Abstract
Understanding the development of the oral microbiota in healthy children is of great importance to oral and general health. However, limited data exist on a healthy maturation of the oral microbial ecosystem in children. Moreover, the data are biased by mislabeling “caries-free” populations. Therefore, we aimed to characterize the healthy salivary and dental plaque microbiome in young children. Caries-free (ICDAS [International Caries Detection and Assessment System] score 0) children (n = 119) and their primary caregivers were followed from 1 until 4 y of child age. Salivary and dental plaque samples were collected from the children at 3 time points (T1, ~1 y old; T2, ~2.5 y old; and T3, ~4 y old). Only saliva samples were collected from the caregivers. Bacterial V4 16S ribosomal DNA amplicons were sequenced using Illumina MiSeq. The reads were denoised and mapped to the zero-radius operational taxonomic units (zOTUs). Taxonomy was assigned using HOMD. The microbial profiles of children showed significant differences (P = 0.0001) over time. Various taxa increased, including Fusobacterium, Actinomyces, and Corynebacterium, while others showed significant decreases (e.g., Alloprevotella and Capnocytophaga) in their relative abundances over time. Microbial diversity and child-caregiver similarity increased most between 1 and 2.5 y of age while still not reaching the complexity of the caregivers at 4 y of age. The microbiome at 1 y of age differed the most from those at later time points. A single zOTU (Streptococcus) was present in all samples (n = 925) of the study. A large variation in the proportion of shared zOTUs was observed within an individual child over time (2% to 42% of zOTUs in saliva; 2.5% to 38% in dental plaque). These findings indicate that the oral ecosystem of caries-free toddlers is highly heterogeneous and dynamic with substantial changes in microbial composition over time and only few taxa persisting across the 3 y of the study. The salivary microbiome of 4-y-old children is still distinct from that of their caregivers.
Keywords: saliva, plaque, caries-free children, caregiver, 16S rRNA gene amplicon sequencing, fungal qPCR
Introduction
The oral microbiome is unique to each individual and comprises a diverse community of microorganisms, including bacteria, archaea, fungi, protozoa, and viruses (Wade 2013). Even among healthy individuals, there are substantial differences in the composition of the resident oral microbiome (Aas et al. 2005). Furthermore, the microbial composition differs across microniches within a healthy oral cavity (Zhou et al. 2013). Equilibrium among the commensal microbiota of the oral ecosystem, interacting with each other and the host, is considered one of the most important factors for maintaining a healthy microbiota (Zaura et al. 2014). In addition, various internal and external factors, such as diet, oral hygiene, use of antibiotics, and others, affect the composition and the stability of the oral microbiome (Dagli et al. 2016).
To date, emphasis has been on describing the differences of caries-affected versus caries-free children (Luo et al. 2012; Jiang et al. 2016), while there is limited knowledge on the “normal” (healthy) microbiome, especially in children. Development and maintenance of the normal microbiome throughout childhood are not well studied due to various limitations, particularly cross-sectional study designs, targeted microbial and clinical diagnostic methods, or small sample sizes (Xin et al. 2013; Lee et al. 2016; Li et al. 2018).
Development of enamel lesions is preceded by microbial ecological shifts toward aciduric and acidogenic microbiota (Marsh 1994). Defining dental caries as a “cavity” leads to a mislabeling of “caries-free” participants in clinical studies where diagnostic thresholds determine what is recorded as “diseased” or “sound” (Pitts 2004). Most studies that focus on the oral microbiome in young children define their “caries-free” populations as a cavity-free group (decayed missing filled surfaces [dmfs] = 0) (Crielaard et al. 2011; Teng et al. 2015; Xu et al. 2015; Li et al. 2018; Xu et al. 2018; Hurley et al. 2019) or fail to report the assessment of the caries status (Papaioannou et al. 2009; Shi et al. 2018) and thus might have included children with clinically detectable enamel lesions in their healthy group.
We aimed to characterize the healthy maturation of the salivary and plaque microbiome in children aged 1 to 4 y. For that, we only included the children who were free of clinical signs of caries at all time points throughout 3 y of the study.
Materials and Methods
This report conforms to the STROBE (Strengthening the Reporting of Observational Studies in Epidemiology) guidelines for cohort studies. Detailed materials and methods of the study are described in the Materials and Methods section in the online Appendix. In brief, this study was part of a larger project with 1,323 children and their primary caregivers (Fontana et al. 2019). Only those participants who were enrolled and passed inclusion criteria of the parental project could participate in the current study. In total, 503 child-caregiver pairs consented to participate in the current study at the baseline visit (time point T1, children ~1 y old). Of these, 321 participant pairs attended and provided samples at the second visit when children were 2.5 y old (T2). Finally, 268 participant pairs attended and provided samples at the third visit at the age of 4 y (T3) (Fontana et al. 2018). Teeth were cleaned, dried, and assessed using the International Caries Detection and Assessment System (ICDAS II) criteria (ICDAS Coordinating Committee 2012) by calibrated dental professionals. A self-reported 53-item questionnaire (DCR-007-Primary Caregiver Questionnaire) was used in the parental study (Eckert et al. 2010; Fontana et al. 2011; Daly et al. 2016; Fontana et al. 2019). Only general questions (demographics, delivery mode, Medicaid) about the child and the caregiver were included in this study.
Current Study Population
In total, 266 children and their primary caregivers completed sample collections at all 3 time points (T1, T2, and T3). Among the 266 participants at the third visit (T3), 127 (47.7%) children had dental caries lesions (ICDAS ≥1), 119 (44.7%) children were caries free (ICDAS = 0) at all time points, and 20 (7.5%) children had experienced remineralization of dental caries lesions. Only children who did not present with clinical signs of dental caries by visual observation until the age of 4 y (n = 119) and their caregivers (n = 116) were included in the current study (Appendix Fig. 1).
Sample Collection and Processing
Saliva samples of children were collected at each visit by swabbing the mouth with sponges in the cheek pouch. From this point onward, a saliva swab sample is referred to as a saliva sample. If teeth were present, a pooled dental plaque sample was taken prior to the ICDAS exam by swabbing all buccal surfaces of the child’s teeth with a sterile microbrush. Unstimulated saliva was collected by drooling from all primary caregivers at T1, while from some caregivers (n = 66), saliva was collected at all 3 visits. Samples were transported on dry ice and stored at −80°C.
After DNA extraction and purification, bacterial DNA concentration was determined by 16S ribosomal DNA (rDNA) quantitative polymerase chain reaction (qPCR) (Ciric et al. 2010). The V4 hypervariable region of the 16S rRNA gene was amplified with barcoded forward and reverse primers (Kozich et al. 2013) and sequenced (MiSeq; Illumina). The sequences are available in the NCBI BioProject database under accession number PRJNA575641.
The reads were denoised using UNOISE3 (Edgar 2016) and mapped to the zero-radius operational taxonomic units (zOTUs). The representative zOTU sequences were assigned a taxonomy using HOMD v14.51 (Chen et al. 2010). The zOTU table was subsampled at 7,000 reads/sample.
The concentration of fungal DNA in the samples was determined using qPCR (Vollmer et al. 2008). The fungal load was calculated as the percentage of fungal DNA over the bacterial DNA.
Statistical Analyses
The zOTU table was log-2 transformed and ordinated by principal component analysis (PCA), and differences in microbial profiles in unrelated samples were assessed with PERMANOVA using PAST v.3.18 (Hammer et al. 2001). PERMANOVA on dependent samples was performed using adonis with permutations restricted within the subject (vegan v.2.4–6; R v.3.4.3; R Core Team 2019). R2 values in adonis give the effect size of the variation in distances that is explained by the variables being tested. Similarity in microbiome profiles was assessed using Bray-Curtis similarity and diversity by Shannon Diversity Index and Species richness using PAST. The Venn diagrams were created using Venny (Oliveros 2007). The linear discriminant analysis effect size (LEfSe) biomarker discovery tool (Segata et al. 2011) was used to identify discriminatory zOTUs. Differences in univariate data were tested using SPSS version 25 (SPSS, Inc.). False discovery rate (FDR) correction of P values for multiple comparisons was performed in R v.3.4.3. The FDR was set to 5%.
Results
Extended results of the study are presented in the Results section of the Appendix file. In total, 119 children who were caries-free at all study time points were included in the study (Table). Children were paired with their own primary caregivers, forming a child-caregiver pair. In case of twins (n = 3 pairs), 1 caregiver was paired with each of his or her twin children separately, creating 2 child-caregiver pairs.
Table.
Demographic and Socioeconomic Data of the Children Who Were Caries Free during All Time Points (n = 119).
| Characteristics | Value |
|---|---|
| Age, median (range), mo | |
| T1 | 11.3 (9–15.9) |
| T2 | 29 (25.1–35.7) |
| T3 | 47 (42.9–53.8) |
| Sex, n (%) | |
| Male | 62 (52.1) |
| Female | 57 (47.9) |
| Race/ethnicity, n (%) | |
| Caucasian not Hispanic | 74 (62.2) |
| African American not Hispanic | 16 (13.4) |
| Hispanic | 14 (11.8) |
| Multirace | 15 (12.6) |
| Medicaid, n (%) | |
| Yes | 54 (45.4) |
| No | 61 (51.3) |
| Unknown | 4 (3.4) |
| Recruitment site, n (%) | |
| Duke University | 13 (10.9) |
| Indiana University | 39 (32.8) |
| University of Iowa | 67 (56.3) |
| Delivery mode, n (%) | |
| Vaginal | 77 (64.7) |
| C-section | 42 (35.3) |
| Caregiver, n (%) | |
| Mother | 110 (92.4) |
| Father | 7 (5.9) |
| Grandmother | 2 (1.7) |
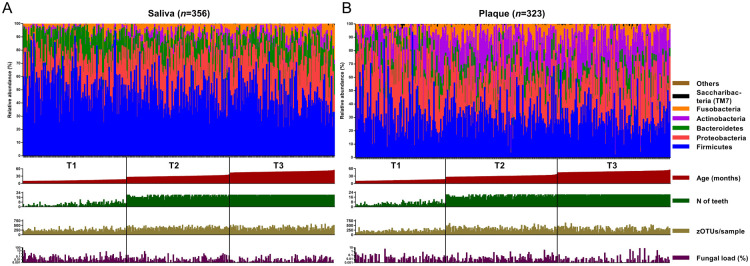
The subsampled data set included 2,320 zOTUs, which were classified in 12 phyla (Fig. 1, Appendix Fig. 2) and in 163 genera or higher taxa (Appendix Table 1A, B). Child saliva samples were dominated by genus Streptococcus, followed by Haemophilus and Neisseria, while most reads in saliva of caregivers were classified as Streptococcus, Prevotella, Veillonella, Haemophilus, and Neisseria. The top 3 genera in child dental plaque samples were Streptococcus, Neisseria, and Actinomyces (Appendix Table 1B). The predominance of the genera mentioned above was consistent among all 3 time points.
Figure 1.
Taxonomic distribution of the relative abundance of reads of major bacterial phyla in (A) salivary and (B) plaque samples of children at all time points (T1, T2, T3). The samples are ordered according to the age of the participants (T1: 9 to 15.9 mo of age; T2: 25.1 to 35.7 mo of age, T3: 42.9 to 53.8 mo of age) at the time of the sample collection (red bars below the taxonomy plots). Green bars indicate the number of teeth present (range, 0 to 20) at the time of the sample collection. Species richness (number of zero-radius operational taxonomic units/sample) of each sample (range, 66 to 629) is indicated with mustard-colored bars, and the fungal load (percentage of fungal DNA/bacterial DNA) of each sample (range, 0 to 10.9) is shown as purple bars below the respective taxonomy plots.
At the phylum level, the salivary microbiome of children changed most from T1 to T2, with significant decreases in relative abundance of Firmicutes and Bacteroidetes and increases in Proteobacteria, Fusobacteria, Actinobacteria, and Saccharibacteria (TM7) (Appendix Fig. 3). In plaque, only Actinobacteria increased significantly with time, while Bacteroidetes and Proteobacteria decreased (Appendix Fig. 3).
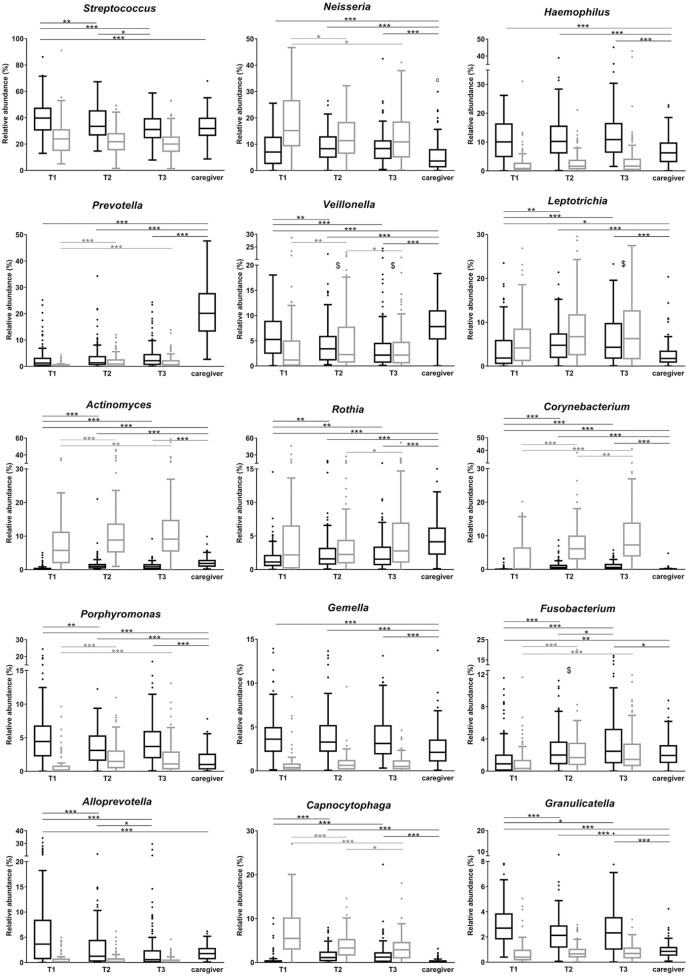
At the genus level, Leptotrichia, Fusobacterium, Actinomyces, Corynebacterium, and Rothia increased significantly with time both in saliva and plaque (Fig. 2). Genera Streptococcus, Veillonella, Granulicatella, Porphyromonas, and Alloprevotella decreased significantly with time in saliva, while in plaque, decreases were observed with Capnocytophaga and Neisseria (Fig. 2).
Figure 2.
The relative abundance of major bacterial genera in salivary (black boxes) and plaque (gray boxes) samples over time. The boxplots are plotted using Tukey’s method. Significant differences over time within the respective sample type are indicated by asterisks: *P < 0.05, **P < 0.01, and ***P < 0.001 (paired samples Friedman test, followed by Wilcoxon rank-sum test, with false discovery rate correction). Lines connect the time points with the respective difference. The pairwise comparisons of 2 types of samples (saliva vs. plaque) were significantly different (P < 0.05) in all but those time points indicated with $.
Changes in Microbial Profiles over Time
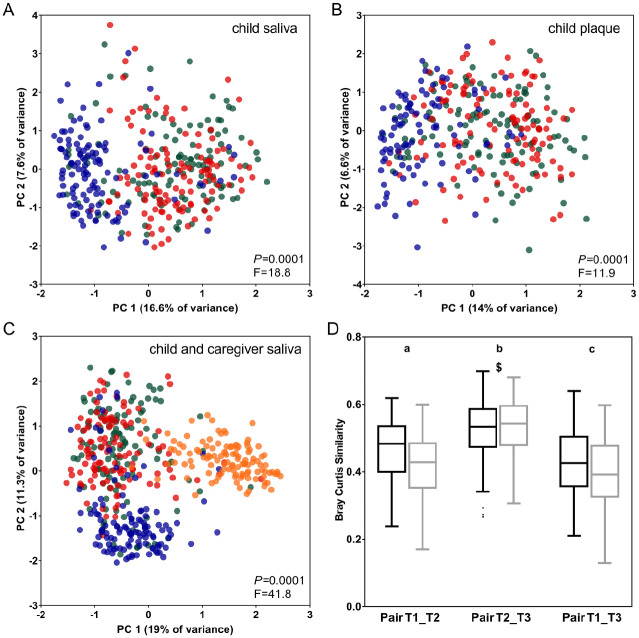
Microbial profile analyses of salivary (Fig. 3A) and dental plaque samples (Fig. 3B) of children showed that samples collected at the earliest time point (T1) formed a separate cluster from the other samples. Although no clear separation was discernible between the later time points, T2 and T3, the differences were significant among all 3 time points (saliva: P = 0.0001, R2 = 0.1; plaque: P = 0.0001, R2 = 0.07, restricted PERMANOVA), also between T2 and T3 in saliva (P = 0.006, R2 = 0.009, Bonferroni corrected, restricted PERMANOVA), and in plaque (P = 0.006, R2 = 0.007, Bonferroni corrected, restricted PERMANOVA). Microbial profiles of the caregivers from whom saliva was collected at all time points (n = 66) changed significantly across the 3 time points (P = 0.01, restricted PERMANOVA) (Appendix Fig. 4A). Explained variation was 0.08% (R2 = 0.008).
Figure 3.
Principal component analysis (PCA) plots displaying the microbial profiles of (A) saliva of children (n = 356 samples), (B) plaque of children (n = 323), and (C) saliva of children and caregivers (n = 471) by time points. Blue dots indicate samples collected at T1 (~1 y of age); red dots, at T2 (~2.5 y of age); and green dots, at T3 (~4 y of age). Orange dots (C) indicate saliva samples of caregivers (n = 115) at T1. Panel C shows a comparison of salivary microbial profiles of children at all time points (T1, T2, T3) and saliva of their caregivers collected at T1. PCA was performed on subsampled and log-2 transformed zero-radius operational taxonomic unit data. Axis shows the first 2 greatest principal components (PCs) explaining the highest intersample variation (percentage of variance). The P and F values indicate the output of PERMANOVA analyses, using the Bray-Curtis similarity measure, in comparing the respective samples. Similarity (D) of salivary (black boxes) and plaque (gray boxes) samples collected at different time points from the same child (pairwise comparison). Saliva (black) and plaque (gray) samples were tested separately and plotted together as they showed the same pattern of changes. Different letters indicate statistically significant differences between the sample pairs (P < 0.05, paired samples Friedman test followed by Wilcoxon rank-sum test). The pairwise comparisons of the 2 types of samples (saliva vs. plaque) were significantly different (P < 0.05) in all but the time point indicated with $.
Comparison of the salivary microbiome profiles of children and their caregivers showed that samples of the caregivers clustered separately from those of children collected at all time points (P = 0.0001, R2 = 0.21, restricted PERMANOVA) (Fig. 3C, Appendix Fig. 4B–D). Although the microbial compositions of children and adults remained significantly different throughout the 3 y of the study, the similarity in salivary microbiome between each child and his or her caregiver increased significantly from the age of 1 (T1) to 2.5 y (T2) (P < 0.01, FDR-corrected general linear model repeated measures test). Within an individual child, the microbial composition changed the most between the first time point and the others (Fig. 3D).
Diversity of both saliva and plaque collected at T1 was significantly lower than at the later time points (T2, T3), while diversity did not change from T2 to T3 (Appendix Fig. 5A, B). The salivary microbiome of the caregivers was significantly more diverse (median, 400 [244 to 641] zOTUs/sample) than that of children at all time points (at T1: 235 [112 to 457], T2: 369 [141 to 518], T3: 359 [161 to 573] zOTUs/sample) (Appendix Fig. 5B).
Individual Microbial Taxa over Time and in Comparison with the Caregivers
A single zOTU, zOTU1, classified as Streptococcus, was present in all saliva and plaque samples (n = 925) of the study (16% of the reads). All saliva samples of children and their caregivers (n = 471) shared 3 zOTUs: zOTU1, zOTU32 (Streptococcus), and zOTU9 (Gemella) with 22%, 0.7%, and 3% of the reads, respectively (Appendix Table 1C). There was 76% overlap in zOTUs in the overall child saliva composition across the 3 time points and 60% overlap overall between the child and caregiver saliva (Appendix Fig. 6A).
A large variation in the proportion of the shared zOTUs was observed within an individual child over time (2% to 42% of zOTUs in saliva; 2.5% to 38% in plaque) or within a child-caregiver pair throughout all time points (2% to 17% of zOTUs in saliva) (Appendix Fig. 6B).
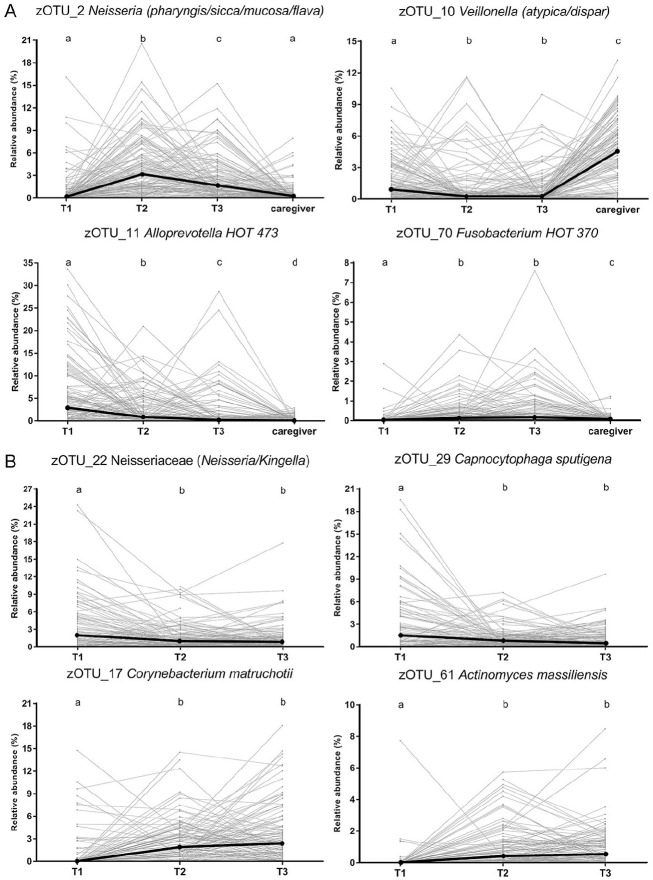
Of the zOTUs included in the analyses, the relative abundance of over 200 zOTUs was significantly higher in saliva of the caregivers compared to the children (Appendix Table 2A). Over 180 and 130 taxa increased in their relative abundance with time in saliva and plaque, respectively, including Fusobacterium, Actinomyces, and Corynebacterium (Fig. 4A, B), while over 40 taxa in saliva and over 20 taxa in plaque showed significant decreases over time (Appendix Table 2A).
Figure 4.
Relative abundance of zero-radius operational taxonomic units (zOTUs) that showed most significant changes over time in (A) saliva or (B) plaque. The individual values at T1, T2, and T3 originating from each individual are connected with a line. Incomplete lines indicate missing samples from the child at one of the time points. Bold black dots connected with a line indicate median of the relative abundance of zOTU at each time point. The presented zOTUs are classified to the highest possible bacterial taxon at a species, genus, or family level. Different letters indicate statistically significant differences (P < 0.05, paired samples Friedman test followed by Wilcoxon rank-sum test with false discovery rate correction). The zOTUs were first identified as discriminatory, using the linear discriminant analysis effect size (LEfSe) (the top 10 zOTUs by their linear discriminant analysis scores are shown in Appendix Table 2A), followed by pairwise tests.
To address if specific microbial taxa (namely, Streptococcus mutans and Porphyromonas gingivalis) traditionally associated with dental caries and periodontal diseases are present in orally healthy children between 1 and 4 y of age, we assessed the presence and relative abundance of these 2 taxa in both child and caregiver samples (Results section of the Appendix file). In brief, 7 zOTUs were classified as S. mutans, found in saliva of 18 and plaque of 21 children. A single zOTU was classified as P. gingivalis, which was found at a very low relative abundance (0.01% to 0.02%) in only 4 children.
Differences between Salivary and Plaque Microbial Composition
We compared the microbial composition of 2 types of samples, unstimulated saliva and pooled dental plaque, over time. As expected, microbial profiles of saliva were significantly different from those of plaque (T1: P = 0.0001, R2 = 0.23; T2: P = 0.0001, R2 = 0.13; T3: P = 0.0001, R2 = 0.15, restricted PERMANOVA), although the compositions of plaque and saliva became more similar from 1 to 2.5 y of age (Appendix Fig. 7A–C). Saliva samples had significantly higher species richness than plaque samples at T1 (P < 0.0001, FDR-corrected Friedman test). Of 214 zOTUs that were at a significantly higher proportion in saliva compared to plaque at T1, zOTUs classified as Streptococcus, Alloprevotella, Neisseria, Haemophilus, and Gemella differed the most (Appendix Fig. 8A). In plaque, 151 zOTUs were higher in their relative abundance than in saliva at T1, with Neisseria, Streptococcus sanguinis, Actinomyces, Capnocytophaga sputigena, Rothia aeria, and Corynebacterium durum being the most discriminatory between the 2 sample types (Appendix Fig. 8B; Appendix Table 2B). These taxa remained in the top 10 most discriminatory zOTUs also at later time points (Appendix Fig. 8).
Microbiome Differences by Age and Tooth Eruption Status of the Children at T1
The age of the children at the start of the study (T1) varied from 9 to 15.9 mo and included both predentate (n = 18) and dentate (n = 101) children (median, 4 teeth; range, 0 to 12) (Table, Fig. 1).
We stratified the children at T1 into 3 subgroups by their age: 1) below 11 mo (n = 54), 2) 11 to 13.9 mo (n = 40), and 3) 14 mo or older (n = 25). Both the salivary and plaque microbiome of the youngest children differed significantly from the 2 older subgroups (Appendix Fig. 9A, B). The diversity of the microbiome of the youngest children (T1) was significantly lower than that of the older children (T2, T3) (Appendix Fig. 9E, F).
The following subgroups were created based on the teeth eruption status at T1: 1) predentate (n = 18), 2) 1 to 4 teeth erupted (n = 56), and 3) 5 to 12 teeth erupted (n = 45). Saliva of the predentate children and children with 1 to 4 teeth differed significantly from the microbial profiles of the saliva from children with 5 or more teeth (Appendix Fig. 9C). These differences were reflected in their alpha and beta diversities (Appendix Fig. 9G, H). Microbial profiles and diversity of plaque from children with 1 to 4 teeth differed significantly from the children with 5 to 12 teeth erupted (Appendix Fig. 9D, G, H).
Salivary and Plaque Microbiome by Demographic Data and Exposure to Antibiotics
We found no differences in the composition of the salivary and plaque microbiome by delivery mode, while plaque at T2 differed by sex (P = 0.007, PERMANOVA) and at T3 by race/ethnicity (Caucasian vs. African American, P = 0.002, PERMANOVA). Microbial profiles of saliva collected at T2 and plaque collected at all 3 time points differed in beta-diversity by Medicaid status of the children (P < 0.01, PERMANOVA). Plaque samples collected at T3 from the Medicaid group showed higher diversity (Shannon Diversity index, P = 0.007, Kruskal-Wallis test) than plaque from the non-Medicaid group. At all time points, multiple zOTUs significantly discriminated between the 2 groups (Appendix Table 2C). Microbial composition of both saliva and plaque samples of children who were exposed to antibiotics within 4 wk of sample collection differed significantly from those who were not (P < 0.05, PERMANOVA).
Total Bacterial and Fungal Load in Saliva and Plaque Samples
Saliva samples collected at T1 and those collected from caregivers had higher bacterial DNA concentrations than saliva from the older children (Appendix Fig. 10A). At T1, fungal DNA was present in the saliva of 98% of the caregivers and 98% of the children, while at T2 and T3, 92% and 89% of the children, respectively, had detectable fungi in their saliva. Over time, saliva collected at T1 had the highest and at T3 the lowest fungal DNA concentration (Appendix Fig. 10B). Salivary fungal load (relative abundance of fungal DNA over bacterial DNA) was also the highest at T1 (median, 0.013%; range, 0% to 11%) (Appendix Fig. 10C).
In plaque, only 66% of samples had detectable fungi at T1, while at T2 and T3, fungi were present in 90% and 68% of the samples, respectively (Appendix Fig. 10B, C).
Discussion
In this study, we focused on development of the healthy oral microbial ecosystem. The children who remained caries free up to age 4 y were followed in time from 1 y of age. Both salivary and plaque microbial composition underwent major changes from 1 until 2.5 y of age, followed by less pronounced microbial shifts up to 4 y of age. However, salivary and plaque microbial composition still did not reach the complexity of the microbiome of their caregivers. At the population level, the majority of the microbial taxa (zOTUs) were shared throughout the 3 y of this study. However, at the level of the individual, large variation in the number of shared zOTUs was observed and only a few taxa were present throughout the 3 y.
Among the longitudinal oral microbiome studies on children, 3 studies, similar to ours, have used the ICDAS = 0 for defining caries-free individuals (Lif Holgerson et al. 2015; Richards et al. 2017; Dzidic et al. 2018). In 2 of these, the sample size of the caries-free group was small (n = 22 in Richards et al. [2017]; n = 11 in Lif Holgerson et al. [2015]). A study by Dzidic et al. (2018) followed the largest group and for the longest period to date; they described the salivary microbiome from 3 mo until 7 y in 90 Swedish children, of whom 45 were caries free at the age of 9 y. Their study provides extensive description of salivary microbiome development in relation to postnatal factors and caries. Unfortunately, no caries assessment was performed before the age of 9 y, precluding the diagnosis of lesions in deciduous teeth other than canines and molars remaining in the mixed dentition at that age. Any dynamics in the caries process evidenced by remineralization of enamel lesions obtained at a very young age also could not be recorded. For example, in 32% of the 2.5-y-old children in our cohort diagnosed with enamel lesions, the lesions were remineralized at the age of 4 y.
Therefore, with the current longitudinal study on 119 individuals, we have provided the first and largest to date extensive description of the maturation of the healthy oral ecosystem in caries-free children aged from 1 to 4 y.
We found an overall increase in microbial diversity with increasing age, supporting the findings of previous studies (Holgerson et al. 2015; Richards et al. 2017; Dzidic et al. 2018; Li et al. 2018). Only a few taxa (several species of genus Streptococcus and Gemella) were present in all samples of children over time, which is in agreement with the study from Sweden where infants from 3 mo until 3 y of age were followed (Holgerson et al. 2015).
Due to the ease of collection, saliva is the most frequently collected sample in oral microbiome studies (Humphrey and Williamson 2001). Due to the young age of the study participants, it was not possible to collect saliva samples using standard saliva collection protocols such as unstimulated saliva by drooling. Instead, a slow and gentle swab over the floor of the mouth with 2 relatively large sponges was performed, allowing salivary absorption. This, however, may have resulted in saliva sample contamination with microbiota residing on the sublingual mucosa. On the other hand, studies comparing different intraoral niches in adults have shown that saliva closely resembles the mucosal surface. Sampling of dental plaque requires a clinical setting and an experienced operator but allows site-specific assessment of the microbiome (Simon-Soro and Mira 2015). Interestingly, despite distinct differences in the composition of these 2 sample types, we observed similar dynamics of the maturation in saliva and plaque during the 3 y of the study.
Relating to the ecological plaque hypothesis in caries etiology, we cannot ensure that some individuals within our cohort, even though diagnosed as enamel caries free at the last time point (aged 4 y), might not, in fact, be experiencing a microbial ecological shift toward caries. A clinical follow-up of the included children at a later age should be performed to confirm their healthy status.
Yeasts, such as Candida albicans, are frequently associated with early childhood caries (Raja et al. 2010), while little is known of fungal communities in health. We found a high prevalence of fungal DNA and especially a high concentration of fungi in the saliva of children at the earliest time point. The low complexity of the bacterial communities, as well as a rather immature immune system during the first year of life, might contribute to enrichment of these microbes (Salvatori et al. 2016). Since their roles in health and in transition to disease remain unclear, this warrants mycobiome assessment in future studies.
In conclusion, the oral ecosystem of caries-free toddlers is highly heterogeneous and dynamic, with substantial changes in microbial composition and only a few taxa persistent across the 3 y of the study. However, the salivary microbiome of 4-y-old children is still distinct from that of their caregivers.
Author Contributions
D. Kahharova, B.W. Brandt, M.J. Buijs, E. Zaura, contributed to data acquisition, drafted and critically revised the manuscript; M. Peters, R. Jackson, G. Eckert, B. Katz, M.A. Keels, S.M. Levy, M. Fontana, contributed to conception, design, and data acquisition, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034519889015 for Maturation of the Oral Microbiome in Caries-Free Toddlers: A Longitudinal Study by D. Kahharova, B.W. Brandt, M.J. Buijs, M. Peters, R. Jackson, G. Eckert, B. Katz, M.A. Keels, S.M. Levy, M. Fontana and E. Zaura in Journal of Dental Research
Acknowledgments
We thank the members of the Caries Risk Grant Group who contributed in data acquisition: Emily Yanca, Susan Flannagan, Barcey Levy, Jeanette Daly, John Warren, Justine Kolker, Alex Kemper, Dennis Clements, Jennifer Talbert, Fredrica Gallack, Brenda Pattison, Beth Patterson, Anderson Hara, Sue Kelly, Jen Tran, Sharon Gwinn, Lorena Galvez, and Lisa Robinson.
Footnotes
A supplemental appendix to this article is available online.
This study was supported by National Health Institute (NIH) grant 5U01DE021412, NIH CTSA grants (UL1-TR000442 [University of Iowa], 2UL1-TR000433 [University of Michigan], and UL1-TR000006 [Indiana University]), Colgate, the University of Michigan, School of Dentistry, and a Consortium of Delta Dental Plans (Delta Dental of Iowa, Delta Dental of Wisconsin, the Renaissance Health Service Corporation for Delta Dental of Michigan). D. Kahharova was supported by Stichting Bevordering Tandheelkundige Kennis with NTvT Onderzoeksbeurs 2017 and by the ACTA Research Institute.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
ORCID iD: M. Peters  https://orcid.org/0000-0003-2050-3978
https://orcid.org/0000-0003-2050-3978
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 43(11):5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric L, Pratten J, Wilson M, Spratt D. 2010. Development of a novel multi-triplex qPCR method for the assessment of bacterial community structure in oral populations. Environ Microbiol Rep. 2(6):770–774. [DOI] [PubMed] [Google Scholar]
- Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. 2011. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagli N, Dagli R, Darwish S, Baroudi K. 2016. Oral microbial shift: factors affecting the microbiome and prevention of oral disease. J Contemp Dent Pract. 17(1):90–96. [DOI] [PubMed] [Google Scholar]
- Daly JM, Levy SM, Xu Y, Jackson RD, Eckert GJ, Levy BT, Fontana M. 2016. Factors associated with parents’ perceptions of their infants’ oral health care. J Prim Care Commun Health. 7(3):180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzidic M, Collado MC, Abrahamsson T, Artacho A, Stensson M, Jenmalm MC, Mira A. 2018. Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 12(9):2292–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert GJ, Jackson R, Fontana M. 2010. Sociodemographic variation of caries risk factors in toddlers and caregivers. Int J Dent. 2010:593487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2016. Unoise2: improved error-correction for Illumina 16S and ITS amplicon sequencing [epub ahead of print 15 Oct 2016]. BioRxiv:081257. [Google Scholar]
- Fontana M, Eckert GJ, Keels MA, Jackson R, Katz B, Levy BT, Levy SM. 2018. Fluoride use in health care settings: association with children’s caries risk. Adv Dent Res. 29(1):24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana M, Eckert GJ, Keels MA, Jackson R, Katz BP, Kemper AR, Levy BT, Levy SM, Yanca E, Kelly S, et al. 2019. Predicting caries in medical settings: risk factors in diverse infant groups. J Dent Res. 98(1):68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana M, Jackson R, Eckert G, Swigonski N, Chin J, Zandona AF, Ando M, Stookey GK, Downs S, Zero DT. 2011. Identification of caries risk factors in toddlers. J Dent Res. 90(2):209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer O, Harper DAT, Ryan P. 2001. Past: paleontological statistics software package for education and data analysis. Palaeontol Electron. 4:1–9. [Google Scholar]
- Humphrey SP, Williamson RT. 2001. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 85(2):162–169. [DOI] [PubMed] [Google Scholar]
- Hurley E, Barrett MPJ, Kinirons M, Whelton H, Ryan CA, Stanton C, Harris HMB, O’Toole PW. 2019. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health. 19(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Caries Detection and Assessment System (ICDAS) Coordinating Committee. 2012. Rationale and Evidence for the International Caries Detection and Assessment System (ICDAS II) [accessed 2019 Oct. 29]. https://pdfs.semanticscholar.org/0478/3d0cfe0a96ffb865c358f780f5227b9baca9.pdf.
- Jiang S, Gao X, Jin L, Lo EC. 2016. Salivary microbiome diversity in caries-free and caries-affected children. Int J Mol Sci. 17(12):E1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 79(17):5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Nam OH, Lee HS, Choi SC. 2016. Diversity and homogeneity of oral microbiota in healthy Korean pre-school children using pyrosequencing. Acta Odontol Scand. 74(5):335–336. [DOI] [PubMed] [Google Scholar]
- Li F, Tao D, Feng X, Wong MCM, Lu H. 2018. Establishment and development of oral microflora in 12–24 month-old toddlers monitored by high-throughput sequencing. Front Cell Infect Microbiol. 8:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lif Holgerson P, Öhman C, Rönnlund A, Johansson I. 2015. Maturation of oral microbiota in children with or without dental caries. PLoS One. 10(5):e0128534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo AH, Yang DQ, Xin BC, Paster BJ, Qin J. 2012. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis. 18(6):595–601. [DOI] [PubMed] [Google Scholar]
- Marsh PD. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 8(2):263–271. [DOI] [PubMed] [Google Scholar]
- Oliveros JC. 2007. VENNY: an interactive tool for comparing lists with Venn diagrams [accessed 1 May 2019]. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Papaioannou W, Gizani S, Haffajee AD, Quirynen M, Mamai-Homata E, Papagiannoulis L. 2009. The microbiota on different oral surfaces in healthy children. Oral Microbiol Immunol. 24(3):183–189. [DOI] [PubMed] [Google Scholar]
- Pitts NB. 2004. Modern concepts of caries measurement. J Dent Res. 83 Spec No C:C43–47. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; https://www.R-project.org/. [Google Scholar]
- Raja M, Hannan A, Ali K. 2010. Association of oral candidal carriage with dental caries in children. Caries Res. 44(3):272–276. [DOI] [PubMed] [Google Scholar]
- Richards VP, Alvarez AJ, Luce AR, Bedenbaugh M, Mitchell ML, Burne RA, Nascimento MM. 2017. Microbiomes of site-specific dental plaques from children with different caries status. Infect Immun. 85(8):e00106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori O, Puri S, Tati S, Edgerton M. 2016. Innate immunity and saliva in Candida albicans–mediated oral diseases. J Dent Res. 95(4):365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Tian J, Xu H, Zhou Q, Qin M. 2018. Distinctions and associations between the microbiota of saliva and supragingival plaque of permanent and deciduous teeth. PLoS One. 13(7):e0200337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Soro A, Mira A. 2015. Solving the etiology of dental caries. Trends Microbiol. 23(2):76–82. [DOI] [PubMed] [Google Scholar]
- Teng F, Yang F, Huang S, Bo C, Xu ZZ, Amir A, Knight R, Ling J, Xu J. 2015. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe. 18(3):296–306. [DOI] [PubMed] [Google Scholar]
- Vollmer T, Stormer M, Kleesiek K, Dreier J. 2008. Evaluation of novel broad-range real-time PCR assay for rapid detection of human pathogenic fungi in various clinical specimens. J Clin Microbiol. 46(6):1919–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade WG. 2013. The oral microbiome in health and disease. Pharmacol Res. 69(1):137–143. [DOI] [PubMed] [Google Scholar]
- Xin BC, Luo AH, Qin J, Paster BJ, Xu YL, Li YL, Yang DQ. 2013. Microbial diversity in the oral cavity of healthy Chinese Han children. Oral Dis. 19(4):401–405. [DOI] [PubMed] [Google Scholar]
- Xu X, He J, Xue J, Wang Y, Li K, Zhang K, Guo Q, Liu X, Zhou Y, Cheng L, et al. 2015. Oral cavity contains distinct niches with dynamic microbial communities. Environ Microbiol. 17(3):699–710. [DOI] [PubMed] [Google Scholar]
- Xu Y, Jia YH, Chen L, Huang WM, Yang DQ. 2018. Metagenomic analysis of oral microbiome in young children aged 6–8 years living in a rural isolated Chinese province. Oral Dis. 24(6):1115–1125. [DOI] [PubMed] [Google Scholar]
- Zaura E, Nicu EA, Krom BP, Keijser BJ. 2014. Acquiring and maintaining a normal oral microbiome: current perspective. Front Cell Infect Microbiol. 4:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI, Nelson DE, et al. 2013. Biogeography of the ecosystems of the healthy human body. Genome Biol. 14(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034519889015 for Maturation of the Oral Microbiome in Caries-Free Toddlers: A Longitudinal Study by D. Kahharova, B.W. Brandt, M.J. Buijs, M. Peters, R. Jackson, G. Eckert, B. Katz, M.A. Keels, S.M. Levy, M. Fontana and E. Zaura in Journal of Dental Research