Abstract
Flowering time is a complex trait and has a key role in crop yield and adaptation to environmental stressors such as heat and drought. This study aimed to better understand the interconnected dynamics of epistasis and environment and look for novel regulators. We investigated 534 spring barley MAGIC DH lines for flowering time at various environments. Analysis of quantitative trait loci (QTLs), epistatic interactions, QTL × environment (Q×E) interactions, and epistasis × environment (E×E) interactions were performed with single SNP and haplotype approaches. In total, 18 QTLs and 2420 epistatic interactions were detected, including intervals harboring major genes such as Ppd-H1, Vrn-H1, Vrn-H3, and denso/sdw1. Epistatic interactions found in field and semi-controlled conditions were distinctive. Q×E and E×E interactions revealed that temperature influenced flowering time by triggering different interactions between known and newly detected regulators. A novel flowering-delaying QTL allele was identified on chromosome 1H (named ‘HvHeading’) and was shown to be engaged in epistatic and environment interactions. Results suggest that investigating epistasis, environment, and their interactions, rather than only single QTLs, is an effective approach for detecting novel regulators. We assume that barley can adapt flowering time to the environment via alternative routes within the pathway.
Keywords: Barley, environmental effect, epistasis, flowering time, MAGIC population, novel QTL, QTL analysis
A novel QTL allele on chromosome 1H of barley is involved in epistatic and environment interactions that influence flowering time, and is associated with delayed flowering.
Introduction
Flowering time indicates the transition from the vegetative to the reproductive phase in plants (Mouradov et al., 2002), and as a major determinant for biomass accumulation and grain filling period length, affects grain yield. It was targeted during crop domestication and breeding to adapt wild ancestors of modern cultivars (Ross-Ibarra et al., 2007). Barley (Hordeum vulgare ssp. vulgare L.) is the fourth most cultivated cereal worldwide and is consumed for food, feed, and the malting process (Schulte et al., 2009). The flowering time regulatory network of barley, as a model for small-grain cereals, is relatively well described (International Barley Genome Sequencing Consortium, 2012). However, there is currently relatively little available information about the genes involved in this network, and their epistatic and environmental interactions, compared with the dicot model plant Arabidopsis thaliana (Blümel et al., 2015). Flowering time regulators belong to a complex network of genes that interact with environmental cues (Putterill et al., 2004). Focusing on the whole system of interacting factors, which includes genes and environment, could offer more informative solutions that provide new insights to identify novel regulators (Blümel et al., 2015; Valentim et al., 2015).
The multiparent advanced generation inter-cross (MAGIC) strategy was designed to improve power and precision in quantitative trait locus (QTL) mapping and overcome limitations of populations derived from biparental crossing systems by providing extensive genetic variance (Cavanagh et al., 2008). MAGIC populations are constructed by linkage-based design and multiple generations of recombinations, which provide high genetic mapping power and resolution (King et al., 2012). Common statistical methods for QTL and epistatic interactions using molecular markers such as single nucleotide polymorphisms (SNPs) have been used successfully with MAGIC populations in A. thaliana (Kover et al., 2009), wheat (Huang et al., 2012), rice (Bandillo et al., 2013), and winter wheat (Mackay et al., 2014). However, the involvement of more than two parents and their possible genetic similarities pose statistical challenges for analyzing the effects of parental alleles (Sannemann et al., 2015). SNP data from progenies can be transformed to a recognizable parental pattern of content, also known as the ‘haplotype phase’, for mapping purposes (Browning and Browning, 2011). Recent efforts have been made to use both single SNP and haplotype-phase analysis to provide sufficient mapping power (Sannemann et al., 2015; N’Diaye et al., 2017; Ogawa et al., 2018). A spring barley MAGIC population was constructed using an eight-way cross of seven barley landraces and one elite cultivar. This population was used to study flowering time QTL (Sannemann et al., 2015) and epistatic interactions (Mathew et al., 2018) in pot experiments, which detected regions harboring major flowering time genes such as Vrn-H1 and Vrn-H3.
Flowering time in barley is regulated by photoperiod and vernalization as well as environment-independent pathways such as earliness per se (EPS) (Cockram et al., 2007). Under long-day conditions, one of the key regulators that responds to photoperiod is PSEUDO-RESPONSE REGULATOR (HvPRR37), also known as PHOTOPERIOD RESPONSE LOCUS1 (Ppd-H1). It functions in the circadian clock oscillator and is orthologous to clock gene PRR7 in Arabidopsis and osPRR37 in rice (Turner et al., 2005). Ppd-H1 interacts with CONSTANS (CO) and promotes flowering by initiating expression of Vrn-H3, a homolog of A. thaliana FLOWERING LOCUS T (FT) (Yan et al., 2006). Vrn-H3 functions at the intersection of three main pathways—vernalization, photoperiod, and circadian clock (Campoli et al., 2012)—and is involved in the development of the reproductive apex and inflorescence (Faure et al., 2007; Digel et al., 2015). Another major gene upstream of Vrn-H3, encoding an APETALA1 family MADS-box transcription factor, Vrn-H1, is a positive regulator of flowering time in response to temperature (Distelfeld et al., 2009) that induces flowering time by promoting the transition from the vegetative to the reproductive phase (Hemming et al., 2008). Epistasis between Vrn-H1 and Vrn-H3, and the effect of lower temperature on promoting Vrn-H1, is an example of flowering time control beyond the effect of single genes (Hemming et al., 2008; Cockram et al., 2015). Epistasis is the term used to describe different levels of interactions among genes, including the functional interaction (protein level), allelic variation affecting the pathway (gene level), and deviations from additivity detected by statistical models (Phillips, 2008). Most mapping studies have successfully introduced novel regulators by focusing on single QTL (Bezant et al., 1996; Pillen et al., 2003; Wang et al., 2010; Alqudah et al., 2014; Sannemann et al., 2015; Maurer et al., 2016). Few reports have described epistatic interactions in the flowering time pathway of barley (Maurer et al., 2015; Mathew et al., 2018). Thus, the effects of epistasis and environment and their collective contribution to flowering time are not well understood. To provide more detailed insights into the flowering time regulation network in barley, using strategies beyond single QTL analysis is crucial.
The main aim of this study was to investigate the effect of epistasis and environment on flowering time in barley to detect novel mediators. Objectives of the study were (i) to investigate epistatic interactions under different environments in field and semi-controlled conditions, (ii) to investigate the effect of environment on the timing of flowering, by analyzing QTL × environment and epistasis × environment interactions, and (3) to shed light on novel flowering time regulator(s) involved in epistatic and/or environment interactions.
Materials and methods
Plant material
The MAGIC population was constructed by inter-crossing eight barley (Hordeum vulgare ssp. vulgare) genotypes, including one plant from each of seven landraces, Ackermanns Bavaria IPK No. HOR 100 (Ack. Bavaria), Ackermanns Danubia IPK No. BCC 1427 (Ack. Danubia), Criewener 403 IPK No. HOR 62, Heils Franken IPK No. BCC 1433, Heines Hanna IPK No. HOR 59, Pflugs Intensiv IPK No. BCC 1441, and Ragusa IPK No. BCC 1359, and the elite cultivar Barke, in an eight-way cross. Then, double haploid (DH) lines were produced as described in Sannemann et al. (2015). Ragusa represents a facultative and the others are spring barley ecotypes. The landraces used in this cross have contributed as founders of German barley cultivars.
Experimental setup and phenotypic data
Data for days to heading were collected under field and semi-controlled conditions. Field trials were conducted in 2016 and 2017 at Campus Klein-Altendorf (50°36′46.6″N, 6°59′39.7″E) of the University of Bonn. The lines were sown on 11 April 2016 (mean temperature 12.14 °C), and 3 April 2017 (mean temperature 8.63 °C). An unreplicated experimental design with a check every third plot (Mangelsdorf, 1953; Warner, 1953; Federer, 1956a) was employed, which is the standard design used for early-generation field trials in breeding programs (Federer, 1956b, 1993). The DH lines were completely randomized. The eight parents and cultivar scarlet were also randomized as controls. Each DH line was sown in one row containing 10 plants. Each plot was 1 m × 1.5 m in size and contained six DH lines, and the space between plots was 1 m. Field trials were subjected to fertilization and pest management following local practices. All DH lines reached the heading stage and the flowered plants for each line were counted; 99% had at least four plants that headed and were considered for data collection. The number of heading plants per DH line was on average six or seven for both years. The setup for semi-controlled conditions for the experiments conducted in 2011 and 2012, which were done under a foil tunnel at Campus Poppelsdorf (50°43′34.1″N, 7°05′14.6″E) of the University of Bonn, is detailed in Sannemann et al. (2015). The sowing dates were 4 April 2011 (mean temperature 11.99 °C), and 3 April 2012 (mean temperature 12.51 °C). Days to heading (BBCH 49; Hack et al., 1992) was scored as the number of days after sowing when at least 50% of plants of each DH line showed 3 cm of awns. The data for each year were analyzed separately. Phenotypic data are provided in Dataset 1 available at Dryad Digital Repository (https://doi.org/10.5061/dryad.g25cm28; Afsharyan et al., 2020). To evaluate the effect of environment, the daily average temperature for 100 days after sowing was measured for the 4 sowing years (2011, 2012, 2016, and 2017) to calculate growing degree-days (GDD), using base temperature 3 °C (Schelling et al., 2003), for each growing season (Dataset 2 at Dryad).
DNA extraction and genotyping
DNA was isolated from each barley MAGIC DH line according to the protocol for Diversity Arrays Technology marker analysis (https://www.diversityarrays.com) and prepared as described by Sannemann et al. (2015). Then, DNA samples were genotyped using the 9k iSelect SNP array (Comadran et al. 2012) at TraitGenetics GmbH (Stadt Seeland OT, Gatersleben, Germany) (Dataset 3 at Dryad). The processing of raw genotypic data was done as described by Sannemann et al. (2015). Genotypes with less than 10% missing values were included and missing data were imputed according to the mean imputation approach (Rutkoski et al., 2013). Then, the genotyping dataset was constructed by eliminating markers with a minor allele frequency (MAF) of less than 1%. Finally, 5199 SNP markers were used for further analysis. Due to the inclusion of more SNPs in this study than in the study of Sannemann et al. (2015), the SNPs were haplotyped in two groups and then collected in one dataset. Construction of haplotype data using SNPs with MAF ≥5% was performed as described by Sannemann et al. (2015). The remaining SNPs (5%>MAF≥1%) were haplotyped by the K-means clustering method using Proc Fastclus in SAS 9.4 (SAS Institute Inc., Cary, NC, USA). The SAS script and sample data are provided in Method 1 at Dryad. Then, manual corrections were made by comparing the phase patterns of parents and DH lines to improve data accuracy and resolution of the haplotype data. Finally, 4557 SNP markers with missing haplotype-phase data ≤15.5% were used for further analysis. In silico analysis was performed using the IPK barley BLAST server (https://webblast.ipk-gatersleben.de/barley_ibsc/) (Colmsee et al., 2015).
Statistical analysis
Analysis of phenotypic data
Descriptive statistics was performed in R software (R Core Team, 2015) by using the following core generic functions in R: summary to calculate minimum, maximum, and mean; std.error, sd, and var to calculate standard error, standard deviation (SD), and coefficient of variation (CV), respectively. Analysis of row and column effect was conducted in R with the lm function from the package stats by considering non-replicated MAGIC DH lines, row, column, and controls as fixed effects. The same R function was used to perform analysis of variance (ANOVA) for days to heading by taking DH lines and year as fixed effect. The GDD accumulation in the 100 days after sowing was compared among years by a paired Student’s t-test. In addition, the relationship between days to heading and the respective GDD (flowering time GDD) of genotypes was tested by using Pearson’s correlation coefficient (r). Variance components were estimated using the PROC VARCOMP procedure (all effects as random) in SAS 9.4. Heritability (h2) was calculated as:
where VG: variance of genotype, VGY: variance of genotype × year, VE: variance of experimental error, and y: number of years.
QTL mapping and QTL × environment interaction models
QTL analysis and QTL × environment interaction through single SNP analysis (single SNP approach; SA) and haplotype analysis (haplotype approach; HA) was conducted, using the PROC MIXED procedure in SAS 9.4, by the following linear model:
where Yij: response variable; μ: general mean; Mi: the fixed effect of the ith marker genotype; Cj: the fixed effect of the jth calendar year; Mi×Cj: the fixed interaction effect of the ith marker genotype with the jth calendar year; and εij: the residual. To reduce the number of detected false positives, the multilocus procedure, as an efficient selection strategy, was implemented within the model (Sillanpää and Corander, 2002; Kilpikari and Sillanpää, 2003; Bauer et al., 2009). This process is composed of a forward selection procedure that inserts the most informative SNP inside the model in each iterative cycle, then uses it to re-analyze the remaining SNPs. The results of each round are considered as the basis for the next round of the forward selection process, and iteration of the multilocus QTL model continues until no other SNP is detected. Additionally, the control of QTL false discovery rate (FDR) was incorporated inside the model, which was conducted by using the PROC MULTTEST procedure in SAS 9.4. A threshold of P-value ≤0.001 with 1000 permutations and FDR value ≤0.05 was determined (Doerge and Churchill, 1996). Due to strong decay in linkage disequilibrium, a confidence interval of 3.5 cM and 15 cM was defined on both sides of the most significant SNP marker in SA and HA, respectively. The model defined QTL intervals by clustering SNPs based on their significance in the first iteration of the multilocus procedure. To test the significance of QTLs, a ‘leave20%out’ cross-validation procedure that randomly left out 20% of the genotypes from the original dataset and re-analyzed the remaining genotypes was performed. This process was executed 20 times and a new P-value was calculated using the mean of all. For analysis of QTL × environment interactions across 4 years, five times cross-validation was used. The SAS script employed for QTL and QTL × environment interaction analysis, including implementation of FDR and cross-validation procedures, is provided in Method 2 at Dryad. Genetic variance explained by a single SNP marker (RM2) was calculated as:
where SQM is the sum of squares of Mi and SQg was calculated as the type I sum of squares of the DH lines in an ANOVA model (von Korff et al. 2006). Finally, the total proportion of explained genetic variance was estimated.
Epistatic interaction and epistasis × environment interaction models
A two-way epistatic interaction multilocus approach and epistasis × environment interaction was performed through SA and HA in SAS 9.4. A threshold of P-value ≤0.001 and FDR value ≤0.05 was set followed by cross-validation using a hierarchical model:
where Yijk: response variable; μ: general mean; M1i and M2j: fixed effects of the ith marker genotype and the jth marker genotype, respectively; M1i×M2j: the fixed interaction effect of the ith M1 marker genotype with the jth M2 marker genotype; Ck: the fixed effect of the kth calendar year; M1i×M2j×Ck: the fixed interaction of the ith M1 marker genotype with the jth M2 marker genotype and the kth calendar year; and εijk: the residual. The SAS script for epistatic interaction and epistasis × environment interaction analysis, including implementation of FDR and cross-validation procedures, is provided in Method 2 at Dryad. Genetic variance explained by a single interaction was analyzed by fitting the model same as genetic variance by a single QTL. Subsequently, the total proportion of genetic variance explained by interactions was calculated.
Results
Flowering time under various environmental conditions
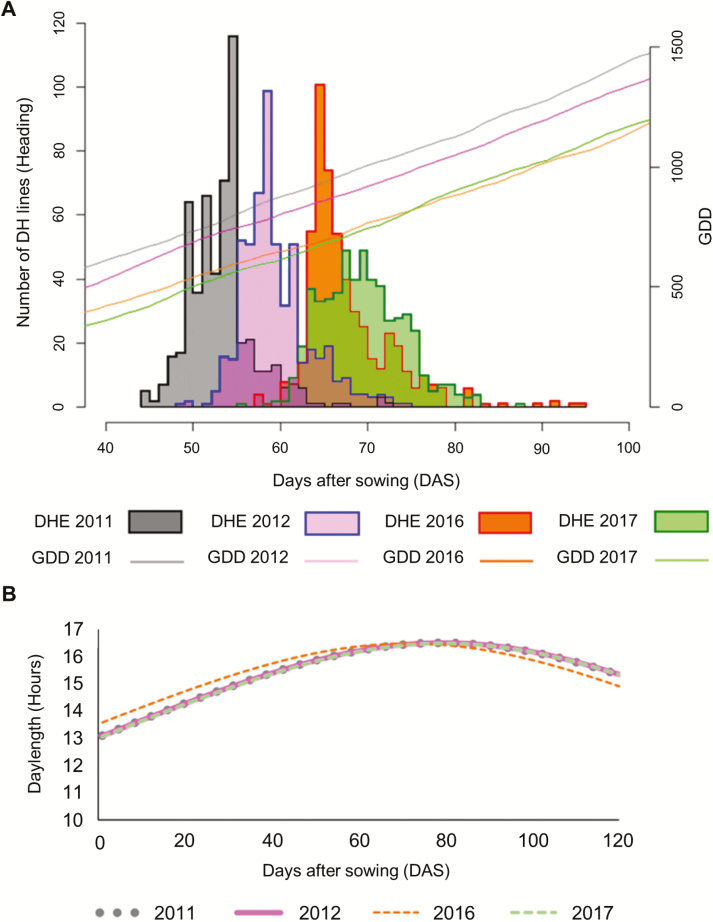
To study flowering time and the effect of environment on it, days to heading was scored in the spring barley MAGIC DH lines grown under field and semi-controlled conditions. Flowering time in the field showed a large phenotypic variation of more than 30 days each year, which was more diverse than that of the parents. Under semi-controlled conditions, the range of flowering time was shorter than under field conditions for both the DH lines and the parents, resulting in a higher CV (%) under field conditions (Table 1). Row and column effects were not significant in all experiments. ANOVA for years revealed highly significant differences (P<0.01) (see Table 1 at Dryad). Evaluation of GDD accumulation for 100 days after sowing by a paired Student’s t-test showed a significant difference between years. GDD accumulated faster in the foil tunnel conditions during the growing seasons, corresponding to an earlier flowering time (Fig. 1A; Table 2 at Dryad). Daily average temperature for the first 2 weeks after sowing was measured in the 4 sowing years to investigate the effect of lack of vernalization (which requires temperatures in the range 4–12 °C), which can delay flowering in some genotypes (Trione and Metzger, 1970). Flowering time was not accelerated in the years in which there was a lower number of days with mean temperature higher than 12 °C. Furthermore, Pearson’s correlation coefficient between days to heading and GDD was >0.99 for each year. The DH lines were subjected to the same day length each year as a result of very similar sowing dates (Fig. 1B).
Table 1.
Descriptive statistics and heritability for days to heading (DHE) for the parental lines and the spring barley MAGIC DH lines under foil tunnel (2011 and 2012) and field (2016 and 2017) conditions
| 2011 | 2012 | 2016 | 2017 | ||
|---|---|---|---|---|---|
| Parents | Min | 50.00 | 55.00 | 65.00 | 61.00 |
| Max | 55.00 | 60.00 | 74.00 | 71.00 | |
| Mean | 52.00 | 57.63 | 68.00 | 64.63 | |
| MAGIC population | Min | 44.00 | 48.00 | 57.00 | 55.00 |
| Max | 73.00 | 75.00 | 95.00 | 88.00 | |
| Mean | 53.73 | 59.91 | 68.00 | 69.00 | |
| SE | 0.16 | 0.17 | 0.22 | 0.20 | |
| SD | 3.85 | 3.96 | 5.02 | 4.69 | |
| CV | 14.82 | 15.64 | 25.19 | 21.98 | |
| h 2 | 0.47 | 0.63 |
CV, Coefficient of variation; h2, heritability (for years 2011 and 2012; 2016 and 2017); Max, maximum; Min, minimum; SD, standard deviation (%); SE, standard error.
Fig. 1.
(A) Comparison of flowering time (days to heading) distribution and GDD values for the years 2011, 2012, 2016 and 2017. (B) Comparison of day length in the same 4 years. (This figure is available in colour at JXB online.)
Identification of QTL for flowering time under field conditions
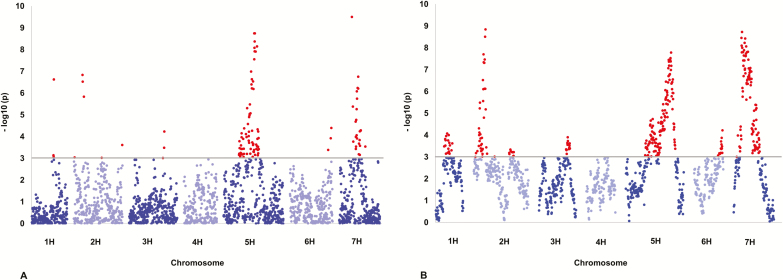
To identify genetic regions that control flowering time, the association between phenotypic and genotypic data was evaluated. The results revealed 11 QTLs by SA (Fig. 2A) and 7 QTLs by HA (Fig. 2B). All seven chromosomal regions found by HA were also detected by SA. One QTL on chromosome 2H was associated with earlier flowering by 8.79 days (Tables 2 and 3). QTLs detected with SA and HA explained in total 48.43% and 52.92% of the genetic variance, respectively. One single parent was the source of five loci mapped by both SA and HA on chromosomes 1H, 2H, 3H, 5H, and 7H (Tables 2 and 3). According to SA, the strongest association with flowering time was detected on chromosome 7H at 34.35 cM (BOPA2-12-30895), and explained 9.96% of the genetic variance (Table 2). For HA, the most significant QTL was mapped to chromosome 2H at 27.69 cM (SCRI_RS_140819), and explained 14.96% of the genetic variance (Table 3). The alleles for both loci originated from the parental line Ragusa.
Fig. 2.
Manhattan plots for (A) the single SNP approach and (B) haplotype approach for the spring barley MAGIC population grown under field conditions. The y axes denote the significance of SNP markers as –log10 (P) for flowering time (days to heading) in the barley population; the chromosomes are denoted on the x axes. The highlighted SNP markers above the cut-off line are significant by a threshold of P≤0.001 with 1000 permutations plus 20 times cross-validation. (This figure is available in colour at JXB online.)
Table 2.
Significant QTLs associated with flowering time in the single SNP approach under field conditions
| QTL | Peak markera | Chrb | cMc | mbpd | Flanking region (cM)e | F | P | FDR | Var (%)f | Parent g | MAFh | SNP1/ effecti | SNP2/ effectj | Genek |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HvHeading-1H-SA | BOPA1_1016_376 | 1H | 71.03 | 480.40 | 70.89–71.030 | 35.76 | 2.33E-07 | 3.89E-05 | 6.37 | 2 | 0.08 | G/3.82 | A/–0.38 | |
| HvHeading-2H-SA | SCRI_RS_233272 | 2H | 18.91 | 27.30 | 18.91–23.80 | 41.19 | 1.43E-07 | 3.05E-05 | 7.22 | 8 | 0.02 | T/–8.79 | G/0.36 | Ppd- H1 |
| HvHeading-2H.2-SA | BOPA1_ABC14531_1_2_91 | 2H | 91.21 | 687.20 | 91.21 | 15.56 | 9.31E-04 | 1.00E-02 | 2.88 | 1, 8 | 0.22 | A/1.52 | G/–0.38 | |
| HvHeading-3H-SA | SCRI_RS_103215 | 3H | 109.21 | 634.07 | 108.85 –109.21 | 24.73 | 5.8E-05 | 3.00E-03 | 4.48 | 3 | 0.1 | A/–0.02 | G/3.17 | denso/ sdw1 |
| HvHeading-5H-SA | SCRI_RS_152347 | 5H | 69.31 | 522.50 | 69.31–74.93 | 18.91 | 1.62E-04 | 6.00E-03 | 3.4 | 8 | 0.14 | A/2.06 | G/–0.11 | |
| HvHeading-5H.2-SA | SCRI_RS_204275 | 5H | 80.21 | 543.30 | 77.52–85.56 | 22.06 | 6.96E-05 | 4.00E-03 | 3.91 | 1, 3, 8 | 0.49 | C/–0.67 | A/1.04 | |
| HvHeading-5H.3-SA | BOPA2_12_21471 | 5H | 122.43 | 595.20 | 118.89–128.54 | 53.97 | 1.78E-09 | 1.39E-06 | 9.22 | 8 | 0.17 | G/2.73 | A/–0.63 | Vrn-H1 |
| HvHeading-6H-SA | SCRI_RS_9648 | 6H | 118.98 | 577.00 | 118.56–118.98 | 23.85 | 4.00E-05 | 3.00E-03 | 4.23 | 2, 5 | 0.31 | A/–1.29 | C/0.73 | |
| HvHeading-7H-SA | BOPA2_12_30895 | 7H | 34.35 | 39.70 | 34.35 | 60.33 | 3.14E-10 | 7.32E-07 | 9.96 | 8 | 0.04 | C/5.79 | G/–0.17 | Vrn-H3 |
| HvHeading-7H.2-SA | BOPA1_1107_392 | 7H | 65.44 | 109.70 | 61.33–70.96 | 42.66 | 1.73E-07 | 3.36E-05 | 7.34 | 8 | 0.13 | A/3.14 | C/–0.35 | HvCO1 |
| HvHeading-7H.3-SA | BOPA1_ABC10040_1_1_238 | 7H | 76.47 | 501.80 | 70.33–76.47 | 26.15 | 5.12E-05 | 3.00E-03 | 4.58 | 8 | 0.10 | G/2.99 | A/–0.26 | |
| Total | 48.43 |
a Most significant marker associated with the QTL.
b The chromosome on which the QTL was located.
c Genetic position of the most significant QTL according to Comadran et al. (2012).
d Physical position of the most significant QTL according to barley pseudomolecules genome assembly (Beier et al., 2017; Mascher et al., 2017).
e The range of the QTL interval according to Comadran et al. (2012).
f Cross-validated proportion of the explained genetic variance of the QTL.
g The parent(s) carrying the allele with lower frequency: 1, Ack. Bavaria; 2, Ack. Danubia; 3, Barke; 4, Criewener; 5, Heils Franken; 6, Heines Hanna; 7, Pflugs Intensiv; 8, Ragusa.
h Minor allele frequency
i Flowering effect of allele with lower frequency (days).
j Flowering effect of allele with higher frequency (days).
k Candidate gene corresponding to the QTL.
Table 3.
Significant QTLs associated with flowering time in the haplotype approach under field conditions
| QTL | Peak markera | Chrb | cMc | Flanking region (cM)d | F | P | FDR | Var (%)e | Parent 1f | Parent 2f | Parent 3f | Parent 4f | Parent 5f | Parent 6f | Parent 7f | Geneg |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HvHeading- 1H-HA | BOPA2_12_30147 | 1H | 66.86 | 60.84–86.47 | 6.72 | 8.27E-05 | 6.72E-04 | 7.79 | 0.59 | 3.82 | –0.44 | –0.73 | –0.38 | –0.05 | 0.11 | |
| HvHeading- 2H-HA | SCRI_RS_140819 | 2H | 27.69 | 3.82–53.75 | 13.61 | 1.42E-09 | 9.77E-07 | 14.96 | –0.22 | 2.31 | –0.39 | –0.58 | 0.19 | 0.40 | –8.79 | Ppd-1 |
| HvHeading- 2H.2-HA | SCRI_RS_160958 | 2H | 92.78 | 91.15–99.26 | 5.25 | 4.54E-04 | 2.24E-03 | 6.25 | 0.66 | –3.37 | –1.04 | –3.33 | –0.17 | –0.05 | 2.45 | |
| HvHeading- 3H-HA | BOPA1_ABC13753- 1-2-167 | 3H | 105.31 | 100.71–109.84 | 6.37 | 1.24E-04 | 9.14E-04 | 7.10 | –1.00 | –0.08 | 2.42 | –0.29 | 0.45 | –0.91 | 0.36 | denso/sdw1 |
| HvHeading- 5H-HA | BOPA2_12_30377 | 5H | 125.76 | 95.90–131.94 | 10.79 | 1.63E-08 | 1.43E-06 | 11.34 | –1.15 | –0.49 | –0.37 | –0.98 | –0.05 | 0.05 | 3.00 | Vrn-H1 |
| HvHeading- 6H-HA | SCRI_RS_144034 | 6H | 119.33 | 116.15–119.33 | 6.07 | 5.96E-05 | 5.21E-04 | 7.67 | 1.48 | –1.29 | 0.19 | 0.99 | –1.14 | –1.14 | 2.34 | |
| HvHeading- 7H-HA | BOPA1_ ConsensusGBS0356-1 | 7H | 37.61 | 23.80–67.42 | 12.63 | 1.87E-09 | 9.77E-07 | 12.65 | –0.73 | –1.07 | –2.31 | 0.63 | 0.22 | 0.70 | 3.91 | Vrn-H3 |
| Total | 52.92 |
a Most significant marker associated with the QTL.
b The chromosome that the QTL was located.
c Genetic position of the most significant QTL according to Comadran et al. (2012)
d The range of the QTL interval according to Comadran et al. (2012)
e Cross-validated proportion of the explained genetic variance of the QTL.
f Effect of allele for each parent (days) in reference to flowering time mean of the population: 1, Ack. Bavaria; 2, Ack. Danubia; 3, Barke; 4, Criewener/Pflugs Intensiv; 5, Heils Franken; 6, Heines Hanna; 7, Ragusa.
g Candidate gene corresponding to the QTL.
Identification of epistatic interactions under field and semi-controlled conditions
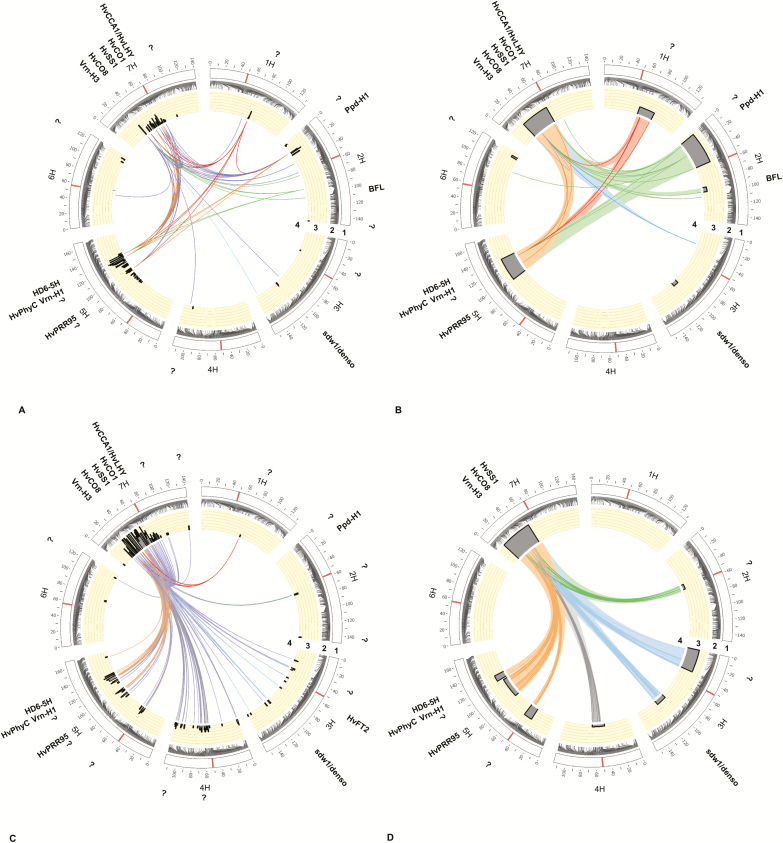
To evaluate how the interaction among genetic loci affects flowering time, genome-wide epistatic interaction analysis was performed. In total, 55 and 27 epistatic interactions were detected (P≤0.1E-15) under field conditions by SA (Fig. 3A; Table 3 at Dryad) and HA (Fig. 3B; Table 4 at Dryad), respectively. The most significant epistatic interaction was between two regions located on chromosomes 7H (34.35 cM, SA; 37.61 cM, HA) and 2H (18.91 cM, SA; 27.69 cM, HA). This interaction affected flowering time by more than 20 days and explained 28.13% and 38.02% of the genetic variation by SA and HA, respectively. Both of the alleles causing the polymorphism originated from the parental line Ragusa; the one on chromosome 2H induced earlier flowering, whereas the one on chromosome 7H delayed flowering time (see Table 4 at Dryad).
Fig. 3.
Genetic composition of flowering time in a spring barley MAGIC population under field conditions, (A) single SNP approach and (B) haplotype approach, and under foil tunnel conditions, (C) single SNP approach and (D) haplotype approach. The candidate genes that might correspond to QTLs and digenic interactions are indicated outside the plots. 1, Barley chromosomes are shown as white bars and centromeres are highlighted within these bars. 2, Genetic position of SNPs on the chromosomes. 3, Probability of QTLs detected with P≤0.001 and 1000 permutations plus cross-validation via multilocus QTL analysis are shown as peak SNPs in SA and peak SNPs/interval (blocks) in HA. 4, Bridges in the center of each circle represent detected digenic interactions between SNP markers with P≤0.1E-15 via cross-validated multilocus epistatic interaction analysis. Question marks indicate loci where no genes for flowering time have been reported so far. Plots were drawn by Circos (Krzywinski et al., 2009). (This figure is available in colour at JXB online.)
Analysis of epistatic interactions under foil tunnel conditions revealed 1139 and 1199 interactions (P≤0.1E-15) using SA (Fig. 3C; Table 5 at Dryad) and HA (Fig. 3D; Table 6 at Dryad), respectively. Both analyses identified the same interacting regions on chromosomes 7H (32.79 cM, SA; 32.79 cM, HA) and 5H (125.49 cM, SA; 118.75 cM, HA) as the most significant ones. This interaction influenced flowering time by more than 4 days in both approaches, and explained 26.94% and 42.01% of the genetic variance by SA and HA, respectively (Tables 5 and 6 at Dryad). The allele on chromosome 5H contributed to earlier flowering whereas the one on chromosome 7H delayed flowering time (Table 6 at Dryad); both originated from the parental line Ragusa.
Detection of HvHeading, a novel flowering-delaying QTL allele
To distinguish the novel QTL, the detected loci in this study were compared with previously reported regions. SA and HA consistently revealed a novel QTL allele with a flowering-delaying effect on chromosome 1H, which explained 6.37% (Table 2) and 7.79% (Table 3) of the genetic variation, respectively. We named this QTL HvHeading. The QTL allele segregated from the parent Danubia and was located within interval 70.89–71.03 cM (BOPA1-1016–376, 71.03 cM) by SA (Table 2) and 60.84–86.47 cM (BOPA2_12_30147, 66.86 cM) by HA (Table 3). Analysis of allelic effect showed that as a single QTL it delayed flowering time by up to 3.82 days compared with the population average.
Several loci were involved in epistatic interactions with HvHeading (Fig. 3). SA revealed that in field conditions, an interaction of HvHeading with one locus on chromosome 7H, 34.35 cM (BOPA2_12_30895) could postpone flowering time by 12.38 days, while its interaction with a locus on chromosome 2H, 18.91 cM (SCRI_RS_233272) could accelerate flowering time by up to 10 days (Table 3 at Dryad). HA showed that interaction of the allele from Danubia with locus 7H, 37.61 cM (SCRI_RS_155061, interval 27.79–67.42 cM) from Ragusa could delay flowering time by 10.83 days, while its interaction with locus 2H, 27.69 cM (SCRI_RS_140819, interval 3.82–53.75 cM) from Ragusa could advance flowering by 10 days (Table 4 at Dryad). According to SA, under foil tunnel conditions HvHeading strongly interacted with several loci (Table 5 at Dryad). The most significant interaction was with the locus at 7H, 32.79 cM (BOPA1_12701_485), which could influence flowering time by 3.36 days.
To perform in silico analysis, the overlapping region between the QTL intervals from SA (7 cM) and HA (30 cM) was determined. The in silico approach revealed 160 annotated genes with predicted function in this region with different Gene Ontology annotations.
Influence of environment on flowering time
To evaluate the effect of environment on flowering time, the interaction of single and epistatic QTLs with the four environments was analyzed. The QTL × environment interaction analysis showed no significant effects under field conditions for both years. However, under foil tunnel conditions one QTL on chromosome 2H was detected by SA (19.90 cM) and HA (23.02 cM) (Tables 7 and 8 at Dryad). Considering all 4 years, SA and HA identified the most prominent region that interacted with environment on chromosome 2H (19.90 cM) and 7H (31.37 cM), respectively (Tables 9 and 10 at Dryad).
Analysis of epistasis × environment interactions using SA revealed 86 interactions with environment under field conditions (Table 11 at Dryad). Position 34.35 cM on chromosome 7H was involved in highly significant interactions (Fig. 4A). Analysis of epistasis × environment interactions in foil tunnel conditions by SA showed that 527 epistatic interactions were affected by environment, including HvHeading. The prominent regions involved in the 10 strongest interactions were located on chromosomes 5H (84.38–122.01 cM) and 7H (28.98–62.18 cM) (Fig. 4B; Table 12 at Dryad). No significant interactions were found when evaluating epistasis × environment interactions via HA. Across all 4 years, strong epistasis × environment interactions were detected (Table 13 at Dryad), which involved the interval located on chromosome 2H at 18.9–23.8 cM (Fig. 4C). Interactions of HvHeading with loci on chromosomes 2H, (SCRI_RS_233272, 18.9 cM), 7H (BOPA1_12701_485, 32.79 cM), and 5H (BOPA2_12_21471, 122.43 cM) were highly affected by environment (Table 14 at Dryad).
Fig. 4.
Epistasis × environment interactions for flowering time in a spring barley MAGIC population by SA in (A) field conditions, (B) foil tunnel conditions, and (C) both field and foil tunnel conditions (4 years). Candidate genes that might correspond to QTLs and digenic interactions are indicated outside the plots. 1, Barley chromosomes are shown as white bars and centromeres are highlighted within these bars. 2. Genetic position of SNPs on the chromosomes. 3, Bridges in the center of each circle represent detected digenic interactions between SNP markers (A and B, P≤0.1E-6; C, P≤0.1E-27) via cross-validated multilocus epistatic interaction analysis. Question marks indicate loci where no genes for flowering time have been reported so far. Plots were drawn by Circos (Krzywinski et al., 2009). (This figure is available in colour at JXB online.)
Discussion
Power of QTL and epistatic interaction analysis in a MAGIC population
Genetic mapping showed that the single SNP approach precisely pinpointed loci that represent prominent flowering time genes. The peak marker BOPA2-12-30895 on chromosome 7H is located in the Vrn-H3 gene (Colmsee et al., 2015). This finding, as well as the detection by both SA and HA of QTLs and epistatic interactions that corresponded to previously described flowering time genes, is a proof of concept for the power of the spring barley MAGIC population and the mixed linear model approach used in this study.
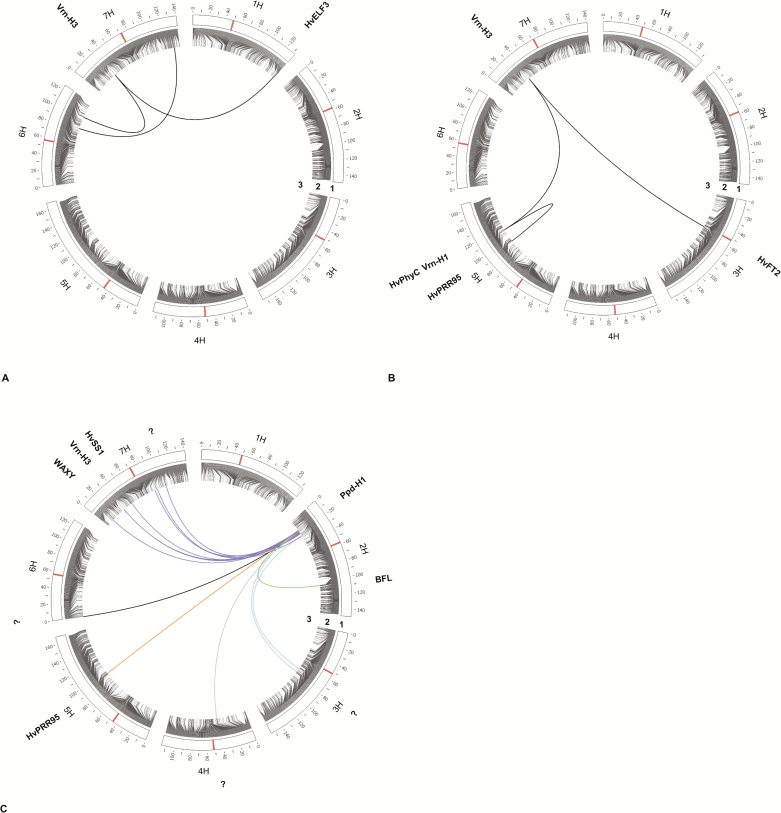
SA has been predominantly performed in QTL studies that used biparental populations or association panels (Xu et al., 2017). However, in multiparental populations, if more than two parents are involved, SA is not able to unambiguously identify the parental origin of a given allele (i.e. SNP). This drawback can be overcome by using HA, which has offered a more informative evaluation of associated loci (Huang and George, 2011; Sannemann et al., 2015; Ogawa et al., 2018). In the present study, HA provided an estimation of allelic effect of each parent and, due to the potential contribution of all SNP information in one haplotype block, it produced smoother P-value plots (Fig. 2B) and created larger QTL intervals (Fig. 5) compared with SA. The presence of more markers in these QTL intervals allowed for the higher explanatory power of HA. This was in agreement with previous reports stating that marker–trait associations based on haplotype-phase data detected more SNPs associated with the trait (Sannemann et al., 2015; N’Diaye et al., 2017; Ogawa et al., 2018).
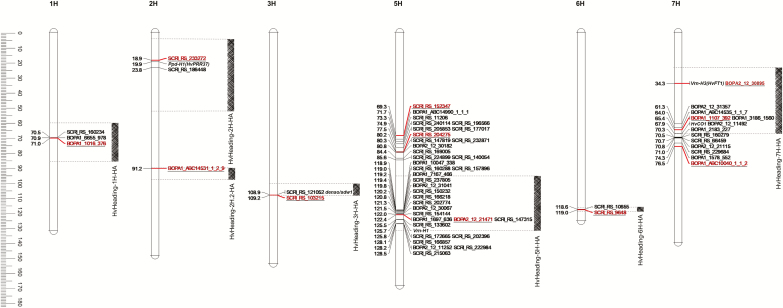
Fig. 5.
Genetic map of QTLs for flowering time in spring barley MAGIC DH lines. Barley chromosomes are represented by white bars. The most significant SNP marker for each QTL according to SA is highlighted and underlined. The position of the haplotype that is associated with the QTL according to HA is shown with grey hatched blocks accompanied by the name of the corresponding QTL. Italicized gene names indicate the position of major flowering time genes as described for the Barke × Morex RILs by Mascher et al. (2013). The ruler on the left shows the chromosome length. (This figure is available in colour at JXB online.)
Higher resolution of ‘haplotype-phasing’ data is mandatory for greater precision in mapping and depends on the linkage disequilibrium of the population, which is affected by the population size, diversity among the parents, population structure, marker density, and recombination frequency (Wang et al., 2002). Considering the nature of the MAGIC population, including the presence of similarity among the founders, more cross-over rounds will not guarantee a greater number of assigned genetic regions (Stadlmeier et al., 2018). One of the challenges during the current research was the construction of high-resolution haplotype data. Performing manual corrections clarified part of the unassigned regions, which shows that the existing algorithms for haplotype-phasing need to be improved to avoid wasting available data. Therefore, besides attempts to increase the number of cross-overs in the analyzed populations, developing an efficient haplotype-phase algorithm to maximize the usability of existing data seems to have a high priority in future studies.
The spring barley MAGIC DH lines supplied unique and diverse genetic material. The lower or higher than expected frequency (12.5%) for some SNP alleles might be due to the limited number of viable seeds obtained from the newly produced DH lines in the first round of cultivation.
Flowering time QTLs in the spring barley MAGIC population
Collectively, 18 QTLs were detected by SA and HA, including the QTL on chromosome 1H, which we named HvHeading, and nine QTLs that correspond to known flowering time genes. Due to the different data resolution of the two analyses, SA and HA found the same reported key genes for flowering time regulation at slightly varying genetic positions, suggesting that the novel region detected by both approaches on chromosome 1H has the same underlying gene. A region was mapped on chromosome 2H (18.91 cM, SA; 27.69 cM, HA) corresponding to the position of Ppd-H1 (HvPRR37) (Alqudah et al., 2014; Maurer et al., 2015). The QTL detected on chromosome 3H (109.21 cM, SA; 105.31 cM, HA) was located in the region harboring the semi-dwarf gene denso/sdw1 (Wang et al., 2010; Maurer et al., 2015, 2016; Sannemann et al., 2015; Alqudah et al., 2016). Furthermore, an interval on chromosome 5H (122.43 cM, SA; 125.76 cM, HA) was mapped to the position of the vernalization-response gene Vrn-H1 (Wang et al., 2010; Alqudah et al., 2014; Maurer et al., 2015; Sannemann et al., 2015), and the detected region on chromosome 7H (34.35 cM, SA; 37.61 cM, HA) matched the position of another major vernalization-response gene, Vrn-H3 (HvFT1) (Wang et al., 2010; Alqudah et al., 2014; Maurer et al., 2015; Sannemann et al., 2015).
Epistatic interactions in field and semi-controlled conditions
Different epistatic interactions were detected in field and semi-controlled conditions. The locus corresponding to Vrn-H3 had the strongest epistatic interaction in both environments irrespective of the genetic approach used for analysis.
In field conditions, regions corresponding to Vrn-H3 and Ppd-H1 had the strongest epistatic interaction. It has been reported that Ppd-H1 advances flowering time under long-day conditions by promoting Vrn-H3 (Yan et al., 2006). The epistatic interactions that involved the Ppd-H1 region from Ragusa accelerated flowering time remarkably, showing an early-flowering haplotype-specific effect. The most significant epistatic interaction in foil tunnel conditions was among regions that corresponded to Vrn-H3 and Vrn-H1. Vrn-H1 seems to respond to low and high temperatures and is known to up-regulate Vrn-H3 (Fu et al., 2005; von Zitzewitz et al., 2005; Gol et al., 2017).
These results support previous descriptions of complex genetic networks in the flowering time pathway in barley (Maurer et al., 2015). Epistatic interaction analysis for the different environments suggested that the flowering time of the MAGIC population was shaped by distinctive digenic interactions that adapt the DH lines to various environments.
HvHeading and participation in epistatic interactions
HvHeading, a novel flowering-delaying QTL allele, originated from the parental line Danubia. HvHeading was involved in significant epistatic interactions with loci that correspond to positions of major genes such as Ppd-H1, Vrn-H1, Vrn-H3, sdw1/denso, HvPRR95, HvPhyC, HvCO8, and HvSS1, indicating that it might have a major role in controlling flowering time in barley.
The closest known candidate gene to this QTL is Ppd-H2 (HvFT3) at 93.l cM (Halliwell et al., 2016), which is involved in the photoperiod response under short-day conditions (Casao et al. 2011). Nevertheless, no association was detected between the region and flowering time in the spring barley MAGIC DH lines.
In silico analysis revealed several genes within the QTL interval that were annotated for families involved in flowering time, such as the MADS-box transcription factor protein (Trevaskis et al., 2003), basic-leucine zipper (bZIP) transcription factor protein (Abe, 2005), FAR1 (Hudson et al., 1999), and OVATE (Wang et al., 2016) families. Further analysis is needed to identify and characterize the gene underlying HvHeading and its role in the flowering time pathway.
Epistasis × environment interactions and involvement of HvHeading
The different times of flowering in field and semi-controlled conditions were linked to environmental factors. The plants in the foil tunnel condition were exposed to higher temperatures compared with those in the field, resulting in a faster accumulation of GDD, which accelerated flowering time.
Analyzing QTL × environment interactions across the four environments (i.e. all 4 years) revealed a strong interaction of QTL regions harboring Ppd-H1 and Vrn-H3. The results of epistasis × environment interaction analysis for the four environments also showed that most of the interactions had Ppd-H1 region in common. Ppd-H1 and Vrn-H3 are both promoted by temperature (Turner et al., 2005; Yan et al., 2006), and recently it was reported that higher ambient temperature triggers Ppd-H1 (Ejaz and von Korff, 2017). The detection of an interaction of Vrn-H3 with the environment suggests that the outstanding effect of Vrn-H3 in epistatic interactions under foil tunnel conditions could be due to the warmer environment. Epistasis × environment interactions showed that under foil tunnel conditions, loci corresponding to Vrn-H1, HvPRR95, and Vrn-H3 were prominent, which supports previous reports that higher temperature triggers Vrn-H1 and the downstream gene Vrn-H3 (Karsai et al., 1997; Yan et al., 2003; von Zitzewitz et al., 2005; Ejaz and von Korff, 2017; Gol et al., 2017), which engage in a positive feedback loop that leads to early flowering under long-day conditions (Distelfeld et al., 2009).
HvHeading showed strong effects in epistasis × environment interactions in all four environments. It was involved in interactions with regions harboring Ppd-H1, Vrn-H3, and Vrn-H1, suggesting that HvHeading might have an effect on flowering time via an interaction with temperature.
Conclusion
Flowering time in barley is ultimately controlled by interactions among genes that can take different routes depending on environmental cues to adapt and fulfill timely flowering. The spring barley MAGIC population provided a genetic depth and richness that was required to study the effect of epistasis and environment interactions on complex traits such as flowering time. The results highlighted flowering time modulators as well as one novel QTL allele, HvHeading, that strongly interacted with regions corresponding to the Vrn-H3, Vrn-H1, and Ppd-H1 genes that are at the intersection of other genetic competitors. Further studies are needed to elaborate the underlying gene (or genes) and decipher its role and function in the pathway by shedding light on its interaction with other genetic and environmental factors.
Data availability
The following data are available at Dryad Digital Repository: https://doi.org/10.5061/dryad.g25cm28.
Dataset 1. Phenotypic data for days to heading (4 years) using MAGIC DH lines.
Dataset 2. Growing degree-days (GDD) during 100 days after sowing (2011, 2012, 2016, and 2017).
Dataset 3. Genotyping data by barley 9k iSelect SNP array using MAGIC DH lines.
Methods 1. SAS script and sample data for haplotype construction by the K-means clustering method.
Methods 2. SAS script for QTL and epistasis mapping process as well as environment interaction analysis including implementation of FDR and cross-validation procedures.
Table 1. Analysis of variance for 534 MAGIC DH lines.
Table 2. Descriptive statistics for growing degree-days (GDD) during 100 days after sowing.
Table 3. Epistatic interactions by single SNP approach under field conditions.
Table 4. Epistatic interactions by haplotype approach under field conditions.
Table 5. Epistatic interactions by single SNP approach under foil tunnel conditions.
Table 6. Epistatic interactions by haplotype approach under foil tunnel conditions.
Table 7. QTL × environment interactions by single SNP approach under foil tunnel conditions.
Table 8. QTL × environment interactions by haplotype approach under foil tunnel conditions.
Table 9. QTL × environment interactions by single SNP approach for 4 years (field and foil tunnel conditions).
Table 10. QTL × environment interactions by haplotype approach for 4 years (field and foil tunnel conditions).
Table 11. Epistasis × environment interactions by single SNP approach under field conditions.
Table 12. Epistasis × environment interactions by single SNP approach under foil tunnel conditions.
Table 13. Epistasis × environment interactions by single SNP approach for 4 years.
Table 14. Epistasis × environment interactions that engaged peak marker for HvHeading (BOPA1_1016_376) by single SNP approach for 4 years.
Acknowledgements
We thank the German Research Foundation (DFG) for funding this research under priority program 1530, ‘Flowering time control: from natural variation to crop improvement’.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Afsharyan NP, Sannemann W, Léon J, Ballvora A. 2020. Data from: Effect of epistasis and environment on flowering time in barley reveals a novel flowering-delaying QTL allele. Dryad Digital Repository. doi: 10.5061/dryad.g25cm28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqudah AM, Koppolu R, Wolde GM, Graner A, Schnurbusch T. 2016. The genetic architecture of barley plant stature. Frontiers in Genetics 7, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqudah AM, Sharma R, Pasam RK, Graner A, Kilian B, Schnurbusch T. 2014. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS One 9, e113120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandillo N, Raghavan C, Muyco PA, et al. . 2013. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer AM, Hoti F, von Korff M, Pillen K, Léon J, Sillanpää MJ. 2009. Advanced backcross-QTL analysis in spring barley (H. vulgare ssp. spontaneum) comparing a REML versus a Bayesian model in multi-environmental field trials. Theoretical and Applied Genetics 119, 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier S, Himmelbach A, Colmsee C, et al. . 2017. Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Scientific Data 4, 170044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezant J, Laurie D, Pratchett N, Chojecki J, Kearsey M. 1996. Marker regression mapping of QTL controlling flowering time and plant height in a spring barley (Hordeum vulgare L.) cross. Heredity 77, 64–73. [Google Scholar]
- Blümel M, Dally N, Jung C. 2015. Flowering time regulation in crops—what did we learn from Arabidopsis? Current Opinion in Biotechnology 32, 121–129. [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2011. Haplotype phasing: existing methods and new developments. Nature Reviews. Genetics 12, 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M. 2012. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. The Plant Journal 69, 868–880. [DOI] [PubMed] [Google Scholar]
- Casao MC, Karsai I, Igartua E, Gracia MP, Veisz O, Casas AM. 2011. Adaptation of barley to mild winters: a role for PPDH2. BMC Plant Biology 11, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Horsnell R, Soh E, Norris C, O’Sullivan DM. 2015. Molecular and phenotypic characterization of the alternative seasonal growth habit and flowering time in barley (Hordeum vulgare ssp. vulgare L.). Molecular Breeding 35, 165. [Google Scholar]
- Cockram J, Jones H, Leigh FJ, O’Sullivan D, Powell W, Laurie DA, Greenland AJ. 2007. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. Journal of Experimental Botany 58, 1231–1244. [DOI] [PubMed] [Google Scholar]
- Cavanagh C, Morell M, Mackay I, Powell W. 2008. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Current Opinion in Plant Biology 11, 215–221. [DOI] [PubMed] [Google Scholar]
- Colmsee C, Beier S, Himmelbach A, Schmutzer T, Stein N, Scholz U, Mascher M. 2015. BARLEX – the barley draft genome explorer. Molecular Plant 8, 964–966. [DOI] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, et al. . 2012. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nature Genetics 44, 1388–1392. [DOI] [PubMed] [Google Scholar]
- Digel B, Pankin A, von Korff M. 2015. Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. The Plant Cell 27, 2318–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J. 2009. Regulation of flowering in temperate cereals. Current Opinion in Plant Biology 12, 178–184. [DOI] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142, 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejaz M, von Korff M. 2017. The genetic control of reproductive development under high ambient temperature. Plant Physiology 173, 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. 2007. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federer WT. 1956a A method for evaluating genetic progress in a sugar cane breeding program. Hawaiian Planters’ Record 55, 177–190. [Google Scholar]
- Federer WT. 1956b Augmented (or hoonuiaku) designs. Hawaiian Planters’ Record 55, 191–208. [Google Scholar]
- Federer WT. 1993. Statistical design and analysis for intercropping experiments. New York: Springer-Verlag. [Google Scholar]
- Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. 2005. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Molecular Genetics and Genomics 273, 54–65. [DOI] [PubMed] [Google Scholar]
- Gol L, Tomé F, von Korff M. 2017. Floral transitions in wheat and barley: interactions between photoperiod, abiotic stresses, and nutrient status. Journal of Experimental Botany 68, 1399–1410. [DOI] [PubMed] [Google Scholar]
- Hack H, Bleiholder L, Buhr L, Meier U, Schnock-Fricke U, Weber E, Witzenberger A. 1992. Einheitliche Codierung der phänologischen Entwicklungsstadien mono- und dikotyledoner Pflanzen. Erweitere BBCH-Skala, Allgemeine. Nachrichtenblatt des Deutschen Pflanzenschutzdienstes 44, 265–270. [Google Scholar]
- Halliwell J, Borrill P, Gordon A, Kowalczyk R, Pagano ML, Saccomanno B, Bentley AR, Uauy C, Cockram J. 2016. Systematic investigation of FLOWERING LOCUS T-like Poaceae gene families identifies the short-day expressed flowering pathway gene, TaFT3 in wheat (Triticum aestivum L.). Frontiers in Plant Science 7, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. 2008. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiology 147, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang BE, George AW. 2011. R/mpMap: a computational platform for the genetic analysis of multiparent recombinant inbred lines. Bioinformatics 27, 727–729. [DOI] [PubMed] [Google Scholar]
- Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR. 2012. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnology Journal 10, 826–839. [DOI] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. 1999. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes & Development 13, 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium 2012. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Karsai I, Mészáros K, Hayes PM, Bedo Z. 1997. Effects of loci on chromosomes 2 (2H) and 7 (5H) on developmental patterns in barley (Hordeum vulgare L.) under different photoperiod regimes. Theoretical and Applied Genetics 94, 612–618. [Google Scholar]
- Kilpikari R, Sillanpää MJ. 2003. Bayesian analysis of multilocus association in quantitative and qualitative traits. Genetic Epidemiology 25, 122–135. [DOI] [PubMed] [Google Scholar]
- King EG, Merkes CM, McNeil CL, Hoofer SR, Sen S, Broman KW, Long AD, Macdonald SJ. 2012. Genetic dissection of a model complex trait using the Drosophila Synthetic Population Resource. Genome Research 22, 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R. 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genetics 5, e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Research 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IJ, Bansept-Basler P, Barber T, et al. . 2014. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 4, 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf AJ. 1953. Sugarcane breeding in Hawaii. Part II. 1921 to 1952. Hawaiian Planters’ Record 54, 101–137. [Google Scholar]
- Mascher M, Gundlach H, Himmelbach A, et al. . 2017. A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. [DOI] [PubMed] [Google Scholar]
- Mascher M, Muehlbauer GJ, Rokhsar DS, et al. . 2013. Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). The Plant Journal 76, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew B, Léon J, Sannemann W, Sillanpää MJ. 2018. Detection of epistasis for flowering time using Bayesian multilocus estimation in a barley MAGIC population. Genetics 208, 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A, Draba V, Jiang Y, Schnaithmann F, Sharma R, Schumann E, Kilian B, Reif JC, Pillen K. 2015. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A, Draba V, Pillen K. 2016. Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. Journal of Experimental Botany 67, 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G. 2002. Control of flowering time: interacting pathways as a basis for diversity. The Plant Cell 14(Suppl), S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N’Diaye A, Haile JK, Cory AT, Clarke FR, Clarke JM, Knox RE, Pozniak CJ. 2017. Single marker and haplotype-based association analysis of semolina and pasta colour in elite durum wheat breeding lines using a high-density consensus map. PLoS One 12, e0170941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamamoto E, Ohtani T, Kanno N, Tsunematsu H, Nonoue Y, Yano M, Yamamoto T, Yonemaru JI. 2018. Haplotype-based allele mining in the Japan-MAGIC rice population. Scientific Reports 8, 4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. 2008. Epistasis – the essential role of gene interactions in the structure and evolution of genetic systems. Nature Reviews. Genetics 9, 855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillen K, Zacharias A, Léon J. 2003. Advanced backcross QTL analysis in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 107, 340–352. [DOI] [PubMed] [Google Scholar]
- Putterill J, Laurie R, Macknight R. 2004. It’s time to flower: the genetic control of flowering time. BioEssays 26, 363–373. [DOI] [PubMed] [Google Scholar]
- R Core Team 2015. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Ross-Ibarra J, Morrell PL, Gaut BS. 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proceedings of the National Academy of Sciences, USA 104(Suppl 1), 8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkoski JE, Poland J, Jannink JL, Sorrells ME. 2013. Imputation of unordered markers and the impact on genomic selection accuracy. G3 3, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannemann W, Huang BE, Mathew B, Léon J. 2015. Multi-parent advanced generation inter-cross in barley: high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Molecular Breeding 35, 86. [Google Scholar]
- Schelling K, Born K, Weissteiner C, Kühbauch W. 2003. Relationships between yield and quality parameters of malting barley (Hordeum vulgare L.) and phenological and meteorological data. Journal of Agronomy and Crop Science 189, 113–122. [Google Scholar]
- Schulte D, Close TJ, Graner A, et al. . 2009. The International Barley Sequencing Consortium—at the threshold of efficient access to the barley genome. Plant Physiology 149, 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää MJ, Corander J. 2002. Model choice in gene mapping: what and why. Trends in Genetics 18, 301–307. [DOI] [PubMed] [Google Scholar]
- Stadlmeier M, Hartl L, Mohler V. 2018. Usefulness of a multiparent advanced generation intercross population with a greatly reduced mating design for genetic studies in winter wheat. Frontiers in Plant Science 871, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. 2003. MADS box genes control vernalization-induced flowering in cereals. Proceedings of the National Academy of Sciences, USA 100, 13099–13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trione EJ, Metzger RJ. 1970. Wheat and barley vernalization in a precise temperature gradient. Crop Science 10, 390–392. [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. [DOI] [PubMed] [Google Scholar]
- Valentim FL, Van Mourik S, Posé D, et al. . 2015. A quantitative and dynamic model of the Arabidopsis flowering time gene regulatory network. PLoS One 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Korff M, Wang H, Léon J, Pillen K. 2006. AB-QTL analysis in spring barley: II. Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp. spontaneum). Theoretical and Applied Genetics 112, 1221–1231. [DOI] [PubMed] [Google Scholar]
- von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TH, Hayes PM, Skinner JS. 2005. Molecular and structural characterization of barley vernalization genes. Plant Molecular Biology 59, 449–467. [DOI] [PubMed] [Google Scholar]
- Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. 2002. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. American Journal of Human Genetics 71, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chang Y, Ellis B. 2016. Overview of OVATE FAMILY PROTEINS, a novel class of plant-specific growth regulators. Frontiers in Plant Science 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Schmalenbach I, von Korff M, Léon J, Kilian B, Rode J, Pillen K. 2010. Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theoretical and Applied Genetics 120, 1559–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JN. 1953. The evolution of a philosophy on sugar cane breeding in Hawaii. Hawaiian Planters’ Record 54, 139–162. [Google Scholar]
- Xu Y, Li P, Yang Z, Xu C. 2017. Genetic mapping of quantitative trait loci in crops. The Crop Journal 5, 175–184. [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. 2006. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences, USA 103, 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. 2003. Positional cloning of the wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences, USA 100, 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following data are available at Dryad Digital Repository: https://doi.org/10.5061/dryad.g25cm28.
Dataset 1. Phenotypic data for days to heading (4 years) using MAGIC DH lines.
Dataset 2. Growing degree-days (GDD) during 100 days after sowing (2011, 2012, 2016, and 2017).
Dataset 3. Genotyping data by barley 9k iSelect SNP array using MAGIC DH lines.
Methods 1. SAS script and sample data for haplotype construction by the K-means clustering method.
Methods 2. SAS script for QTL and epistasis mapping process as well as environment interaction analysis including implementation of FDR and cross-validation procedures.
Table 1. Analysis of variance for 534 MAGIC DH lines.
Table 2. Descriptive statistics for growing degree-days (GDD) during 100 days after sowing.
Table 3. Epistatic interactions by single SNP approach under field conditions.
Table 4. Epistatic interactions by haplotype approach under field conditions.
Table 5. Epistatic interactions by single SNP approach under foil tunnel conditions.
Table 6. Epistatic interactions by haplotype approach under foil tunnel conditions.
Table 7. QTL × environment interactions by single SNP approach under foil tunnel conditions.
Table 8. QTL × environment interactions by haplotype approach under foil tunnel conditions.
Table 9. QTL × environment interactions by single SNP approach for 4 years (field and foil tunnel conditions).
Table 10. QTL × environment interactions by haplotype approach for 4 years (field and foil tunnel conditions).
Table 11. Epistasis × environment interactions by single SNP approach under field conditions.
Table 12. Epistasis × environment interactions by single SNP approach under foil tunnel conditions.
Table 13. Epistasis × environment interactions by single SNP approach for 4 years.
Table 14. Epistasis × environment interactions that engaged peak marker for HvHeading (BOPA1_1016_376) by single SNP approach for 4 years.