed by using FlowJo 7.6.1 software.
Interaction between activated BMDCs and T cells.
Splenocytes from nonimmunized BALB/c mice were acquired as previously described (33). Naive CD4+ T cells and CD8+ T cells were sorted using anti-mouse CD4 magnetic particles and anti-mouse CD8a particles (BD Pharmingen) according to the manufacturer’s instructions. The sorted CD4+ and CD8+ T cells were cocultured with activated BMDCs from each group in P24 dishes at a 1:1 cell ratio. After 48 h, the IFN-γ concentration in supernatants was detected using an ELISA kit in accordance with the manufacturer’s instructions. The cells from each group were then collected, and intracellular cytokine staining (ICS) was performed. PerCP-Cy5.5-conjugated antibodies for CD3e, FITC-conjugated antibodies for CD4, APC-Cy7-conjugated antibodies for CD8, and PE-conjugated anti-IFN-γ and PE-Cy7-conjugated anti-CD107a (BD Pharmingen) antibodies were used to label CD4+ T cells or CD8+ T cells. Individual isotype control antibodies (BD Pharmingen) were also included (34).
Mouse immunization and the challenge study.
Specific-pathogen-free (SPF) BALB/c and NOD/Lt-SCID female 5- to 6-week-old mice were obtained from Beijing HFK Bioscience Co., Ltd., China. SPF embryonated chicken eggs from Beijing Merial Vital Laboratory Animal Technology Co., Ltd., China, were used. In detail, 40 BALB/c mice were randomly separated into five groups, including a PBS control group, a 409p′ group, an HA group, an aCD11c-HA group, and an inactivated vaccine group. The immunization protocols were performed in accordance with a previously described method (33). The inactivated vaccine (Keqian, Wuhan, China) group mice were intramuscularly injected as a positive control on day 10 in accordance with the manufacturer’s instructions. The three mice from each group were sacrificed to detect immune indexes and specific antibody levels on day 26. Subsequently, mice anesthetized with 100 ml of 15 mg/ml mebubarbital (Sigma-Aldrich) were intranasally inoculated with a lethal dose (0.05 ml) of H1N1 influenza virus (an ∼10× 50% lethal dose [LD50]) at day 27. Mice were observed for 14 days, and weight loss and survival rates were recorded every day after H1N1 influenza virus infection. All applicable international and national guidelines for the care and use of animals were followed.
Flow cytometry assay.
Single-cell suspensions were collected from the spleen cells, MLNs, and PPs and adjusted to 1.0 × 106 cells/100 μl or 2.0 × 106 cells/100 μl. Anti-CD11c, anti-CD80, and anti-CD83 antibodies were then used in the FCM analysis of DC phenotypes in the spleen (1.0 × 106/100 μl) as previously described (17).
ICS was carried out according to a previously described method (35). Splenic or MLN cells (2.0 × 106/100 μl) were cocultured with 20 μg/ml purified HA protein and 2 μg/ml anti-CD28 antibody (BD Pharmingen) for 8 h at 37°C. Golgi-Plug protein transport inhibitor (BD Pharmingen) was added 4 h before ICS. PerCP-Cy5.5-conjugated antibodies against CD3e, FITC-conjugated antibodies against CD4, and APC-Cy7-conjugated antibodies against CD8, were added to stain the cells. PP cells were stained with an APC-conjugated anti-B220 antibody to directly detect B cells. These cells were then washed, fixed, permeabilized, and stained with PE-conjugated anti-IFN-γ, PE-Cy7-conjugated anti-CD107a for splenic and MLN cells and FITC-conjugated anti-IgA for PP cells. The cells were detected by using FCM (36).
T cell proliferation assay.
The proliferation of T cells was evaluated. In detail, the murine splenocytes were stained with CFSE (Invitrogen) and then incubated with an anti-CD28 antibody and purified HA protein for 72 h; the detailed methods were as described by Yang et al. (29). Antibodies, including PerCP-Cy5.5-conjugated antibodies against CD3e, PE-conjugated antibodies against CD4, and APC-Cy7-conjugated antibodies against CD8, were used to label splenocytes. The cells were analyzed by FCM.
Evaluation of antibody levels.
To evaluate the HA-specific antibody levels of IgG in serum and secretory IgA (sIgA) in feces and BALF at day 26 after the last immunization, ELISA was performed; the endpoint titers are presented as described previously (2, 33). For the serum neutralization assay, 10-day-old embryonated SPF eggs were used to assess the level of neutralizing antibody, as described by Wen et al. (37). Neutralizing antibody titers were calculated using the Reed and Muench method (39).
Histopathological examination.
Pulmonary samples were acquired from all mice after influenza virus infection, fixed using 4% paraformaldehyde, and then embedded using paraffin. The samples were cut and stained with hematoxylin and eosin (H&E). The stained sections were observed at ×200 magnification on an Aperio CS2 microscope (Leica, Germany).
Isolation of CD8+ T cells and transfer to NOD/Lt-SCID mice.
Twenty BALB/c mice were purchased and randomly separated into four groups: the PBS control group, the 409p′ group, the HA group, and the aCD11c-HA group. The mice were immunized according to the above immune procedures. At day 26 after immunization, the splenocytes from each group were aseptically prepared and acquired as described above. CD8+ T cells were sorted using anti-mouse CD8a particles (BD Pharmingen) according to the manufacturer’s instructions. After collection, the purity of the CD8+ T cells was verified, and the cells were adoptively transferred into SPF NOD/Lt-SCID mice at a dose of 2 × 106 cells/mouse by tail vein injection. All NOD/Lt-SCID mice were inoculated under anesthesia with 0.5× the LD50 of H1N1 influenza virus (A/PR/8/34) after 24 h. After challenge, the mice were observed daily until the end of the study, and the body weights and survival were recorded (29).
Statistical analysis.
Data were analyzed using GraphPad Prism 5.0 software. Statistical significance was measured using one-way analysis of variance (ANOVA [Dunnett’s multiple-comparison test]). All data are shown as the means ± standard errors of the mean (SEM) of at least three independent experiments.
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China (2017YFD0501000 and 2017YFD0501200), the National Natural Science Foundation of China (31602092 and 31672528), the Science and Technology Program of Jilin Province Educational Ministry (JJKH20180647KJ), and the Science and Technology Development Program of Jilin Province (20170204034NY and 20180201040NY).
We declare that no competing financial interests exist.
REFERENCES
- 1.Mohebbi A, Fotouhi F, Jamali A, Yaghobi R, Farahmand B, Mohebbi R. 2019. Molecular epidemiology of the hemagglutinin gene of prevalent influenza virus A/H1N1/pdm09 among patient in Iran. Virus Res 259:38–45. doi: 10.1016/j.virusres.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Yang WT, Yang GL, Wang Q, Huang HB, Jiang YL, Shi CW, Wang JZ, Huang KY, Jin YB, Wang CF. 2017. Protective efficacy of Fc targeting conserved influenza virus M2e antigen expressed by Lactobacillus plantarum. Antiviral Res 138:9–21. doi: 10.1016/j.antiviral.2016.11.025. [DOI] [PubMed] [Google Scholar]
- 3.Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, Griffin J, Baggaley RF, Jenkins HE, Lyons EJ, Jombart T, Hinsley WR, Grassly NC, Balloux F, Ghani AC, Ferguson NM, Rambaut A, Pybus OG, Lopez-Gatell H, Alpuche-Aranda CM, Chapela IB, Zavala EP, Guevara DM, Checchi F, Garcia E, Hugonnet S, Roth C, The WHO Rapid Pandemic Assessment Collaboration . 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng L, Roose K, Job ER, De Rycke R, Van Hamme E, Goncalves A, Parthoens E, Cicchelero L, Sanders N, Fiers W, Saelens X. 2017. Oral delivery of Escherichia coli persistently infected with M2e-displaying bacteriophages partially protects against influenza A virus. J Control Release 264:55–65. doi: 10.1016/j.jconrel.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Te Velthuis AJ, Fodor E. 2016. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wohlbold TJ, Krammer F. 2014. In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 6:2465–2494. doi: 10.3390/v6062465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson BE, Brett IC. 2007. Changing perspective on immunization against influenza. Vaccine 25:3062–3065. doi: 10.1016/j.vaccine.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 8.de Castro CP, Drumond MM, Batista VL, Nunes A, Mancha-Agresti P, Azevedo V. 2018. Vector development timeline for mucosal vaccination and treatment of disease using Lactococcus lactis and design approaches of next-generation food-grade plasmids. Front Microbiol 9:1805. doi: 10.3389/fmicb.2018.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeCureux JS, Dean GA. 2018. Lactobacillus mucosal vaccine vectors: immune responses against bacterial and viral antigens. mSphere 3:00061-18. doi: 10.1128/mSphere.00061-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corthesy B, Boris S, Isler P, Grangette C, Mercenier A. 2005. Oral immunization of mice with lactic acid bacteria producing Helicobacter pylori urease B subunit partially protects against challenge with Helicobacter felis. J Infect Dis 192:1441–1449. doi: 10.1086/444425. [DOI] [PubMed] [Google Scholar]
- 11.Kuczkowska K, Kleiveland CR, Minic R, Moen LF, Overland L, Tjaland R, Carlsen H, Lea T, Mathiesen G, Eijsink VGH. 2017. Immunogenic properties of Lactobacillus plantarum producing surface-displayed Mycobacterium tuberculosis antigens. Appl Environ Microbiol 83:02782-16. doi: 10.1128/AEM.02782-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mobergslien A, Vasovic V, Mathiesen G, Fredriksen L, Westby P, Eijsink VG, Peng Q, Sioud M. 2015. Recombinant Lactobacillus plantarum induces immune responses to cancer testis antigen NY-ESO-1 and maturation of dendritic cells. Hum Vaccin Immunother 11:2664–2673. doi: 10.1080/21645515.2015.1056952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang WT, Yang GL, Yang X, Shonyela SM, Zhao L, Jiang YL, Huang HB, Shi CW, Wang JZ, Wang G, Zhao JH, Wang CF. 2017. Recombinant Lactobacillus plantarum expressing HA2 antigen elicits protective immunity against H9N2 avian influenza virus in chickens. Appl Microbiol Biotechnol 101:8475–8484. doi: 10.1007/s00253-017-8600-2. [DOI] [PubMed] [Google Scholar]
- 14.Yang WT, Yang GL, Shi SH, Liu YY, Huang HB, Jiang YL, Wang JZ, Shi CW, Jing YB, Wang CF. 2017. Protection of chickens against H9N2 avian influenza virus challenge with recombinant Lactobacillus plantarum expressing conserved antigens. Appl Microbiol Biotechnol 101:4593–4603. doi: 10.1007/s00253-017-8230-8. [DOI] [PubMed] [Google Scholar]
- 15.Michon C, Christophe M, Kuczkowska K, Langella P, Eijsink VGH, Mathiesen G, Chatel J-M. 2015. Surface display of an anti-DEC-205 single-chain Fv fragment in Lactobacillus plantarum increases internalization and plasmid transfer to dendritic cells in vitro and in vivo. Microb Cell Fact 14:95. doi: 10.1186/s12934-015-0290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejaz A, Ammann CG, Werner R, Huber G, Oberhauser V, Hörl S, Schimmer S, Dittmer U, von Laer D, Stoiber H, Bánki Z. 2012. Targeting viral antigens to CD11c on dendritic cells induces retrovirus-specific T cell responses. PLoS One 7:e45102. doi: 10.1371/journal.pone.0045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Yang G, Gao X, Zhang Z, Liu Y, Yang X, Shi C, Liu Q, Jiang Y, Wang C. 2019. Immunomodulatory properties of Lactobacillus plantarum NC8 expressing an anti-CD11c single-chain Fv fragment. J Microbiol Biotechnol 29:160–170. doi: 10.4014/jmb.1809.09017. [DOI] [PubMed] [Google Scholar]
- 18.Casadevall A, Dadachova E, Pirofski LA. 2004. Passive antibody therapy for infectious diseases. Nat Rev Microbiol 2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Li H, Xue Y, Zhao S, Li C, Qu L, Zhang Y, Liu M. 2017. Characterization of monoclonal antibodies against HA protein of H1N1 swine influenza virus and protective efficacy against H1 viruses in mice. Viruses 9:209. doi: 10.3390/v9080209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson AM, Baranowska-Hustad M, Braathen R, Grodeland G, Bogen B. 2018. Simultaneous targeting of multiple hemagglutinins to APCs for induction of broad immunity against influenza. J Immunol 200:2057–2066. doi: 10.4049/jimmunol.1701088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, Kent SJ. 2013. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol 190:1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 23.Terajima M, Cruz J, Co MD, Lee JH, Kaur K, Wrammert J, Wilson PC, Ennis FA. 2011. Complement-dependent lysis of influenza a virus-infected cells by broadly cross-reactive human monoclonal antibodies. J Virol 85:13463–13467. doi: 10.1128/JVI.05193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berthoud TK, Hamill M, Lillie PJ, Hwenda L, Collins KA, Ewer KJ, Milicic A, Poyntz HC, Lambe T, Fletcher HA, Hill AV, Gilbert SC. 2011. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin Infect Dis 52:1–7. doi: 10.1093/cid/ciq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nussing S, Sant S, Koutsakos M, Subbarao K, Nguyen THO, Kedzierska K. 2018. Innate and adaptive T cells in influenza disease. Front Med 12:34–47. doi: 10.1007/s11684-017-0606-8. [DOI] [PubMed] [Google Scholar]
- 26.Koutsakos M, Illing PT, Nguyen THO, Mifsud NA, Crawford JC, Rizzetto S, Eltahla AA, Clemens EB, Sant S, Chua BY, Wong CY, Allen EK, Teng D, Dash P, Boyd DF, Grzelak L, Zeng W, Hurt AC, Barr I, Rockman S, Jackson DC, Kotsimbos TC, Cheng AC, Richards M, Westall GP, Loudovaris T, Mannering SI, Elliott M, Tangye SG, Wakim LM, Rossjohn J, Vijaykrishna D, Luciani F, Thomas PG, Gras S, Purcell AW, Kedzierska K. 2019. Human CD8+ T cell cross-reactivity across influenza A, B, and C viruses. Nat Immunol 20:613–625. doi: 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
- 27.Topham DJ, Tripp RA, Doherty PC. 1997. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol 159:5197–5200. [PubMed] [Google Scholar]
- 28.Grodeland G, Fredriksen AB, Loset GA, Vikse E, Fugger L, Bogen B. 2016. Antigen targeting to human HLA class II molecules increases efficacy of DNA vaccination. J Immunol 197:3575–3585. doi: 10.4049/jimmunol.1600893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang WT, Shi SH, Yang GL, Jiang YL, Zhao L, Li Y, Wang CF. 2016. Cross-protective efficacy of dendritic cells targeting conserved influenza virus antigen expressed by Lactobacillus plantarum. Sci Rep 6:39665. doi: 10.1038/srep39665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells JM, Mercenier A. 2008. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat Rev Microbiol 6:349–362. doi: 10.1038/nrmicro1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai R, Jiang Y, Yang W, Yang W, Shi S, Shi C, Hu J, Gu W, Ye L, Zhou F, Gong Q, Han W, Yang G, Wang C. 2016. Surface-displayed IL-10 by recombinant Lactobacillus plantarum reduces Th1 responses of RAW 264.7 cells stimulated with poly(I:C) or LPS. J Microbiol Biotechnol 26:421–431. doi: 10.4014/jmb.1509.09030. [DOI] [PubMed] [Google Scholar]
- 32.Perone MJ, Larregina AT, Shufesky WJ, Papworth GD, Sullivan ML, Zahorchak AF, Stolz DB, Baum LG, Watkins SC, Thomson AW, Morelli AE. 2006. Transgenic galectin-1 induces maturation of dendritic cells that elicit contrasting responses in naive and activated T cells. J Immunol 176:7207–7220. doi: 10.4049/jimmunol.176.12.7207. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Yang G, Gao X, Zhang Z, Liu Y, Liu Q, Chatel JM, Jiang Y, Wang C. 2019. Recombinant invasive Lactobacillus plantarum expressing fibronectin binding protein A induce specific humoral immune response by stimulating differentiation of dendritic cells. Benef Microbes 10:589–604. doi: 10.3920/BM2018.0157. [DOI] [PubMed] [Google Scholar]
- 34.Shi SH, Yang WT, Yang GL, Cong YL, Huang HB, Wang Q, Cai RP, Ye LP, Hu JT, Zhou JY, Wang CF, Li Y. 2014. Immunoprotection against influenza virus H9N2 by the oral administration of recombinant Lactobacillus plantarumNC8 expressing hemagglutinin in BALB/c mice. Virology 464–465:166–176. doi: 10.1016/j.virol.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med 13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 36.Lee S, Miller SA, Wright DW, Rock MT, Crowe JE Jr.. 2007. Tissue-specific regulation of CD8+ T-lymphocyte immunodominance in respiratory syncytial virus infection. J Virol 81:2349–2358. doi: 10.1128/JVI.01910-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen F, Yu H, Yang FR, Huang M, Yang S, Zhou YJ, Li ZJ, Tong GZ. 2014. Efficacy of a high-growth reassortant H1N1 influenza virus vaccine against the classical swine H1N1 subtype influenza virus in mice and pigs. Arch Virol 159:2957–2967. doi: 10.1007/s00705-014-2151-y. [DOI] [PubMed] [Google Scholar]
- 38.Sorvig E, Mathiesen G, Naterstad K, Eijsink VG, Axelsson L. 2005. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 151:2439–2449. doi: 10.1099/mic.0.28084-0. [DOI] [PubMed] [Google Scholar]
- 39.Reed LJ, Muench HA. 1938. A simple method of estimating fifity per cent endpoints. Am J Trop Med Hyg 27:493–497. [Google Scholar]