Abstract
Sigma factors are dissociable subunits of bacterial RNA polymerase that ensure efficient transcription initiation from gene promoters. Owing to their prokaryotic origin, chloroplasts possess a typical bacterial RNA polymerase together with its sigma factor subunit. The higher plant Arabidopsis thaliana contain as many as six sigma factors for the hundred or so of its chloroplast genes. The role of this relatively large number of transcription initiation factors for the miniature chloroplast genome, however, is not fully understood. Using two Arabidopsis T-DNA insertion mutants, we show that sigma factor 1 (SIG1) initiates transcription of a specific subset of chloroplast genes. We further show that the photosynthetic control of PSI reaction center gene transcription requires complementary regulation of the nuclear SIG1 gene at the transcriptional level. This SIG1 gene regulation is dependent on both a plastid redox signal and a light signal transduced by the phytochrome photoreceptor.
Keywords: Chloroplast gene transcription, light acclimation, photosynthesis, photosystem stoichiometry adjustment, phytochrome, redox signaling, sigma factor 1, transcriptional control
Sigma factor 1 initiates transcription of a specific subset of chloroplast genes in Arabidopsis. Photosynthetic control of psaAB transcription requires a light quality-mediated regulation of the nuclear sigma factor 1 gene.
Introduction
Sigma factors are subunits of bacterial RNA polymerases. They enable efficient transcription of bacterial genes by their three distinct activities: by imparting a promoter recognition property to the RNA polymerase; by melting the double-stranded promoter regions into transcription-competent, single-stranded open complexes; and by interacting with other DNA-binding transcription factors for regulated gene expression (Paget and Helmann, 2003; Davis et al., 2017). Chloroplasts are cytoplasmic organelles in which photosynthesis takes place in plants and algae. By virtue of their cyanobacterial ancestry, chloroplasts contain a small transcriptionally active genome and a bacterial gene expression machinery (Keeling, 2010). The chloroplast genome typically contains 100–300 genes, which are mostly organized in polycistronic operons as in bacteria. Most chloroplast genes have bacterial-type –10 and –35 promoter elements, which are recognized and transcribed by a multisubunit eubacterial RNA polymerase (Weihe and Börner, 1999). The core subunits of this bacterial-type polymerase are encoded in the plastid genome and hence named as the plastid-encoded polymerase (PEP). Like the bacterial polymerase, the core PEP enzyme requires the reversibly binding, nuclear-encoded sigma factor subunit for efficient transcription. In flowering plants and moss, one or more single-subunit phage-type RNA polymerases, known as the nuclear-encoded polymerases (NEPs), transcribe a small subset of chloroplast genes from distinct promoter elements (Börner et al., 2015). These NEP-transcribed genes include rpoB, encoding the β-subunit of PEP, and a few tRNA genes. Some chloroplast genes contain both PEP and NEP promoters and are transcribed by both polymerases in all stages of plastid development and all plant tissue types (Börner et al., 2015).
The chloroplast proteome contains ~3000 proteins, of which only <5% are encoded in the chloroplast genome (Martin et al., 2002). The rest are products of nuclear genes. The diminutive chloroplast genome is nevertheless over-represented by genes that encode core subunits of the photosynthetic electron transport complexes (Allen et al., 2011). Two large pigment protein complexes, PSII and PSI, form the functional units of photosynthesis. They undertake the light harvesting and the primary light-driven electron transport reactions of photosynthesis. A cytochrome b6f complex (Cyt b6f) connects the two photosystems in series. These three protein complexes together carry out the linear electron transport from water to NADP+, generating the electron-rich NADPH molecule and a proton motive force that drives the synthesis of ATP through the ATP synthase enzyme. The NAD(P)H dehydrogenase-like complex (NDH), a homolog of respiratory complex I, forms a further electron transport complex in chloroplasts. The function of NDH is the cycling of electrons around PSI in order to build a proton motive force that drives the synthesis of additional ATP molecules (Shikanai, 2016; Strand et al., 2017; Laughlin et al., 2019). The core protein subunits of all five complexes are products of chloroplast genes. Some of these chloroplast-encoded subunits act as dominant assembly factors for de novo complex biogenesis and thus set the pace of complex formation in the photosynthetic thylakoid membrane (Choquet and Vallon, 2000; Nickelsen and Rengstl, 2013). Regulation of the gene expression of these dominant assembly factors could therefore adjust the relative abundance of electron transport complexes, optimizing electron flux and photosynthetic efficiency in changing light conditions (Pfannschmidt et al., 1999).
Chloroplast sigma factors belong to the large sigma 70 family of sigma factors (Chi et al., 2015). The model higher plant Arabidopsis thaliana contain as many as six chloroplast sigma factors (Schweer et al., 2010a). Biochemical and molecular genetic studies reveal a certain degree of functional specialization among chloroplast sigma factors, with each seemingly possessing a unique set of target genes that are transcribed in an environment- and plant development-dependent fashion (Lerbs-Mache, 2011; Yagi and Shiina, 2014; Chi et al., 2015). Sigma factor 1 (SIG1) appears to be a major housekeeping sigma factor in plant chloroplasts, but its target genes are not yet fully identified in Arabidopsis (Chi et al., 2015). The recombinant Arabidopsis SIG1 binds to psbA and rbcL gene promoters in vitro, albeit with a lower affinity (Privat et al., 2003). In rice, knocking out the SIG1 gene results in decreased transcript accumulation of mostly PSI (psa) genes, which is accompanied by a reduction in the amount of PSI (Tozawa et al., 2007). Transcript accumulation of psbB and psbE operons, encoding PSII reaction center core polypeptides, also decreases in the rice sig1 knockout, but to a lesser degree compared with the psa genes. A similarly altered chloroplast transcription is seen in a SIG1 knockout mutant of the liverwort Marchantia (Ueda et al., 2013). ChIP of Arabidopsis SIG-bound promoter regions shows enrichment of multiple chloroplast operons including genes for both photosystems and the Rubisco large subunit (RbcL) (Hanaoka et al., 2012). Arabidopsis SIG1 has further been shown to become phosphorylated with important regulatory implications for PSI gene transcription (Shimizu et al., 2010). To further understand the role of SIG1 in basal and regulated transcription of chloroplast genes, we analyzed two independent Arabidopsis SIG1 T-DNA insertion mutants.
Materials and methods
Plant growth conditions
Arabidopsis thaliana (Col-0) wild-type and mutant plants were grown from seeds on soil at 23 °C under a photon flux density of 150 µmol m–2 s–1 with an 8 h light and 16 h dark photoperiod, unless otherwise specified. For the light switch time-course experiment, wild-type Arabidopsis, sig1-1, sig1-2, and phyB null mutant (phyB-9; harboring a premature stop codon) seedlings were grown in white light (150 µmol m–2 s–1; 16 h light) for 7 d and then transferred to light 1 (L1) or light 2 (L2) cabinets and allowed to acclimate for 4 d. At the end of the fourth day, the lights were switched (i.e. from L1 to L2, or vice versa). Leaf samples were collected before the light switch (zero time) and at 4, 8, 24, 28, and 32 h after the light switch.
Light 1 and 2
L1 was provided with Narva 18 W/015 red fluorescent strip lamps wrapped with a layer of plasa red filter (LEE 029). White fluorescent lamps (Philips Master TL-D 18 W/827) covered with a layer of burnt yellow filter (LEE 770) produced L2. Photon flux density at the leaf level was ~6 µmol m–2 s–1 in L1 and ~12 µmol m–2 s–1 in L2. L1 and L2 were made available on a photoperiod of 16 h light and 8 h dark. The specific action of L1 and L2 on PSI and PSII, respectively, were confirmed with measurement of the room temperature variable fluorescence yield associated with state transitions (data not shown).
Genotyping of SALK T-DNA lines
The SALK lines used in this study were genotyped for homozygous T-DNA insertion by using genomic and T-DNA cassette primers. Genomic DNA were isolated from plant leaf tissues by a protocol adapted from the Dellaporta method (Dellaporta et al., 1983). A reverse transcription–PCR (RT–PCR) was further used to check SIG1 transcript accumulation in the T-DNA lines. Amplification of the housekeeping gene Actin8 served as a control for RNA integrity. Total RNA was isolated from leaves by using the TRIzol reagent (Invitrogen) as per the manufacturer’s instructions. The cDNA was synthesized from 1 µg of total RNA with the RevertAid First Strand cDNA synthesis kit (Fisher Scientific) using an oligo(dT)18 primer. SIG1 and Actin8 transcripts were further detected by a Taq PCR using gene-specific primers. Sequences of all primers used are provided in Supplementary Table S1 at JXB online.
SIG1 protein quantification
The level of SIG1 protein was analyzed by a polyclonal antibody raised against the Arabidopsis SIG1. Total leaf protein was extracted from the wild type and sig1 mutants, and the protein concentration was determined with the Pierce BCA kit (Thermo Scientific). Equal amounts of protein samples were subjected to 11.5% (w/v) SDS–6 M urea–PAGE and electroblotted onto a polyvinylidene fluoride (PVDF) membrane (Immobilon-P, Millipore). The membrane was then blocked with 5% (w/v) non-fat dry milk (Bio-Rad) overnight at 4 °C, washed, and probed with the SIG1 primary antibody for 90 min at room temperature. Horseradish peroxidase-conjugated anti-rabbit secondary antibody (NA934, GE Healthcare) was used in the immunodetection of SIG1. Immunoreactive bands were visualized on a ChemiDoc MP imager (Bio-Rad) using a chemiluminescence detection reagent (Clarity Western ECL Substrate, Bio-Rad). A monoclonal plant actin antibody (A0480, Sigma) was used as a loading control and was detected using an anti-mouse secondary antibody (NA931, GE Healthcare). Band intensities of SIG1 and actin were analyzed by the ImageJ software.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from the leaves of 11- to 14-day-old light switch samples by using the TRIzol reagent. RNA was treated with RNase-free DNase (New England Biolabs) to eliminate possible DNA contamination. qRT-PCR was performed with a one-step QuantiTech SRBR Green RT-PCR kit from Qiagen in a StepOnePlus thermocycler (Applied Biosystems). The amplification efficiency of each primer pair (Supplementary Table S1) was checked by a 64-fold serial dilution of the template, and the R2 value of each primer pair was found to be ≥0.99. The expression values of target genes were normalized to both total RNA and endogenous Actin8 control. The relative changes in gene expression were analyzed by a 2–ΔΔCt method.
For qRT-PCR of DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea]-treated samples, excised leaves from 5- to 6-week-old wild-type plants were vacuum infiltrated with 10 µM DCMU and incubated under 150 µmol m–2 s–1 white light for 6 h while being floated in an isotonic buffer containing 0.4 M sorbitol, 20 mM tricine (pH 8.4), 10 mM EDTA, 10 mM NaHCO3, 0.15% (w/v) BSA, and 10 µM DCMU. At the end of the DCMU treatment, total RNA was isolated and qRT-PCR was done as before.
SIG1 complementation
SIG1 was complemented in the SALK_147985c line using Agrobacterium-mediated gene transfer. The coding sequence of SIG1 was cloned into a customized pCC2134_BAR expression vector (see Supplementary Table S1 for the primers used) and the resulting gene construct was delivered to Arabidopsis plants by the floral dip method. Transformants were subsequently screened by the Basta (bar) selection marker. qRT-PCR of selected nuclear and chloroplast genes was performed in complemented lines as before.
Results
Characterization of the SIG1 T-DNA insertion mutants
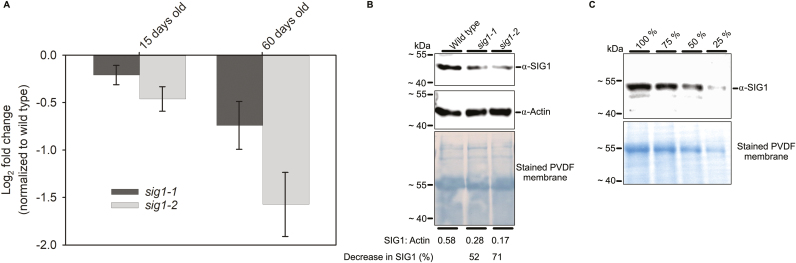
Two Arabidopsis T-DNA lines harboring insertions in the SIG1 gene locus At1g64860 were obtained from the ABRC. These mutant lines, SALK_147985c and CS371990, are hereafter simply referred to as sig1-1 and sig1-2, respectively. Figure 1A shows SIG1 transcript abundance in these mutants as quantified by qRT-PCR. Both mutants show decreased accumulation of the SIG1 transcript. The SIG1 transcript reduction is, however, more marked in sig1-2. sig1-1 is a confirmed homozygous SALK line with a T-DNA insertion in the 3'-untranslated region (UTR) of the SIG1 gene, while sig1-2 is a confirmed homozygous GABI-Kat line that carries an insertion within the penultimate exon (Supplementary Figs S1, S2). The different extent of SIG1 transcript reduction in the two mutants (Fig. 1A) is consistent with their unique T-DNA insertion sites. A T-DNA within the coding sequence is expected to disrupt transcript accumulation more severely, as seen in sig1-2, than an insertion within the 3'-UTR as in sig1-1. The varying degrees of transcript reduction further manifest at the SIG1 protein level in both mutants, as quantified by immunoblotting with an anti-SIG1 antibody (Fig. 1B). Though the SIG1 protein level decreases more noticeably in sig1-2, it is interesting to note that both mutants, regardless of T-DNA insertion, are able to accumulate apparently full-length SIG1 proteins. For sig1-1, this observation is consistent with its 3'-UTR location of T-DNA. For sig1-2, the T-DNA insertion is predicted to shorten the SIG1 protein by 62 amino acids (~6 kDa). A genotyping RT–PCR, designed to amplify the coding sequence of the mature SIG1 protein, however, shows that this does not happen (Supplementary Fig. S1D). The sig1-2 mutant indeed accumulates a faint amplificate of the right size, indicating that it is somehow able to splice out the T-DNA from the precursor mRNA and produce the full-length SIG1 protein—albeit less efficiently (Fig. 1; Supplementary Fig. S1D). sig1-1 and sig1-2 thus appear not to be true knockout mutants of SIG1 but rather knockdown lines.
Fig. 1.
T-DNA insertional mutagenesis decreases SIG1 transcript and protein levels. (A) SIG1 transcript abundance in sig1-1 and sig1-2 mutants as quantified by qRT-PCR. The log2 fold change after normalization with the wild type is shown. Error bars represent ±SE of the mean of four biological replicates. (B) SIG1 protein level as estimated by immunoblotting. Representative SIG1 and actin blots are shown with the corresponding stained PVDF membrane. Both SIG1 and actin are detected on the same membrane. Numbers below each lane denote the ratio of SIG1 to actin band intensity. The percentage decreases in SIG1 relative to the wild-type control are also given. The full uncropped versions of SIG1 and actin immunoblots are given in Supplementary Fig. S5. (C) An immunoblot of SIG1 with serial dilutions of the wild-type sample. The corresponding stained membrane is also shown. Molecular weight markers are indicated on the left. The mature SIG1 protein has a predicted mol. wt of 54 kDa. The SIG1 protein, however, runs on an 11.5% (w/v) SDS–6 M urea–PAGE gel with an apparent mol. wt of ~49 kDa.
Chloroplast gene targets of SIG1
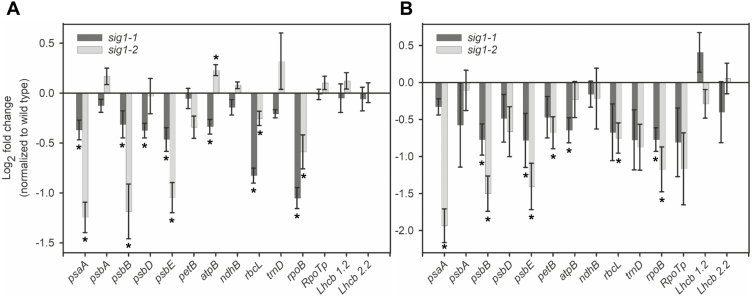
Since expression of chloroplast genes may correlate with the expression of their corresponding sigma factors, we analyzed transcript accumulation of all six sigma factor genes in leaves taken from different developmental stages of Arabidopsis. Plants were grown in short-day conditions to promote vegetative growth and to delay flowering. SIG1 seems to be maximally expressed in 60-day-old plants (Supplementary Fig. 3). Other sigma factors also show unique development-specific expression profiles, the implications of which for SIG1 function and chloroplast transcription in general are discussed further on. In our analysis of SIG1 gene targets, we have thus included a 60-day-old sample in addition to the 15-day-old young plants. Transcript abundance of all major chloroplast gene operons were quantified in wild-type, sig1-1, and sig1-2 plants by qRT-PCR. In 15-day-old sig1-1, transcript levels of the following chloroplast operons were significantly decreased compared with the wild type (P-value <0.05): psaA, psbD, psbE, rbcL, and rpoB (Fig. 2A). If we reanalyze the gene expression data using a one-tailed t-test distribution with the expectation that the lack of SIG1 affects chloroplast genes only in one direction (i.e. lower than the wild type), then the psbB and atpB operons also join the list of down-regulated genes (P-value <0.05). The rpoB gene, encoding the β-subunit of PEP, has been shown to be transcribed predominantly by the major chloroplast NEP isoform RpoTp in multiple plant species (Silhavy and Maliga, 1998; Zhelyazkova et al., 2012). We therefore examined whether the observed reduction in rpoB transcript arises from a corresponding decrease in RpoTp transcript. This, however, seems not to be the case as the RpoTp transcript level remains unaffected in 15-day-old sig1-1 (Fig. 2).
Fig. 2.
Abundance of selected chloroplast and nuclear gene transcripts in sig1 mutants. Transcript abundance in 15- (A) and 60-day-old plants (B). The log2 fold change after normalization with the wild type is shown. The asterisks indicate statistically significant changes (P-value <0.05 on a one- or two-tailed distribution). Error bars represent ±SE of the mean of four biological replicates.
The activities of chloroplast sigma factors SIG2 and SIG6 have been shown to generate distinct plastid to nuclear retrograde signals for the regulation of photosynthesis-associated nuclear genes (PhANGs) (Woodson et al., 2013). To examine whether the SIG1 activity is the source of any retrograde signal, the expression of two canonical PhANG genes Lhcb 1.2 and Lhcb 2.2 was checked in sig1 mutants. The transcript levels of these two genes were unchanged in 15-day-old sig1-1 (Fig. 2A). The 15-day-old sig1-2 mutant shows a similar set of chloroplast transcripts, with the levels of psaA, psbB, psbE, and rpoB significantly decreased, more so than in the sig1-1 mutant (Fig. 2A). A one-tailed t-test (P-value <0.05) would add rbcL to the list of down-regulated genes in sig1-2, but the psbD and atpB transcripts seem to decrease only in sig1-1. None of the nuclear gene transcripts shows any change in sig1-2, as with sig1-1 (Fig. 2A). The 60-day-old sig1-1 shows decreased accumulation of psbB, psbE, atpB, and rpoB transcripts on a two- or one-tailed t-test distribution (P-value <0.05), while the similarly aged sig1-2 mutant reveals reduction in psaA, psbB, psbE, petB, rbcL, and rpoB transcripts (P-value <0.05; two-tailed t-test) (Fig. 2B). As with 15-day-old plants, neither of the 60-day-old mutant plants shows any change in nuclear gene transcripts (Fig. 2B).
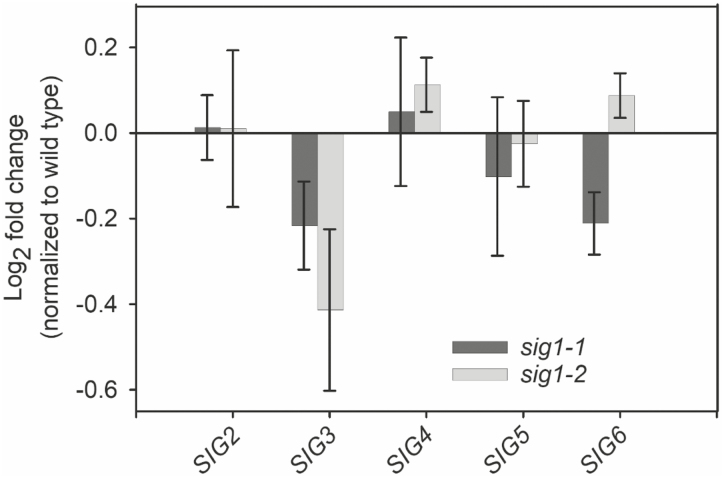
Overall, the magnitude of the reduction in chloroplast gene transcripts parallels the SIG1 protein level in each mutant (Fig. 1), with less SIG1 protein leading to lower accumulation of chloroplast transcripts in both 15- and 60-day-old mutants (Fig. 2). To check whether the decreased accumulation of SIG1 protein triggers any pleiotropic or compensatory response on the gene expression of other sigma factors, we analyzed their transcript abundance in 15-day-old sig1-1 and sig1-2 mutants (Fig. 3). Compared with the wild type, neither of the mutants shows any statistically significant change in transcript accumulation of SIG2, SIG3, SIG4, SIG5, and SIG6 (Fig. 3).
Fig. 3.
Unchanged SIG2–SIG6 expression in 15-day-old sig1 mutants. The sig1 mutation does not result in any statistically significant changes in the transcript abundance of other sigma factors. The log2 fold change after normalization with the wild type is shown. Error bars represent ±SE of the mean of four biological replicates.
Light quality acclimation involves transcriptional control of the nuclear SIG1 gene
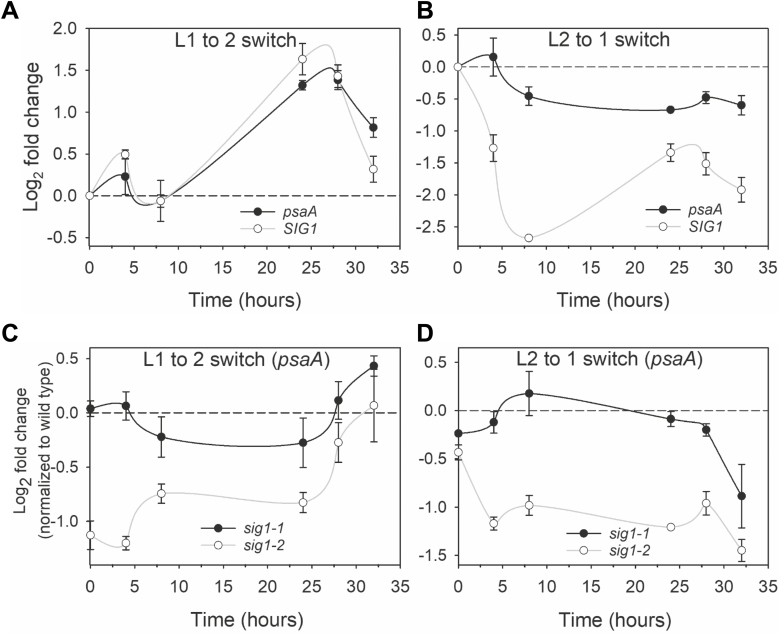
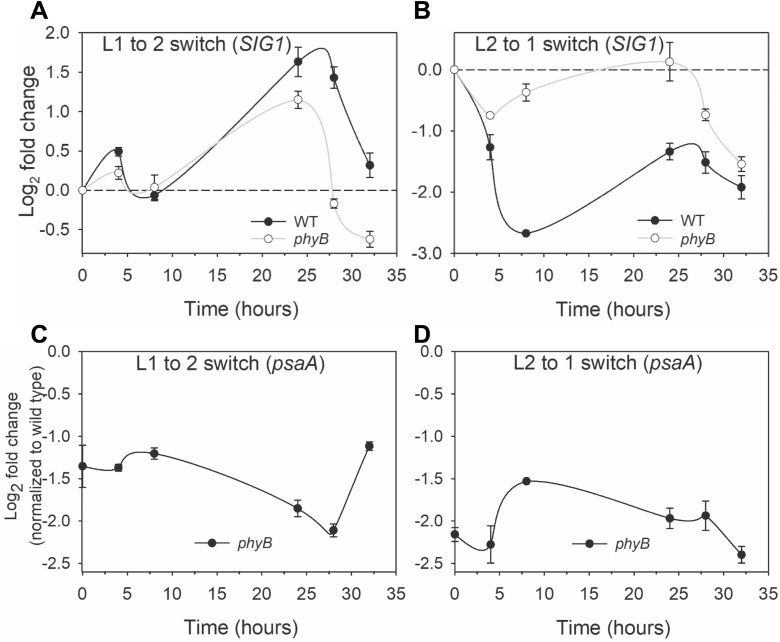
L1, enriched in far red, preferentially excites PSI and thereby oxidizes the plastoquinone (PQ) pool. An oxidized PQ pool leads to phosphorylation of SIG1 and the phospho-SIG1 represses psaA gene transcription (Shimizu et al., 2010). L2 is a short wavelength light that selectively excites PSII and thus reduces the PQ pool. A reduced PQ pool releases the repression on psaA gene transcription by dephosphorylating and/or degrading phosphorylated SIG1 (Shimizu et al., 2010). To further understand the involvement of SIG1 in the transcriptional control of psaA, we measured the kinetics of psaA and SIG1 transcript accumulation in the wild type in response to changes in light quality. Figure 4A shows psaA transcript accumulation kinetics of the wild type in response to an L1 to L2 switch. As expected, the psaA transcript level increases in PSII-specific L2. Interestingly, the SIG1 gene shows similar transcript accumulation kinetics to psaA (Fig. 4A). In the opposite light switch, namely when the PSII-specific L2 is replaced with PSI-specific L1, the amount of psaA transcript decreases as the light begins to favor PSI (Fig. 4B). The SIG1 transcript shows a similar reduction in L1 but the magnitude of its decrease is much higher than that of psaA (Fig. 4B).
Fig. 4.
Transcript accumulation kinetics of psaA and SIG1 genes. (A and B) Transcript accumulation kinetics of psaA and SIG1 in the wild type as quantified with qRT-PCR. Experimental conditions are switch from light 1 (L1) to light 2 (L2), and vice versa. The log2 fold change in transcript abundance is plotted against time. The time point at which lights are switched is taken as zero time and the concomitant up- or down-regulation is calculated by taking the expression at the time of light switch (zero time) as baseline. (C and D) Transcript accumulation kinetics of psaA in sig1 mutants. Each data point represents log2 fold change relative to the wild-type control. Error bars represent ±SE of the mean of three biological replicates.
To shed further light on the significance of concerted psaA and SIG1 transcript accumulation, we monitored psaA transcript accumulation kinetics in sig1 knockdown mutants. Figure 4C shows psaA transcript accumulation in response to an L1 to L2 switch. Compared with the wild type, sig1-2 has significantly less psaA transcript at the end of L1 illumination (time zero). After switch to L2, the psaA transcript level begins to increase but it takes nearly 32 h for the mutant to reach the wild-type level. The slower psaA transcriptional response of sig1-2 is consistent with its substantially lower SIG1 protein amount and its inability to further increase the SIG1 protein level promptly in L2. Interestingly, psaA transcript accumulation kinetics of the sig1-1 mutant were similar to those of the wild type except at 8 h and 24 h after the light switch. This nearly wild-type-like response of sig1-1 is compatible with its weaker SIG1 T-DNA allele. In the reciprocal light shift (i.e. L2 to L1 switch), the psaA transcript decreases more than that of the wild type in both mutants at 32 h after the light switch (Fig. 4D). However, the level of psaA transcript before the light switch (i.e. at time zero in L2) is also lower than that of the wild type in both mutants (Fig. 4D).
Redox- and light quality-dependent regulation of SIG1 gene transcription
To identify the factors that allow coordinated expression of the nuclear SIG1 gene with its chloroplast psaA target gene, we first checked the role of the PQ pool redox state. This is based on two considerations: (i) the psaA transcriptional response is governed by the PQ pool redox state (Pfannschmidt et al., 1999); and (ii) the PQ pool redox signal affects transcription of multiple nuclear genes (Fey et al., 2005). To test the role of the PQ pool, we first chemically altered its redox state with the electron transport inhibitor DCMU. DCMU binds to the QB-site of PSII and thus prevents reduction of PQ by PSII. The PQ pool therefore exists in a more oxidized state in DCMU-treated plants. Leaves from mature wild-type plants were infiltrated with DCMU and illuminated. The SIG1 transcript level in DCMU-treated samples decreased by 0.34±0.06-fold on a log2 scale (Supplementary Fig. S4), which is equivalent to a reduction of 21.2±4.0% relative to the untreated sample. Both DCMU and L1 (Fig. 4B; Supplementary Fig. S4) thus appear to decrease SIG1 transcript abundance. The effect of DCMU, however, seems modest compared with the large drop in SIG1 transcript abundance in L1 (Fig. 4B; Supplementary Fig. S4), raising the possibility that other factors besides the PQ pool redox state may also regulate the nuclear SIG1 gene expression.
Since phytochromes mediate many plant responses to light, we further checked SIG1 transcript accumulation kinetics in a phytochrome mutant (phyB). Figure 5A shows the response of the phyB mutant to the L1 to L2 switch. The level of SIG1 transcript in phyB fails to reach wild-type levels at 24, 28, and 32 h after the light switch. The aberrant SIG1 transcriptional response of phyB is more noticeable in the opposite L2 to L1 switch, where the initial rapid fall component of the response is nearly absent in the mutant (Fig. 5B). Significant differences from the wild type are also apparent at other time points (Fig. 5B). To determine how this impaired SIG1 transcriptional response affects psaA transcript accumulation kinetics in phyB, we quantified its psaA transcript level (Fig. 5C, D). The psaA transcript level in phyB is significantly lower than in the wild type in both L1 to L2 and L2 to L1 conditions (Fig. 5C, D). It is notable that the psaA transcript hardly changes after 32 h in L2 or L1 from the level at zero time (before the light switch) (Fig. 5C, D). This stands in contrast to sig1 knockdown mutants, which are able to catch up with the wild type eventually (Fig. 4C, D).
Fig. 5.
Light quality regulation of nuclear SIG1 and chloroplast psaA genes. (A and B) SIG1 transcript accumulation kinetics in the wild type and phyB mutant. The wild-type data are replotted from Fig. 4. (C and D) The log2 fold change in transcript accumulation of psaA in the phyB mutant is shown relative to the wild-type control. Error bars represent ±SE of three biological replicates.
Discussion
SIG1 initiates transcription of a specific subset of chloroplast genes
With a eubacterial RNA polymerase, six sigma factors, and two phage-type RNA polymerases, the transcriptional machinery of chloroplasts seems an overelaboration for just a handful of genes. It nevertheless appears that this elaborate cellular machinery forms the basis of a complex transcriptional program in chloroplasts that is both developmentally attuned and environmentally responsive. This transcriptional robustness seems to be built on an intricate division of labor among chloroplast sigma factors. SIG2 is required for transcription initiation of psbA, psaJ, and several tRNAs, including glutamyl-tRNA (trnE). trnE is a precursor for the biosynthesis of tetrapyrroles—chlorophylls and hemes—and therefore SIG2 knockout mutants show a pale phenotype (Hanaoka et al., 2003; Nagashima et al., 2004). Because of its involvement in trnE transcription and, as a result, tetrapyrrole metabolism, SIG2 activity seems to be the source of a chloroplast to nuclear retrograde signal (Woodson et al., 2013).
Available data indicate that SIG3 initiates transcription of only psbN and atp (ATPase) genes (Zghidi et al., 2007; Malik Ghulam et al., 2012), and SIG4, likewise, has a marginal role, transcribing only the ndhF gene encoding the F subunit of the NDH complex (Favory et al., 2005). SIG5 has a 2-fold function in chloroplast transcription. Binding to a blue light-responsive promoter, it transcribes the psbD gene that encodes the D2 protein of PSII (Tsunoyama et al., 2004). SIG5 further functions as a nuclear-encoded timing signal that generates a circadian rhythm in chloroplast gene transcription (Noordally et al., 2013). The null mutant of SIG6 exhibits a cotyledon-specific pale green phenotype with delayed chloroplast development (Ishizaki et al., 2005). Transcripts of several photosynthetic genes are decreased in the sig6 mutant. The SIG6 activity, like that of SIG2, seems to generate a nuclear gene regulatory retrograde signal (Woodson et al., 2013). The age-dependent expression profiles of nuclear sigma factors (Supplementary Fig. S3) broadly reflect their division of labor in plastid transcription. Out of the six factors, SIG3 shows the lowest expression in all stages of development, probably consistent with its limited functional role. The other sigma factors exhibit relatively high expression, peaking especially in mature plants, in line with their major transcriptional roles in chloroplasts. An exception is SIG4, which seems to act only at the ndhF gene promoter and yet displays high expression in all growth stages. It is possible that SIG4 has a wider functional role than currently realized with as yet unknown target genes.
The current understanding of the role of Arabidopsis SIG1 is largely based on a ChIP sequencing (ChIP-Seq) study and on inference from the rice sig1 knockout mutant (Tozawa et al., 2007; Hanaoka et al., 2012). The sig1-1 and sig1-2 mutants described in the current study have been used before with the assumption that they are complete knockout mutants (Lai et al., 2011; Woodson et al., 2013). As we show here, these mutants are rather knockdown lines of SIG1 (Fig. 1). The sig1-2 mutant, which contains the T-DNA insert within the coding sequence, appears to be a stronger knockdown allele of SIG1 than the sig1-1 mutant that harbors the insertion in the 3'-UTR (Fig. 1). The transcript abundance of chloroplast genes in sig1-1 and sig1-2 suggests that SIG1 is one of the sigma factors that initiate transcription of psaA, psbB, psbE, rbcL, and rpoB operons in Arabidopsis (Fig. 2). The psbD and petB transcripts also show reduction, but only in one out of the four samples analyzed (Fig. 2). Likewise, the atpB transcript seems to decrease only in sig1-1 (Fig. 2). The sig1-1 mutant shows a slight variation between 15- and 60-day-old samples in the set of significantly affected chloroplast genes, consistent with its weaker T-DNA allele. Interestingly, our list of SIG1 gene targets overlaps significantly with the operons identified in the ChIP-Seq study, which shows enrichment of SIG1 in the gene promoters of psaA, psbB, psbE, rbcL, and clpP (Hanaoka et al., 2012). The ClpP gene, which encodes the chloroplast ClpP protease, was not analyzed in our study. Our gene expression data go beyond the ChIP-Seq study by identifying the rpoB gene as a likely target of SIG1 in Arabidopsis (Fig. 2).
The plant rpoB gene is mostly transcribed by the NEP isoform RpoTp (Hricova, 2006). Given the unchanged RpoTp gene expression in mutants (Fig. 2), it is unclear how the deficiency in SIG1, a PEP subunit, would cause this phenotype. Two scenarios might explain this odd observation. The first is an intriguing possibility that RpoTp utilizes SIG1 for initiation of rpoB transcription. It has been suggested that the plant phage-type polymerases, similar to yeast and human mitochondrial phage-type polymerases, require additional factors to melt gene promoters. None has, however, been identified in plants thus far (Börner et al., 2015). Interestingly, multiple sigma factors including SIG1 and SIG5 seem to be targeted to plant mitochondria and are co-purified with the mitochondrial phage-type polymerase (Beardslee et al., 2002). It is thus possible that SIG1 functions as an accessory subunit of RpoTp for the transcription initiation of rpoB in plant chloroplasts. A second explanation for the decreased rpoB transcription is that the rpoB operon contains an as yet uncharacterized SIG1-dependent PEP promoter. Such a PEP promoter may create a positive feedback loop in PEP β-subunit expression, allowing rapid developmental and environmental acclimation of chloroplast gene transcription.
Since rpoB encodes a core subunit of PEP, it should be considered whether a decrease in its transcription and in turn the PEP content explains the observed reduction of chloroplast transcripts in sig1 mutants (Fig. 2). The unchanged expression of many PEP-dependent operons in sig1-1 and sig1-2, however, rules out this possibility (Fig. 2), as a deficiency in PEP would affect the overall chloroplast transcription rather than that of a specific subset of chloroplast genes. In fact, a general decrease in chloroplast transcription, including that of the exclusively PEP-transcribed psbA and rbcL genes, has been reported in an Arabidopsis RpoTp null mutant. This was attributed to a diminished PEP level resulting from the decreased transcription of rpoB (Courtois et al., 2007). The RpoTp knockout mutant that produces very little rpoB transcript and our sig1 knockdown mutants, which show only a 2-fold reduction in rpoB, thus seem to possess two distinct chloroplast transcriptional phenotypes.
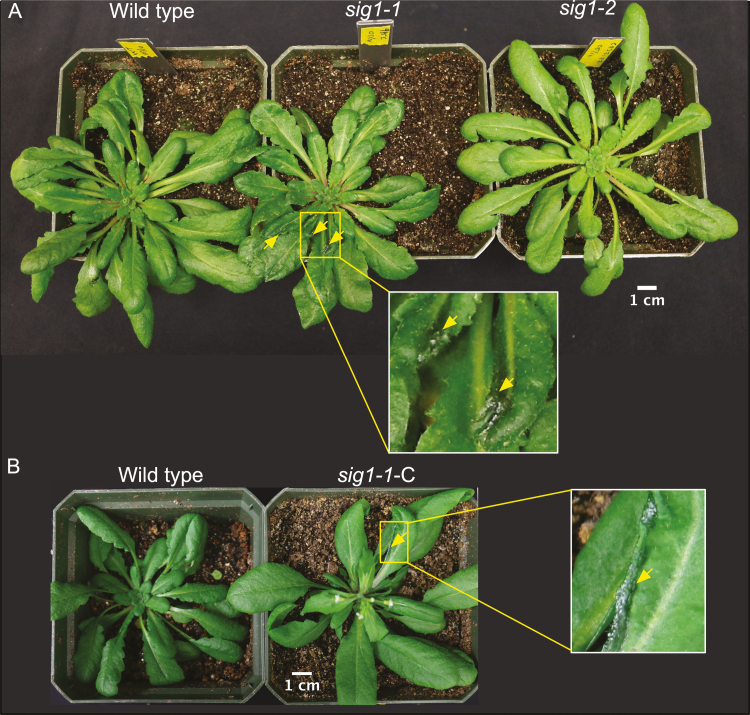
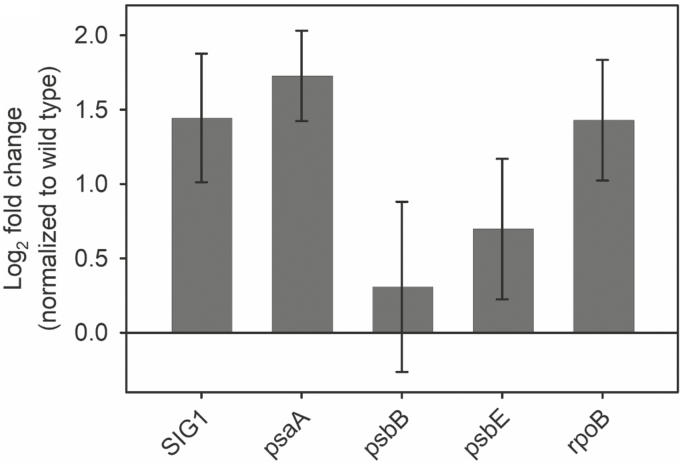
Curiously, the mature sig1-1 plants show a unique leaf phenotype in which some leaves become curly and deformed (Fig. 6). This phenotype, however, must be due to a second T-DNA insertion in a separate gene locus since the sig1-2 mutant does not show any such abnormal leaves (Fig. 6A). To further test whether the ‘curly’ leaf phenotype has anything to do with SIG1, we complemented the SIG1 gene in sig1-1 on a Cauliflower mosaic virus (CaMV) 35S promoter. The ‘curly’ leaf phenotype, however, could not be complemented (Fig. 6B), supporting the distinct genetic nature of this anomaly. SIG1 overexpression instead seems to complement the chloroplast transcriptional phenotype of sig1-1, with some gene targets of SIG1 showing increased transcript accumulation compared with the wild type (Fig. 7).
Fig. 6.
The sig1-1 mutant shows a curly leaf phenotype that is unrelated to SIG1 deficiency. (A) RGB images of mature wild-type, sig1-1, and sig1-2 plants. The curly, deformed leaf areas in sig1-1 are indicated by yellow arrowheads. The inset shows an enlarged view of the lesion. (B) Images of the wild type and complemented sig1-1 mutant (sig1-1-C). Scale bars are shown.
Fig. 7.
Abundance of selected gene transcripts in 60-day-old sig1-1-C. The log2 fold change relative to the wild type is shown. Error bars represent ±SE of four biological replicates.
SIG1 in photosynthetic control of PSI gene transcription
Besides their intrinsic preference for certain gene promoters, a property common to all sigma factors, chloroplast sigma factors further possess a unique regulatory feature in the form of phosphorylation. Protein phosphorylation has been shown to modulate the activity of SIG1 and SIG6 (Schweer et al., 2010b; Shimizu et al., 2010). SIG6 is phosphorylated by a chloroplast casein kinase known as the plastid transcription kinase (PTK). Phosphorylated SIG6 initiates transcription of the atpBE operon from its PEP promoter. The functional context of this regulation, however, remains unclear. Phosphorylation of SIG1, on the other hand, is associated with the transcriptional control of the PSI reaction center operon psaAB during a remarkable light quality acclimatory response known as photosystem stoichiometry adjustment. Changes in the wavelength of light that selectively excites PSII and PSI, and therefore the redox state of the PQ pool located between the two photosystems, initiates stoichiometric changes in the two photosystems so as to correct any imbalance in light energy conversion at either photosystem. It has been recognized that the adjustment of photosystem stoichiometry occurs mostly through changes in PSI amount and that the transcriptional regulation of psaAB is critical to this regulation (Murakami et al., 1997a, b; Pfannschmidt et al., 1999). The phosphorylation of SIG1 seems to provide a mechanism to connect the PQ pool redox state with transcription of the psaAB gene (Shimizu et al., 2010). The oxidized PQ pool, as under PSI-specific far-red light, leads to the phosphorylation of SIG1, which then represses psaAB transcription initiation and, as a result, lowers the amount of PSI. In the opposite light condition (i.e. under PSII-specific short wavelength illumination), the PQ pool becomes more reduced. Under this condition, phospho-SIG is thought to be either dephosphorylated or degraded. This releases the repression on psaAB transcription, increasing the amount of PSI. A key observation that links SIG1 phosphorylation with photosystem stoichiometry adjustment is the demonstration that the phosphorylation site mutant of SIG1 is unable to repress psaAB in PSI light.
An Arabidopsis knockout mutant of the chloroplast sensor kinase (CSK), a modified bacterial-type two-component sensor kinase, is similarly unable to repress psaAB in PSI light (Puthiyaveetil et al., 2008). CSK has therefore been suggested as the SIG1 kinase (Puthiyaveetil et al., 2013). Many key aspects of the CSK–SIG1 signaling pathway, however, remain unclear. For example, in csk knockout mutants, the amount of psaAB transcript reaches the wild-type level after nearly 24 h of misregulation (Puthiyaveetil et al., 2008). This suggests that SIG1 phosphorylation by CSK could be just one facet of photosystem stoichiometry adjustment and that there are other as yet unknown layers of regulation in this important photosynthetic light acclimatory response. Towards this, we identify that the psaAB transcriptional control requires a complementary regulation of nuclear SIG1 gene transcription (Fig. 4). It thus appears that psaAB gene regulation during photosystem stoichiometry adjustment requires not only post-translational control of SIG1 activity through phosphorylation but also regulation of the amount of SIG1 protein via changes in SIG1 gene transcription. The apparent emphasis on regulation of the SIG1 protein level further raises the question of whether the phosphorylation is indeed a tag for degradation of SIG1. If correct, this would suggest that both phosphorylation and SIG1 transcriptional control act in concert to decrease the SIG1 protein level and, as a result, psaAB transcription in PSI light (Fig. 4).
The use of the electron transport inhibitor DCMU shows that the redox state of the PQ pool, the very signal that initiates psaAB transcriptional changes, also controls nuclear SIG1 gene transcription. The PQ redox state is a well-known retrograde signal affecting nuclear photosynthetic genes and it is not surprising that it may also regulate the SIG1 gene (Pfannschmidt et al., 2001). The true extent of the PQ control of SIG1 transcription remains to be seen as we find only a modest effect in our experimental conditions (Supplementary Fig. S4). The phyB mutant nevertheless shows that light quality per se is the most effective signal driving SIG1 transcription, with the phyB mutant lacking some crucial aspects of SIG1 and psaA gene regulation (Fig. 5). The dual redox and light control is not unique to SIG1, as the transcription of SIG5 is regulated in part by redox- and phytochrome-mediated light signaling (Mellenthin et al., 2014). Some of the complex kinetics in psaA and SIG1 transcript accumulation are probably the result of both redox and light signals exerting their influence simultaneously (Figs 4, 5). These redox and light effects seem further superimposed on an endogenous rhythm in transcription as driven by nuclear and chloroplast circadian timekeepers (Figs 4, 5). Finally, it is pertinent to ask what purpose these seemingly redundant pathways of chloroplast psaAB gene regulation serve. It is likely that the redox-controlled and CSK-mediated SIG1 phosphorylation is a fast-acting regulatory mechanism, while the regulation through nuclear SIG1 transcriptional control is a long-term strategy. The mostly light quality-dependent regulation of SIG1 gene transcription (Fig. 5) may further provide a crucial, fail-safe input into the predominantly redox-controlled psaAB gene transcription.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Genotyping of sig1-1 and sig1-2 mutants.
Fig. S2. T-DNA insertion sites in sig1-1 and sig1-2 mutants.
Fig. S3. Developmental profile of sigma factor gene expression.
Fig. S4. The effect of DCMU on SIG1 gene expression.
Fig. S5. Full uncropped western blots of SIG1 and actin.
Table S1. Primers used in this study.
Acknowledgements
We thank Purdue University Biochemistry Department and Showalter Trust for funding, Clint Chapple for the phyB-9 mutant seeds, and Xiangying Mao for assistance with SIG1 complementation. The SIG1 antibody is a kind gift from Mitsumasa Hanaoka.
References
- Allen JF, de Paula WB, Puthiyaveetil S, Nield J. 2011. A structural phylogenetic map for chloroplast photosynthesis. Trends in Plant Science 16, 645–655. [DOI] [PubMed] [Google Scholar]
- Beardslee TA, Roy-Chowdhury S, Jaiswal P, Buhot L, Lerbs-Mache S, Stern DB, Allison LA. 2002. A nucleus-encoded maize protein with sigma factor activity accumulates in mitochondria and chloroplasts. The Plant Journal 31, 199–209. [DOI] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV. 2015. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochimica et Biophysica Acta 1847, 761–769. [DOI] [PubMed] [Google Scholar]
- Chi W, He B, Mao J, Jiang J, Zhang L. 2015. Plastid sigma factors: their individual functions and regulation in transcription. Biochimica et Biophysica Acta 1847, 770–778. [DOI] [PubMed] [Google Scholar]
- Choquet Y, Vallon O. 2000. Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82, 615–634. [DOI] [PubMed] [Google Scholar]
- Courtois F, Merendino L, Demarsy E, Mache R, Lerbs-Mache S. 2007. Phage-type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis. Plant Physiology 145, 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MC, Kesthely CA, Franklin EA, MacLellan SR. 2017. The essential activities of the bacterial sigma factor. Canadian Journal of Microbiology 63, 89–99. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. 1983. A plant DNA minipreparation: version II. Plant Molecular Biology Reporter 1, 19–21. [Google Scholar]
- Favory JJ, Kobayshi M, Tanaka K, Peltier G, Kreis M, Valay JG, Lerbs-Mache S. 2005. Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Research 33, 5991–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey V, Wagner R, Braütigam K, Wirtz M, Hell R, Dietzmann A, Leister D, Oelmüller R, Pfannschmidt T. 2005. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. Journal of Biological Chemistry 280, 5318–5328. [DOI] [PubMed] [Google Scholar]
- Hanaoka M, Kanamaru K, Takahashi H, Tanaka K. 2003. Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Research 31, 7090–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Kato M, Anma M, Tanaka K. 2012. SIG1, a sigma factor for the chloroplast RNA polymerase, differently associates with multiple DNA regions in the chl oroplast chromosomes in vivo. International Journal of Molecular Sciences 13, 12182–12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricová A, Quesada V, Micol JL. 2006. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiology 141, 942–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. 2005. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. The Plant Journal 42, 133–144. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. 2010. The endosymbiotic origin, diversification and fate of plastids. Philosophical Transactions of the Royal Society B: Biological Sciences 365, 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. 2011. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. The Plant Cell 23, 3824–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin TG, Bayne AN, Trempe JF, Savage DF, Davies KM. 2019. Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566, 411–414. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. 2011. Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Molecular Biology 76, 235–249. [DOI] [PubMed] [Google Scholar]
- Malik Ghulam M, Zghidi-Abouzid O, Lambert E, Lerbs-Mache S, Merendino L. 2012. Transcriptional organization of the large and the small ATP synthase operons, atpI/H/F/A and atpB/E, in Arabidopsis thaliana chloroplasts. Plant Molecular Biology 79, 259–272. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proceedings of the National Academy of Sciences, USA 99, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellenthin M, Ellersiek U, Börger A, Baier M. 2014. Expression of the Arabidopsis sigma factor SIG5 is photoreceptor and photosynthesis controlled. Plants (Basel, Switzerland) 3, 359–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A, Fujita Y, Nemson JA, Melis A. 1997a Chromatic regulation in Chlamydomonas reinhardtii: time course of photosystem stoichiometry adjustment following a shift in growth light quality. Plant & Cell Physiology 38, 188–193. [Google Scholar]
- Murakami A, Kim SJ, Fujita Y. 1997b Changes in photosystem stoichiometry in response to environmental conditions for cell growth observed with the cyanophyte Synechocystis PCC 6714. Plant & Cell Physiology 38, 392–397. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Motohashi R, Seki M, Shinozaki K, Kanamaru K, Takahashi H, Tanaka K. 2004. DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Bioscience, Biotechnology, and Biochemistry 68, 694–704. [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B. 2013. Photosystem II assembly: from cyanobacteria to plants. Annual Review of Plant Biology 64, 609–635. [DOI] [PubMed] [Google Scholar]
- Noordally ZB, Ishii K, Atkins KA, et al. 2013. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 339, 1316–1319. [DOI] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. 2003. The sigma70 family of sigma factors. Genome Biology 4, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Allen JF, Oelmuller R. 2001. Principles of redox control in photosynthesis gene expression. Physiologia Plantarum 112, 1–9. [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. 1999. Photosynthetic control of chloroplast gene expression. Nature 397, 625–628. [Google Scholar]
- Privat I, Hakimi MA, Buhot L, Favory JJ, Mache-Lerbs S. 2003. Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant molecular biology 51, 385–399. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Ibrahim IM, Allen JF. 2013. Evolutionary rewiring: a modified prokaryotic gene-regulatory pathway in chloroplasts. Philosophical Transactions of the Royal Society B: Biological Sciences 368, 20120260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kavanagh TA, Cain P, Sullivan JA, Newell CA, Gray JC, Robinson C, van der Giezen M, Rogers MB, Allen JF. 2008. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proceedings of the National Academy of Sciences, USA 105, 10061–10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Kolpack A, Link G. 2010a Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription—recent lessons from Arabidopsis thaliana. European Journal of Cell Biology 89, 940–946. [DOI] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Link B, Link G. 2010b AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. The Plant Journal 62, 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. 2016. Chloroplast NDH: a different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochimica et Biophysica Acta 1857, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Kato H, Ogawa T, Kurachi A, Nakagawa Y, Kobayashi H. 2010. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proceedings of the National Academy of Sciences, USA 107, 10760–10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D, Maliga P. 1998. Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Current Genetics 33, 340–344. [DOI] [PubMed] [Google Scholar]
- Strand DD, Fisher N, Kramer DM. 2017. The higher plant plastid NAD(P)H dehydrogenase-like complex (NDH) is a high efficiency proton pump that increases ATP production by cyclic electron flow. Journal of Biological Chemistry 292, 11850–11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozawa Y, Teraishi M, Sasaki T, Sonoike K, Nishiyama Y, Itaya M, Miyao A, Hirochika H. 2007. The plastid sigma factor SIG1 maintains photosystem I activity via regulated expression of the psaA operon in rice chloroplasts. The Plant Journal 52, 124–132. [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. 2004. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proceedings of the National Academy of Sciences, USA 101, 3304–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Takami T, Peng L, Ishizaki K, Kohchi T, Shikanai T, Nishimura Y. 2013. Subfunctionalization of sigma factors during the evolution of land plants based on mutant analysis of liverwort (Marchantia polymorpha L.) MpSIG1. Genome Biology and Evolution 5, 1836–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A, Börner T. 1999. Transcription and the architecture of promoters in chloroplasts. Trends in Plant Science 4, 169–170. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J. 2013. Sigma factor-mediated plastid retrograde signals control nuclear gene expression. The Plant Journal 73, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Shiina T. 2014. Recent advances in the study of chloroplast gene expression and its evolution. Frontiers in Plant Science 5, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghidi W, Merendino L, Cottet A, Mache R, Lerbs-Mache S. 2007. Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Research 35, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T. 2012. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. The Plant Cell 24, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.