Abstract
Background
Root system architecture (RSA) traits are of interest for breeding selection; however, measurement of these traits is difficult, resource intensive, and results in large variability. The advent of computer vision and machine learning (ML) enabled trait extraction and measurement has renewed interest in utilizing RSA traits for genetic enhancement to develop more robust and resilient crop cultivars. We developed a mobile, low-cost, and high-resolution root phenotyping system composed of an imaging platform with computer vision and ML based segmentation approach to establish a seamless end-to-end pipeline - from obtaining large quantities of root samples through image based trait processing and analysis.
Results
This high throughput phenotyping system, which has the capacity to handle hundreds to thousands of plants, integrates time series image capture coupled with automated image processing that uses optical character recognition (OCR) to identify seedlings via barcode, followed by robust segmentation integrating convolutional auto-encoder (CAE) method prior to feature extraction. The pipeline includes an updated and customized version of the Automatic Root Imaging Analysis (ARIA) root phenotyping software. Using this system, we studied diverse soybean accessions from a wide geographical distribution and report genetic variability for RSA traits, including root shape, length, number, mass, and angle.
Conclusions
This system provides a high-throughput, cost effective, non-destructive methodology that delivers biologically relevant time-series data on root growth and development for phenomics, genomics, and plant breeding applications. This phenotyping platform is designed to quantify root traits and rank genotypes in a common environment thereby serving as a selection tool for use in plant breeding. Root phenotyping platforms and image based phenotyping are essential to mirror the current focus on shoot phenotyping in breeding efforts.
Keywords: RSA, Root, Phenotyping, Phenomics, Computer vision, Machine learning, Breeding, Soybean, Time series, Image analysis
Background
Over the past century, classical and technology-driven breeding techniques have aimed to achieve higher seed yield in major crops. The increase in seed yield comes concomitantly with a focus on improving the agronomic, disease protection and other perceptible traits that are expressed and observable above ground. Root system architecture (RSA), or the spatial arrangement of the root and its components [1], functions to provide water and nutrient acquisition, nutrient storage, anchorage and to foster plant–microbe interactions such as nodulation in N-fixing crops, which are relatively inconspicuous yet fundamental to plants’ performance and are indirectly selected traits in breeding programs particularly for non-tuber or root crops [2]. Root structure also correlates to environmental advantages, such as nutrient acquisition [1, 3], drought [4–7], flood tolerance [8], and lodging resistance [9].
Plant breeders have continually modified the above-ground features of the plant as these have been the easier to select; however, the hidden-half of the plant warrants further investigation for major agronomic crops including soybean (Glycine max L. Merr.), maize (Zea mays L.), wheat (Triticum aestivum L.) and rice (Oryza sativa L.) [5, 10]. This limitation in selecting for root phenotypes in soybean and other pulse, oilseed and cereal crop species arises from the difficulty of root trait measurement, and therefore the inability to study and utilize root architecture, morphology, topology, distribution within the soil, response to environmental stimuli and growth over time [1, 11–14]
Root structure correlates to environmental advantages, such as nutrient acquisition [1, 3], drought [4–7], flood tolerance [8], and lodging resistance [9]. Root phenotyping and further research is hindered due to wide technological gap in our ability to collect, observe and quantify important root trait data which is exacerbated by trait genetic complexity [15–18], phenotypic expression complexity [19], morphometric nature of their expression [20], and environmental interaction including soil structure [20], nutrient availability [3], temperature [21], water [22], interactions with other plants [2, 23] and microbes [24].
Researchers have generally taken one of three strategies to approach root phenotyping: including (1) controlled laboratory methods [18, 25, 26], (2) moderately controlled greenhouse methods [27, 28] and (3) minimally controlled field methods [29–32]. While the complexity of environment becomes more relevant to field scale production and physiological relevance with field methods, controlled laboratory methods are amenable to large scale phenotyping and throughput; therefore, researchers continue to explore ways to bridge the gap of lab versus field methods [11]. The existing major impediment is the high labor and time costs in the field for root trait phenotyping [29, 32]. This motivates our research to enable automation and increase throughput of root trait studies. The ability to study larger sample sizes will provide exciting opportunities to understand the role of RSA and its and application in future research.
Breeding for root system architecture (RSA) traits
Root system architecture is a complex of polygenic traits consisting of sub-root system parameterizations such as root growth habit, total root length, primary root length, root number, root angle, root thickness, root length density (root extension and distribution), root surface area, and are paramount in improving plant performance and seed yield [2, 23]. Monocot and dicots have distinct morphological parameters that are used to classify their roots into fibrous and taproot growth types, respectively. Due to the difficulty associated with the measurement of RSA traits and the high level of morphological plasticity of roots in soil [33–36], breeding programs rarely utilize RSA traits as a method of selection [5]. Furthermore, RSA traits remain elusive in plant breeding selection practices due to the RSA plasticity caused by environmental variation, lack of cost effective field plot root extraction protocols, and limited appropriate phenotyping platforms and tools [5, 37]. Identification of genes which control QTL (quantitative trait loci) for RSA has come with minimal success demonstrating that further genetics research is needed [38, 39].
Researchers have noted diversity of RSA within crop species such as maize [40], soybean [5, 41–44], common bean (Phaseolus vulgaris L.) [45], rice [26, 46] and wheat [47]. It is important to note that the North American soybean genepool is very narrow [48], therefore the expected gain from RSA traits could be high and rewarding, but will rely on the identification and incorporation of genetic diversity [49–51].
Before plant breeders can select for RSA traits, available genetic and phenotypic diversity needs to be explored and characterized. Therefore, accurate and efficient quantification of root architecture traits and diversity as well as associated physiological processes, is a pertinent requirement for addressing breeding objectives. Fortuitously, in the current era of phenomics and big data there is a continual advancement in high-throughput phenotyping (HTP) methods that can enhance researcher’s ability to assess above and below ground organs and traits. New technological innovation in computers, sensors, robotics and data analytics, including computer vision [52, 53], automation [54], remote sensing [55], ML [56] and deep learning (DL) [57] have allowed breeders and researchers to capture high resolution and high dimensional attributes of diverse phenotypic data non-destructively on a vast spatio-temporal scale [56, 58, 59]. These include primarily above ground traits [60–65] and to a lesser extent, root related traits [66–68]. However, continual efforts are needed to decipher the genetics of root traits to realize the genetic potential of root trait driven breeding. With phenomic information on both root and shoot traits, plant scientists will be empowered to deploy above and below ground phenotypes optimized to targeted climactic conditions and agronomic management techniques.
Technological challenges in RSA trait phenotyping can be divided into two major components: (1) root extraction from soil (for review, see [69]), and (2) imaging and computer aided feature (trait) extraction [70]. This dictates a need for advances in imaging protocols, computer vision and ML for trait extraction. Conventional approaches for root examination include field extractions [32], soil coring [13, 71] and minirhizotrons [72], but advances in X-ray computed tomography [68, 73, 74], magnetic resonance imaging (MRI) and positron emission tomography (PET) [21, 75], and 3D imaging approaches [26, 76, 77] have helped obtain higher resolution root trait data. The low throughput and high cost often prevent integration of these approaches in large scale genetic material screening [11, 12, 59, 69, 78]. At the onset, a reduction of cost and time are imperative to scaling plant phenotyping methodologies and require standardized protocols. There is little standardization on physical platforms (hardware) used for image based root phenotyping. However, for image analytics, several software tools are currently available that extract data through analyses of high resolution digital images with advanced computer analysis. This non-exhaustive list includes: archiDART [79], ARIA [80], DART [81], DIRT [82], EZ-Rhizo [83], GiA Roots [84], GLO-Roots [85], RhizoChamber-Monitor [86], RootNav [87], RootReader2D, RootSystemAnalyzer [88], RooTrak [68], RootTrace [89], SmartRoot [66]. These freeware such as ARIA (Automatic Root Image Analysis), have been developed to be faster and more adaptable to the alternative industry standard software WinRHIZO [80]. Advances in computer vision and image analytics have made feature extraction efficient, effective, accurate, and potentially non-destructive. The recent software are also multi-functional due to their ability to perform fast processing based on digital images, generation of information on various traits, with higher throughput [80].
Recent coupling of computer vision with ML has facilitated the generation of software tools that include automated learning for image preprocessing, image processing and feature extraction that will aid to reduce measurement variability and remove subjectivity and biases. Alone, computer vision enables software to identify objects and structures within images; while ML has been deployed to learn and classify those objects or structures [90]. In recent root architecture studies, researchers trained their model to recognize and differentiate root tips from 2D images in an automated process [91]. Other studies used a random forest based approach to replace missing trait values in highly noisy root images [92]. Nevertheless, with the strides being made in software, data processing, and phenotyping protocols, a methodology is needed that is low-cost, scalable, and robust to diverse phenotypes and experimentation to begin standardizing RSA trait acquisition.
In this paper, we describe hardware, software and analytical solutions for an end-to-end controlled environment soybean root phenotyping pipeline. The main objectives were to develop: (1) a low barrier to entry system facilitating the growth and imaging of hundreds of plants, (2) a computer vision program to automate image capture and curation, (3) image segmentation using heuristic and ML approaches and (4) a software tool to automate the extraction of a multitude of seedling RSA traits. The final product is an end-to-end pipeline with a fully automated software complete with tunable image thresholding and image based trait extraction. To summarize, the pipeline provides non-destructive evaluation of a large number of soybean genotypes in controlled conditions in a rapid manner at lowered cost of phenotyping alleviating the phenotyping bottleneck thus enhancing research and breeding progress related to RSA. We envision that this combination of phenotyping platform and data analytics will meet the needs of various users regardless of technical experience.
Methods
Plant material
For this study, 292 genotypes comprising a subset of the USDA soybean core collection and a subset of Soybean Nested Association Mapping (SoyNAM) parents were selected and were previously genotyped [93, 94]. For the purpose of this manuscript, we restrict our analyses and results presentation to 115 maturity group II (MG 2) genotypes to target the local Iowa environment for within maturity group comparisons. These genotypes consisted of a wide range in geographical origin (12 countries) and growth habit (determinate, semi-determinate, indeterminate) along with various other morphological and seed quality traits to meet our requirement for a diverse set of lines to test during hardware and software development.
Growing protocol
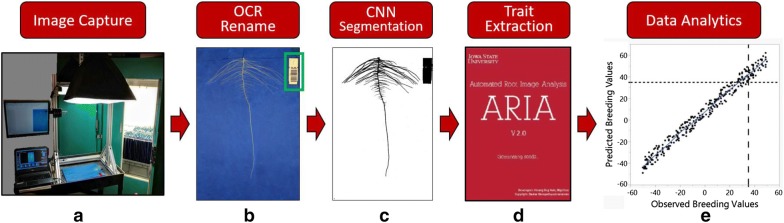
Motivated by previous research, we present a hardware system that is affordable and simple to construct, requiring few materials [95–98]. Seedlings are grown on the pouch-and-wick system [95] consisting of flat blue blotter germination paper (Anchor Paper Co., Minneapolis, MN), which creates high contrast with the yellow roots facilitating higher quality computer based root identification and segmentation [96]. A total of 4,088 seedlings were grown on blue blotter germination paper, suspended from the rungs of the shelving platform in a standard 1.75 m2 growth chamber during the course of the experiment (Fig. 1). The seedlings were phenotyped at three time points leading to 12,264 images that were a part of the overall experiment and a basis of software development for image processing and feature extraction.
Fig. 1.

Root phenotyping platform. a 10 seeds per genotype rolled into germination paper. b Plants germinate in growth chamber and removed at 5 days. c Two representative seedlings are selected from each roll for transplantation onto labeled moist blue germination paper. d Single, transplanted seedlings are covered with moist brown germination paper and sandwiched together creating one experimental unit. e Experimental units are bound with binder clips, each placed between the metal rungs of a growth chamber with the bottom 2.5 cm submerged in water. f At 6 days, 9 days and 12 days, experimental units are individually removed, split, imaged, automatically rotated, renamed via an image processing algorithm and saved to the server database, and replaced into the growth chamber
The standard 175 cm by 100 cm growth chamber (Controlled Environments Ltd, Winnipeg, Canada) contains standard metal grate shelves (1.3 cm by 35 cm slots) which double as a support framework for suspending the germination paper above a water reservoir allowing for 2.5 cm of each paper unit to be submerged. The growth chamber could house up to 400 seedlings in 200 slots (corresponding to 228 seedlings m2). The growth chambers were set at 25 °C during a 16 h day, 22 °C for an 8 h night. Light intensity of the growth chambers was measured at 300 and 350 µmol photons m−2 s−1 respectively, as measured by a Li-250A light meter (Li-Cor Biosciences, Lincoln, NE, USA).
Ten seedlings were germinated using a paper roll assay (Fig. 1a, b) [99], of which two representatives of healthy seedlings were chosen at 5 days after germination and transplanted to the blue blotter germination system (Fig. 1c) [100] minimizing the variability associated with seed source of plant introduction lines [101] and the effect of poor or delayed germination [95]. When working with such diverse soybean plant introduction landraces seed uniformity and viability can be a hurdle. Specifically germination differences within genotypes was often substantial as displayed in Fig. 1c. To reduce variability across the experiment, 14 seedlings were grown and phenotyped for each genotype during the duration of this study. The apparatus and methodology of the germination paper roll assay is further presented in Additional file 1: Video S1 [102].
A blue blotter germination paper sheet cut to 30 cm × 45 cm with perforations made at 2.5 cm from top of the page was used in the experiment. The large sized germination paper allowed for undisturbed root growth for up to 12 days. Each sheet of blue paper was wetted and subsequently folded along the perforation to place one selected 5-day old seedling allowing for shoot penetration through the perforation. Thereafter, a wetted brown germination sheet was placed on top of the blue blotter paper and emerging radicle to isolate and adhere to each root to retain moisture (Fig. 1d). The thin brown paper is non-porous, preventing root penetration and allowing for easy removal prior to imaging. This procedure was repeated for the second seedling, after which the two seedlings of one genotype were affixed together using two binder clips. Each group of blue paper and brown germination paper combination housing two separated seedlings (hereon called, growth pouch unit) was suspended vertically via binder clips in slots between the labeled grates of the growth chamber with the lower 2.5 cm of blue paper submerged into water (Fig. 1e). Additional water was manually added to the reservoir as needed. Image capture began 6 days after germination, 1 day after transplanting, with consecutive images captured at 9 days and 12 days. The apparatus and methodology of the transplantation from brown to blue germination paper is further presented in Additional file 2: Video S2 [103].
Imaging platform and protocol
The imaging platform consists of a utility cart, framework for camera mounting, and computer connectivity for image storage and file management. The imaging stage was fabricated using rugged, adjustable 80/20 aluminum T-slot extrusion (80/20 Inc., Columbia City, IN) (Fig. 1f) to provide a customizable, rigid structural framework including camera and light mounting to provide consistent image quality. Sensors included an 18 megapixel Canon Rebel T5i digital SLR camera (Lens: EF-S 18–55 mm f/3.5–5.6 IS II) (Canon USA, Inc, Melville, NY) mounted to a gimbal tripod head affixed to a T-slot extrusion crossbeam 60 cm directly above the imaging stage with the camera’s frame set to a consistent position at each imaging day. Additionally, the camera was positioned at a sufficient distance to capture the maximum length of a root at 12 days, as identified in preliminary experiments. The camera was set at a consistent white balance, focal length and maximum resolution to ensure high image quality (100 pixels per cm). The USB cable connected the camera to computer allowing for direct image transfer and live view of the imaging stage. To provide consistent illumination, two softbox photography lights (with four bulbs: 70 watts, 5500 K CFL) (Neewer; Shenzen, China) extending out from the stage at a height of 90 cm from the cart top base were directed at the imaging stage from opposite sides. The imaging platform was constructed on an Uline utility cart (Uline, Pleasant Prairie, WI), creating a compact mobile imaging station. The phenotyping platform consisted of off-the-shelf material and the total cost (excluding cameras and laptop) was less than $200. The remote capture software, Smart Shooter 3 [104] on a Dell Latitude E7470 laptop (Dell, Round Rock, TX), was used for a live view of the stage followed by triggering the camera for image capture. Plastic labels were affixed to each paper which included a unique barcode for each plant (Fig. 2).
Fig. 2.
Time series growth of a single soybean plant with images taken at a 6 days, b 9 days and c 12 days after germination. Images were captured remotely via a laptop computer using software automating the image file renaming via the in-frame barcode. Smart Shooter 3 optimized the system’s throughput by renaming each image at acquisition using Object Character Recognition (OCR), reducing time and eliminating user input and human error. Image files were directly saved to a cloud-based database system. An additional computer monitor was affixed to the platform to facilitate manual inspection of captured images
Images were captured remotely via laptop computer using software automating the image file renaming via the in-frame barcode and current date. Smart Shooter 3 optimized the system’s throughput by renaming each image at acquisition using Object Character Recognition (OCR), reducing time and eliminating user input labor and human error [104]. Image files were directly saved to a cloud-based database system. An additional computer monitor was affixed to the platform to facilitate manual inspection of captured images. A list of system components can be found in Additional file 3: Table S1.
To capture images, each growth pouch unit was removed individually from the growth chamber grates, binder clips were removed, and blue paper and seedling combination were separated (Additional file 4: Video S3) [105]. Individual seedlings were placed on a 30 cm × 50 cm blue acrylic sheet for easier handling. Brown germination paper top sheet was removed and slight user adjustment of some roots was required to ensure that the computer algorithm could detect differences between multiple roots converging in parallel. A thin stainless steel laboratory spatula was used to lift and separate two side-by-side growing roots with an effort to reduce the movement and thus morphological change of the root and eliminate root damage. Root images were captured using a fixed digital SLR camera and a laptop computer as seen in Additional file 4: Video S3 [105]. After imaging, the growth pouch unit was reassembled and placed back into the growth chamber for the time series imaging pipeline. Seedling RSA analysis was conducted using a revised version of ARIA software (Table 1) [80]. To image approximately 300 seedlings, 3 to 5 h were needed dependent on number and experience of technical staff and growth stage of the seedling. The handling and imaging steps exposed the root to light up to 3 min.
Table 1.
Root system architecture (RSA) traits captured by ARIA 2.0 software
| Trait name | Symbol | Unit | Trait description |
|---|---|---|---|
| Total root length | TRL | cm | Cumulative length of all the roots in centimeters |
| Primary root length | PRL | cm | Length of the Primary root in centimeters |
| Lateral root length | LRL | cm | Cumulative length of all lateral roots in centimeters |
| Mean lateral root length | MSL | cm | Mean length of all lateral roots in centimeters |
| TRLUpper | TRLUpper | cm | Total root length of the upper one third |
| TRLLower | TRLLower | cm | Total root length of the lower two third |
| Perimeter | PER | cm | Total number of network pixels connected to a background pixel |
| Depth | DEP | cm | The maximum vertical distance reached by the root system |
| Width | WID | cm | The maximum horizontal width of the whole RSA |
| Diameter | DIA | cm | Diameter of the primary root |
| Lateral root branches | LRB | Count | Number of lateral root branches |
| Nodes of lateral roots | NLR | Count | Number of nodes of lateral roots |
| Independent root branches | IRB | Count | Number of independent lateral root branches |
| Lateral root tip | RTA | Count | Number of lateral root tips |
| Median | MED | Count | The median number of roots at all Y-location |
| MaximumR | MAX | Count | The maximum number of roots at all Y-location |
| Maximum number of roots | MNR | Count | The 84th percentile value of the sum of every row |
| Network area | NWA | Count | The number of pixels that are connected in the skeletonized image |
| Convex area | CVA | cm2 | The area of the convex hull that encloses the entire root image |
| RhizoArea | RHZO | cm2 | Length of 2 mm surrounding the TRL |
| TRArea | TRArea | cm2 | Area of the RSA as observed in the 2D projected view |
| Primary root surface area | PRA | cm2 | Surface area of the primary root |
| TRAUpper | TRAUpper | cm2 | Total root area of the upper one third |
| TRALower | TRALower | cm2 | Total root area of the lower two third |
| Volume | VOL | cm3 | Volume of the primary root |
| Lateral root branching angle | LBA | Angle | Lateral root branching angle near the primary root node |
| Lateral root angles | LRA | Angle | Root angles along the extent of all lateral roots |
| Lateral root tip angle | RTA | Angle | Root angle at lateral root tips |
| Width/depth ratio | WDR | Ratio | The ratio of the maximum width to depth |
| Solidity | SOL | Ratio | The fraction equal to the network area divided by the convex area |
| Bushiness | BSH | Ratio | The ratio of the maximum to the median number of roots |
| Length distribution | LED | Ratio | TRLUpper/TRLower |
| LRL by PRL | LSLPL | Ratio | Number of the Lateral root per unit length of the Primary root |
| Center of mass | COM | Ratio | Center of gravity of the root/Depth |
| Center of point | COP | Ratio | Absolute center of the root regardless of root length/Depth |
| Center of mass (Top) | CMT | Ratio | Center of gravity of the top 1/3 of the root (Top)/Depth |
| Center of mass (Mid) | CMM | Ratio | Center of gravity of the middle 1/3 root (Middle)/Depth |
| Center of mass (Bottom) | CMB | Ratio | Center of gravity of the bottom 1/3 root (Bottom)/Depth |
| Center of point (Top) | CPT | Ratio | Absolute center of the root regardless of root length (Top)/Depth |
| Center of point (Mid) | CPM | Ratio | Absolute center of the root regardless of length (Middle)/Depth |
| Center of point (Bottom) | CPB | Ratio | Absolute center of the root regardless of root length (Bottom)/Depth |
Image processing
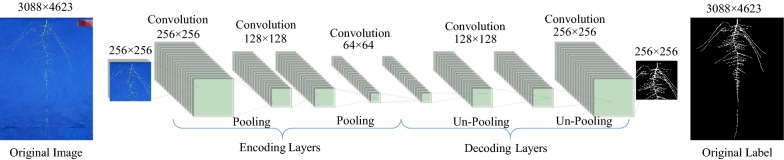
Our platform was constructed to allow for post-capture automation, eliminating the requirement of image cropping and other image-preprocessing steps. An enhancement over the original release, ARIA 2.0 automatically detects the primary root allowing automated batch processing requiring minimal user input. In addition, ARIA 2.0 provides heuristic, k-means and convolution neural network (CNN) based segmentation functionality. The color segmentation method is based on a heuristic approach on the HSV (hue, saturation, value) color space (Additional file 5: Video S4). ARIA 2.0 provides a graphic user interface allowing for an optional quality check, manual adjustment and subsequent identification of problematic images. In our experience, the heuristic color segmentation was not successful for all images in a batch due to subtle differences caused by light reflection from infrequent over saturation of the blue germination paper. A ML approach was implemented to overcome the constraint of problematic images while providing full automation. The CNN based segmentation was built on a convolutional auto-encoder (CAE) architecture and implemented via MATLAB 2018b Deep Learning Toolbox (MathWorks, Inc., Natick, MA). CAE have been used as a robust method to segment features of object from a complex and cluttered background [89, 107]. To train our auto-encoder, we utilized the manual color segmentation method to generate a training data set. A set of randomly selected ~ 2450 images (20% of the dataset, including problematic and non-problematic images) across the three time points, were segmented and used to train the CAE (Fig. 3). The encoder segment of the final network architecture comprises three convolutional layers (32 feature maps of size 3 × 3 for each layer). In addition, two pooling layers of size 2 × 2 were deployed for downsampling the features and reducing the computational load. The Rectified Linear Unit (ReLU) function was used as the activation function. The learning rate was initialized with 0.001 using Adam optimization. Training was performed using a total of ~ 2100 samples with an additional 100 randomly selected validation samples, and testing was conducted on 250 random samples. We trained the model using a NVIDIA Tesla K20 installed on the CyEnce computing cluster at Iowa State University. Using a combination of binary cross entropy and Jaccard loss as loss function, the model was validated by fivefold cross-validation resulting in F1 score and IOU of 0.8824 and 0.8725, respectively. To further validate the model, a subset of 298 images segmented by both the heuristic and CAE methods resulted in a mean correlation of 0.91 across 23 ARIA 2.0 extracted root traits (Additional file 3: Table S2).
Fig. 3.
Convolutional Auto-Encoder with 32 feature maps of size 3 × 3 for each layer with two pooling layers of size 2 × 2 that were deployed for downsampling
Analysis
The original release of ARIA software measures length, surface area, volume and was validated through correlation with WinRHIZO Pro 9.0 (Regent Instruments, Quebec, Canada) [80]. Updated validations were conducted using ImageJ (https://imagej.nih.gov/ij/) and GiARoots [84] software. To assess broad sense heritability, 115 soybean genotypes belonging to soybean maturity group II with fourteen replications (genotypes were randomly assigned to the growth chamber) were grown in growth chambers and RSA traits were measured. Outliers that fell outside the interquartile range were identified for each trait for each genotype and were eliminated prior to calculating best linear unbiased predictors (BLUPs). The model used (Eq. 1) where yik is the response variable of the ith genotype at the kth block (i.e., growth chamber used), μ is the total mean, gi is the genetic effect of the ith genotype, bk is the block effect, and eik, is a random error following N(0, σ2e). All factors were considered random effects. Broad sense heritability was calculated on an entry-mean basis using Eq. 2, where σ2g is the genotypic variance, n is the number of replications = 14. Tukey’s Honest Significance Difference (HSD) groupings were calculated from the experimental data (alpha = 0.05, rep = 14 per genotype) where q = the relevant critical value of the studentized range statistic and n* is the number of scores used in calculating the group means of interest using the HSD.test function of the agricolae package in R (Eq. 3). Genetic coefficient of variation (CVG) for each trait was calculated using Eq. 4. Pearson’s correlation coefficients between traits were calculated using the ‘stats’ package in R. A Kolmogorov–Smirnov test, a nonparametric test of continuous probability distributions, was used to test statistical differences in directionality on root branching angle at each of the three time points after germination.
| 1 |
| 2 |
| 3 |
| 4 |
Calculating lateral root branch count and measurement of root angles
RSA traits added to ARIA 2.0 include automated selection of the taproot, completely eliminating user-input, root measurement of root angles and lateral root branch count, which has been suggested as an important topological trait [1, 106]. Root branch counts were conducted using three different methods including: (1) lateral root branch count (LRB), nodes of lateral roots (NLR), and independent lateral root branches (IRB) (Fig. 4a–c) to determine the most informative and accurate way to study this trait. Lateral root branch count (LRB) was determined by first taking the skeletonized root in which the primary root is first removed. A sliding window with a five pixel width was moved across the root. The maximum number of individual root segments from each group (left or right of the primary root) were recorded. Nodes of lateral roots (NLR) were identified on the root skeleton using pixels that have more than two neighboring pixels (network analysis). The original black and white image is then dilated by 10 pixels, false or spurious branch points were identified and removed. The number of branch points after removing the false points was outputted.
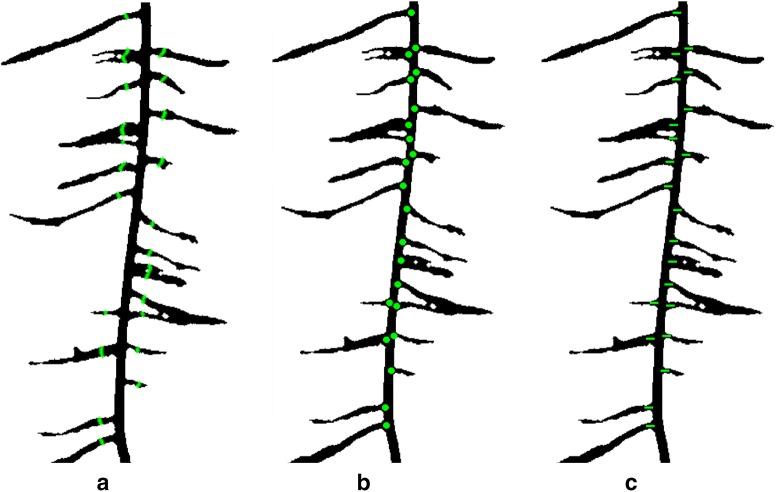
Fig. 4.
Lateral root branch count measured using three different methods a lateral root branch count (LRB), b count of nodes of lateral roots (NLR), and c independent lateral root branch count (IRB). The LRB method showed better correlation to ground truth data (R2 = 0.88 (LRB), R2 = 0.79 (NLR) and R2 = 0.76 (IRB))
Three algorithms of root branching angle were automatically quantified: (1) the lateral root branching angle (LBA), (2) lateral root angle (LRA), and (3) lateral root tip angle (RTA). The spatial distribution of the angles were shown with normalized vertical (soil) depth. The depth was normalized with the total root length. Measurement of root angles were generated within a 100 × 100 pixel window using a Fourier transform (to reduce noise), then a Hough transform algorithm was used to measure the angle of each segment (Fig. 5). The mean angle of each segment was placed into one of 45 bins of 2° increments from 0° to 90° (0° being vertical). For each algorithm, the lateral root angle of the highest frequency within each 100 × 100 pixel window was reported. ARIA 2.0 data output consisted of a tally of root segments for each algorithm allowing for simple visualization.
Fig. 5.
Three methods to identify lateral root angle including a lateral root branching angle (LBA), b lateral root angle along the entirety of each branch (LRA), and c lateral root tip angle (RTA)
Root shape classification
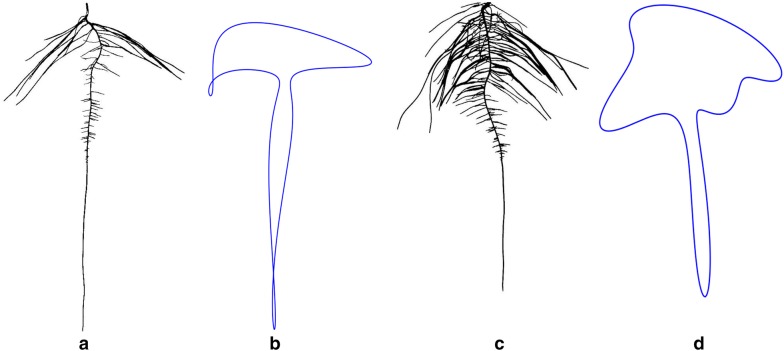
Two approaches were taken to extract root profiles from segmented root images, (1) mean boundary distance and (2) convex hull boundary. This dimension reduction processes distills images into simple numbers to allow for further phenotyping based applications. Fourier coefficients at multiple harmonics from 1 to 100 (Additional file 6: Figure S1) were used to create root shape profiles enabling the user to select which profile was most appropriate [107].
Biomass weights
Shoot and root biomass weight was collected from plants at 12 days after being dried at 70 °C for 48 h. For each genotype, 100-seed weight (g) was recorded from the seed sources used for these experiments, and was obtained by counting random 100 seed for each genotype.
Clustering algorithms (PCA and LDA)
Principal components analysis (PCA) and linear discriminant analysis (LDA) were used to visualize possible clusters of genotypes that form based on RSA traits. Linear discriminant analysis (LDA) finds a linear combination of RSA traits (explanatory variables) to discriminate between genotypes, the response variable. Principal components analysis (PCA) and LDA were performed using JMP ver. 13.1 (SAS Institute).
Results
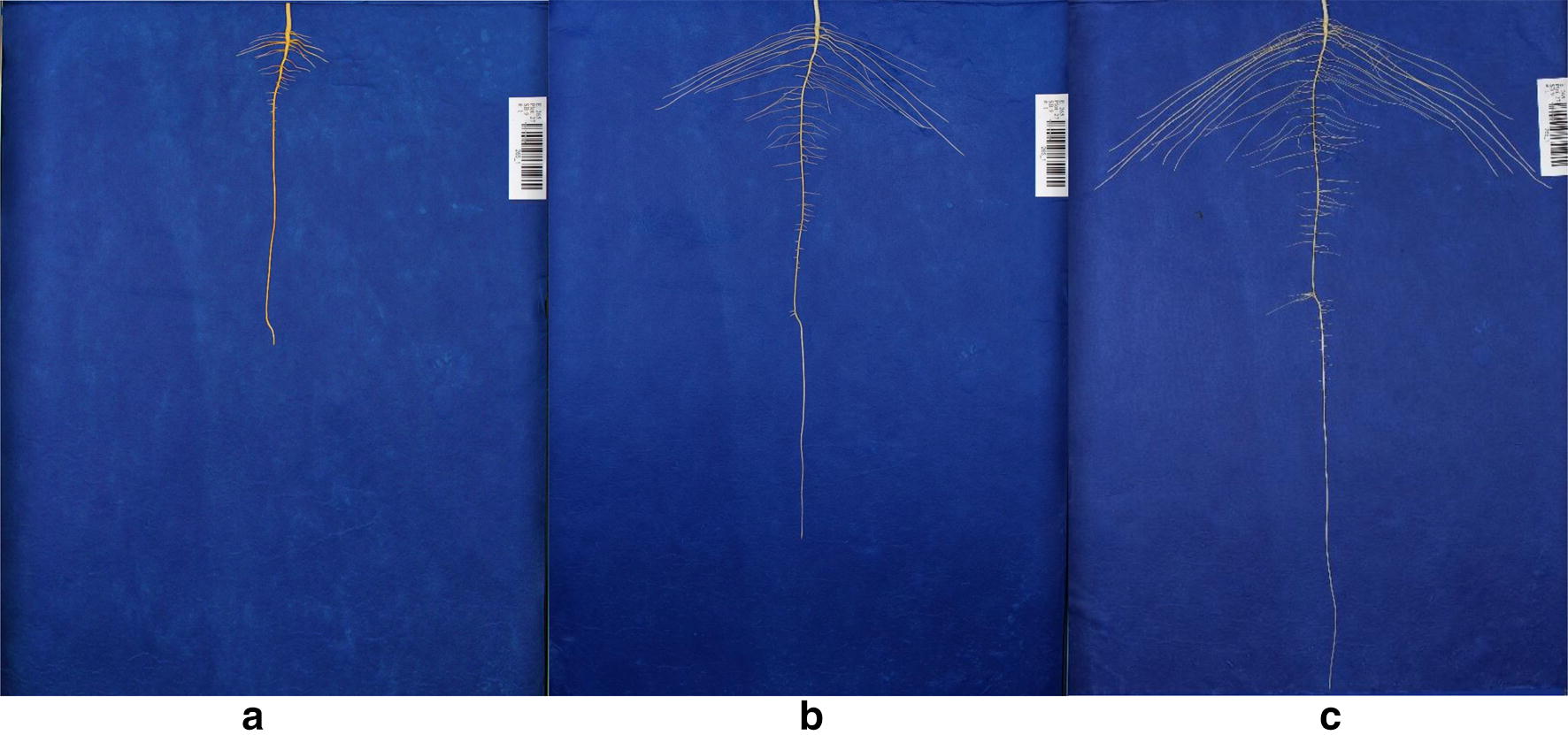
The end-to-end phenotyping pipeline consisted of the following main components: (1) a simple phenotyping platform capable of growing hundreds of plants, (2) automated image processing and curation system, (3) high fidelity image segmentation using both heuristic and ML approaches, and (4) root trait extraction software workflow and demonstrated through data analysis of 115 diverse soybean breeding lines. Using this pipeline 12,264 images were generated from 4088 plants. The image acquisition and processing rate varied depending on the number of technical persons available. This platform was used in single or multiple user modes, from individuals to teams of four, providing flexibility in time and labor management. Duration of image capture for 292 roots ranged from 3 to 5 h with throughput increased with younger roots and experienced technical staff. We present the capabilities of this root trait phenotyping pipeline to investigate the RSA diversity using test case samples consisting of diverse soybean genotypes from MG 2 using the heuristic segmentation approach and more in-depth study of three genotypes (PI 417,138, PI 643,146, PI 479718B) using the CAE segmentation approach for various RSA traits and visualize results for this diversity for the main traits (Fig. 6).
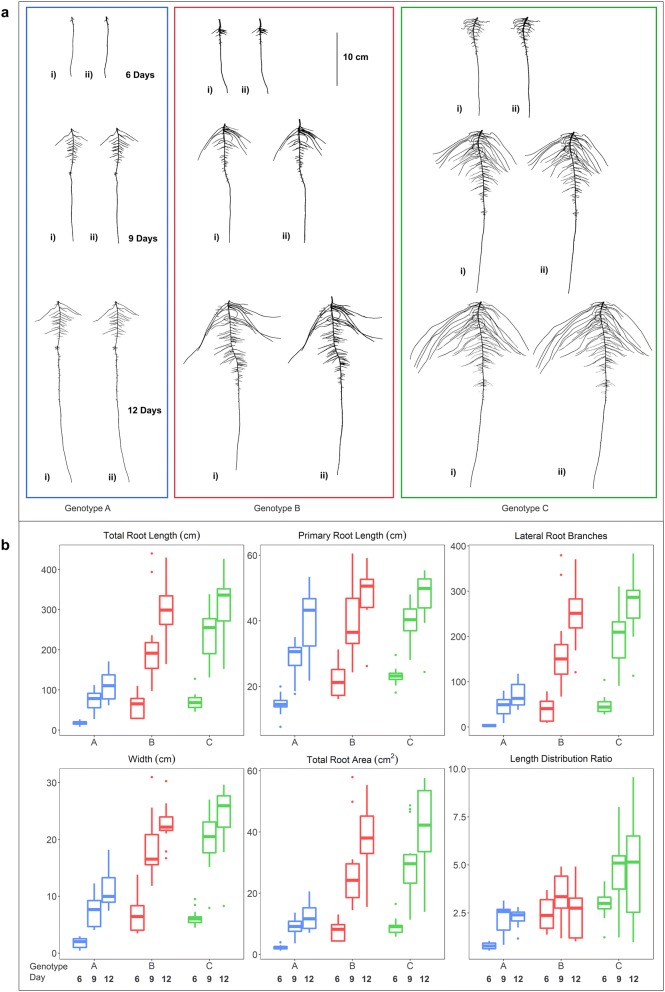
Fig. 6.
a Heuristic (i) and CAE (ii) based segmented root images of genotype A (blue), B (red) and C (green) at 6 (top), 9 (center) and 12 (bottom) days after germination. b Boxplot of displaying RSA traits of genotypes A (PI 417,138; blue), B (PI 643,146; red) and C (PI 479718B; green). TRL, PRL, LRB, WID, TRArea and LED were automatically calculated from the CAE segmented images by ARIA 2.0 (n = 14)
We evaluated the relationship between root traits and genotype descriptors including country of origin, stem termination and genetic diversity (Additional file 3: Table S3). A PCA plot was created using the genomic SNP data to further explore the associations between the continuous root trait data and discrete metadata (Additional file 6: Figure S2). Genotype was a significant source of variation for each trait (Additional file 3: Table S4). Heritability estimates increased concurrently with additional replicates and were highest for root and shoot dry weights (0.99) (Additional file 6: Figure S3).
Validation of measurements
We reevaluated the updated ARIA framework by conducting manual benchmark validation. The fidelity of the measured traits depends on the accurate count of the pixels on the images, which was validated through primary root length measurements reported by ARIA 2.0 and confirmed manually with ImageJ software (R2 = 0.999) (Additional file 6: Figure S4). Furthermore, there was high correlation between traits extracted from a set of 300 heuristically segmented root images using GiARoots [84] software and ARIA 2.0 ranging between R2 = 0.659 (BSH) to R2 = 0.998 (CVA) with a mean of R2 = 0.916 (Additional file 3: Table S5).
ARIA 2.0 software was used to compare three methods of counting lateral root branches. The LRB method showed better correlation to ground truth data (R2 = 0.88 (LRB), R2 = 0.79 (NLR) and R2 = 0.76 (IRB)). Correlation between ImageJ manually extracted angles and ARIA 2.0 measurement of root tip angles was conducted to validate our approach after outliers were removed with a R2 = 0.9025 (Additional file 6: Figure S5). Minimal root angle diversity was noted among the three genotypes (Additional file 6: Figure S6). A Kolmogorov–Smirnov test was used to detect statistical differences in directionality on root branching angle at each of the three time points. When comparing between genotypes, significant differences (p < 0.05) were seen at 6 days between genotypes A and B and genotypes A and C (Additional file 3: Table S6).
Using root images segmented with the CAE approach, three genotypes PI 417,138 (genotype A), PI 643,146 (genotype B), PI 479718B (genotype C)) were further assessed. These three genotypes displayed diversity in several biologically relevant RSA traits (TRL, PRL, LRB, WID, TRArea, and LED (length distribution)) (Additional file 6: Figure S7). Genotype A was distinct from B and C for each of the six traits except for LED (a root trait based on a ratio of TRLUpper/TRLLower) it did not differ from genotype B and 9 days and 12 days (Table 2). Genotypes B and C differed from each other at 6 days for LED, at 9 days for all six traits, and at 12 days for TRL, WID, TRArea and LED. Shoot dry weight and root dry weight BLUPs were highly correlated (R = 0.86; genotypes = 115). The majority of RSA traits had significant correlation with both shoot and root dry weight (Additional file 3: Table S7).
Table 2.
RSA trait mean values obtained from CAE segemented images, Tukey’s Honest Significant Difference (HSD) test groupings and growth rate day−1 for genotypes A, B and C for TRL (total root length), PRL (primary root length), LRB (lateral root branching count), WID (root width), Area (total root area) and LED (length distribution, total root length of the upper 1/3 of the root image divided by the total root length in the lower 2/3 of the root image
| Day | TRL (cm) | PRL (cm) | LRB | WID (cm) | TRArea (cm2) | LED | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 9 | 12 | 6 | 9 | 12 | 6 | 9 | 12 | 6 | 9 | 12 | 6 | 9 | 12 | 6 | 9 | 12 | |
| Genotype A | ||||||||||||||||||
| BLUP | 33.5 | 121 | 184.9 | 16.9 | 32.1 | 42.7 | 18.7 | 51.4 | 84.4 | 3.4 | 10 | 13.8 | 3.1 | 9.3 | 14.6 | 1.1 | 2.0 | 2.1 |
| HSD grouping | b | c | c | b | c | b | c | c | b | b | c | c | b | c | c | c | b | b |
| Growth day−1 | 29.2 | 21.3 | 5.1 | 3.5 | 10.9 | 11.0 | 2.2 | 1.3 | 2.1 | 1.8 | 0.3 | 0.0 | ||||||
| Genotype B | ||||||||||||||||||
| BLUP | 67.2 | 216.2 | 342.9 | 21.3 | 36.2 | 48.1 | 33.5 | 71.5 | 121.3 | 6.5 | 16.8 | 22.3 | 6.5 | 19.5 | 27.6 | 1.9 | 2.5 | 2.2 |
| HSD grouping | a | b | b | a | b | a | b | b | a | a | b | b | a | b | b | b | b | b |
| Growth day−1 | 49.7 | 42.2 | 5.0 | 4.0 | 12.7 | 16.6 | 3.4 | 1.8 | 4.3 | 2.7 | 0.2 | ‒ 0.1 | ||||||
| Genotype C | ||||||||||||||||||
| BLUP | 76.1 | 255.2 | 393.3 | 21.5 | 37.2 | 47.6 | 39.1 | 75.8 | 103.8 | 5.9 | 18.3 | 25 | 6.8 | 23.6 | 36 | 2.1 | 3.4 | 3.1 |
| HSD grouping | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a |
| Growth day−1 | 59.7 | 46.0 | 5.2 | 3.5 | 12.2 | 9.3 | 4.1 | 2.2 | 5.6 | 4.1 | 0.4 | ‒ 0.1 | ||||||
Root shape classification
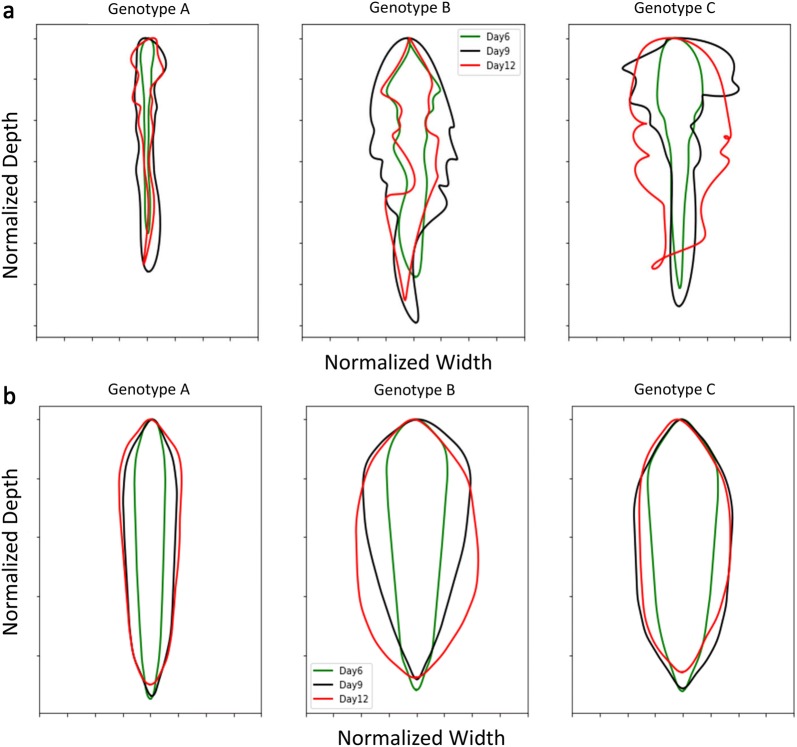
Root profiles were extracted from images and using Fourier coefficients were expanded into a shape spectrum. Figure 7a displays pseudo-outline (i.e., mean boundary) of normalized values from three genotypes to highlight root shape variation. A similar approach was taken using convex hull area (CVArea) as an input (Fig. 7b). Mean boundary and convex hull boundary analyses identified interesting divergences in root shape between genotypes. PCA and LDA were used to evaluate the contributions of RSA traits between genotypes at multiple growth stages using the output of ARIA 2.0 at 6 days, 9 days, and 12 days. LDA revealed distinct clustering patterns, where observations of three genotypes at three separate time points after germination created nine groupings (Additional file 6: Figure S8a). The PCA based on root shape defining traits at 9 days and 12 days creates three clusters while the results at 6 days are not as definitive (Additional file 6: Figure S8b).
Fig. 7.
a Mean boundary based on five harmonic Fourier descriptors of genotypes A (PI 417,138) (left), B (PI 643,146) (middle), and C (PI 479718B) (right) at 6 days (green), 9 days (black), 12 days (red). b Convex hull boundary of root shape developed from Fourier analysis (five harmonic descriptors) of the three genotypes at 6 days (green), 9 days (black), 12 days (red) (n = 14)
Discussion
In this paper, we describe end-to-end hardware and software solutions for a soybean root phenotyping pipeline (Fig. 8). This phenotyping platform provides a non-destructive evaluation pipeline with high repeatability, ease of use and scalability, capacity for hundreds genotypes in a short period of time at a lowered cost and a level of automation that will meet the needs of plant breeding. The seedling growth apparatus requires minimal supplies, expense, and experience to set up. ARIA 2.0 graphic based user interface is simple, straight-forward and builds on previous work [80]. Improvements to ARIA include additional functionality including more RSA traits, including root shape and multiple segmentation approaches. Despite the original release of ARIA being an alternative option to established programs such as WinRHIZO Pro 9.0, the application was limited to binary images and batch processed root images in a semi-automatic manner requiring user-input to identify the taproot. ARIA 2.0 was developed to address deficiencies and build additional functionality, customization and automation. The high-throughput root imaging system and fully autonomous batch processing of thousands of images with ARIA 2.0 allow for an automated imaging pipeline. In an effort to capture the essence of a root, we integrated a holistic approach which identified root shape as a trait in the phenotyping framework using Fourier transformations. We created a color segmentation package that is user-customizable for specific applications. The outcome served as a training set which fed into our ML segmentation application, reducing time through automated handling of problematic images. ARIA 2.0 software is an open-source application with simple installation, requiring no other software to operate. Multiple methods of root branching angle and counts were added to the original software’s extracted root traits. The low barrier (cost and technical expertise) to entry and rapid image capture and data processing time make this phenotyping platform suitable for large scans of diverse genetic material, genetic mapping studies, RSA trait studies and selection strategies in breeding.
Fig. 8.
Proposed root phenotyping pipeline. a Root phenotyping platform. a Image stage fabricated from aluminum, softbox lights, Canon T5i, laptop computer and, LCD monitor to evaluate images quality and image database. b Software scans and renames image automatically using barcode. c CNN framework identifies and segments root from background. d ARIA 2.0 extracts RSA traits from root images. e Data analytics (genomic selection, GWAS) are performed
Low-cost imaging platform
As research in roots has expanded, technologies such as high resolution digital cameras and computational power have increased along with specialized technicians. While we limit the presentation of blue paper for imaging background, our preliminary studies were done using gellan gum, hydroponics and brown paper cigar rolls (data not presented). Pouch-and-wick systems using blue germination paper have been routinely used to analyze and record root growth in previous studies [25, 95–98]. The correlation to root growth between different systems or media such as germination paper, hydroponic or gellan-gum and soil suggests there are differences that affect the growth of the roots; however, since all genotypes in this study were tested under the same system we were able to make comparisons between genotypes. The blue germination paper approach was deemed most suitable as it allowed simultaneous growing and imaging of hundreds of plants nondestructively in a time series manner with a minimal person-hour requirement. For soybean, at 12 days post germination, the taproot of a limited number of genotypes outgrew the blue paper system; longer lengths of paper can be used if the intent is to perform post 12 day imaging.
The imaging platform’s efficiency was reduced by root convergence as previously identified by Dupuy et al. [97]. Slight user adjustment of some roots was required to ensure that the computer vision algorithm could detect differences between multiple roots converging in parallel. Our preliminary studies concluded that convergence was inconsistent between root and genotypes and could reduce TRL up to 18% (data not shown) due to the software’s inability to identify individual roots out of a mass. Therefore, our protocol was developed with minimal manual adjustment to reduce convergence. Removal of the technician adjustment would decrease the duration of image acquisition. Further experiments are needed to solve the issue of convergence without adding a root processing step.
While our system shows promise for a time-series root trait data collection, continual work is needed to expand the ability to phenotype large number of genotypes in a wider time-series manner and also establishing controlled environment and field grown trait relationship.
ML based image preprocessing and image analysis
Plant phenotyping can vary in number of experimental treatments and genotypes and thus degrees of complexity [108]. While alternate root image analysis software tools exist, our platform implements additional RSA traits alongside automated machine learning segmentation to make it feasible for use with large data sets. In a step towards full automation from image acquisition through analysis, user input is minimized, removing the interaction between the user and individual root images (such as determining anchor points with ARIA (original release) [80] or SmartRoot [66]). The system is capable of scaling to both large root systems and large quantities of root systems. Alternative software, such as Root System Analyzer, are capable of using image sequences to track growth. However, even the fully automated system requires substantial user intervention (data not presented) and is more amenable for smaller datasets. Traditional image segmentation methods are not generalizable, as users’ needs to trial and error to identify the best segmentation model and model input parameter (heuristically). More importantly, it is our experience that while some (traditional) segmentation models work on a good fraction of images, no segmentation model works on (nearly) all the images. The end-user then has to either make a sub-optimal choice or perform exhaustive quality control. This is where ML approaches like the CAE model become attractive. With a small amount of annotated data, they can be specifically trained for a specialized application with minimal subsequent user input. We present the end-to-end pipeline we included both hardware and software advances for root trait imaging and analytics without exclusively focusing on ARIA 2.0 improvement. ML has become a critical tool to improve analysis and quantification of data in plant phenomics [56]. Until recently, the use of ML in root phenomics has been limited to root tip identification [91, 109] and data prediction [92]. Using a CAE for image preprocessing, is a new approach in root phenomics, and overcomes current challenges in image preprocessing.
RSA trait measurements
The ideal root architecture is dependent on breeding objectives as desired architecture may be determined by crop, environment, fertility, and water availability however are often not well described [1]. Using computer vision tools, the creation and collection of RSA traits is nearly endless. What is important however, is to collect biologically relevant traits. The system described in this paper delivers as much information to the user as possible so that the user can then determine the biological usefulness of each trait as per the objectives of their studies.
ARIA 2.0 software was used to compare three improved methods of counting first-order lateral root branches, which were validated using manual assessments of 68 random plants from the three time points (6 days, 9 days, and 12 days). Automated identification of second-order lateral roots could be an addition of future versions of ARIA. Lateral root branch number (LRB) had the strongest correlation to the ground truth results. NLR and IRB often overestimated the number of roots compared to manual counts likely due to the misidentification of pixel spurs. Misidentification of pixels spurs, an erroneous grouping of pixels on the boundary of the blue paper which is a product of the segmentation process, as roots resulted in false positives. Multiple roots growing together in parallel were liable to be counted as one, as color-based image segmentation was unable to isolate individual roots. One particular improvement to ARIA is the measurement of root angles, with respect to the direction of gravity, taken in three locations, near the primary root, at the root tip and throughout the root system as a whole. Previous studies have shown that root angle in rice [39, 110], chickpea (Cicer arietinum L.) [111] and sorghum (Sorghum bicolor L.) [112] is correlated to drought tolerance and root depth. The correlation between root angle and drought tolerance was identified in rhizotrons [85] because water-deficient Arabidopsis roots grow at a steeper angle than well-watered roots [85, 113, 114]. Aside from root count, primary root volume and surface area can also be found in ARIA 2.0. Thick roots have been shown to penetrate deeper through soil-layers [115, 116]. Thin, fibrous roots have shown plasticity in response to drought. Large, thick roots act as a conduit pipe and serve a purpose in anchorage however, it is the fine secondary and tertiary roots that make up the vast network of absorbing area. Plants that optimize root absorption area while minimizing biological cost are desirable [36]. The aforementioned traits can be identified using the ARIA 2.0 seedling phenotyping pipeline, demonstrating the relevance of ML and computer vision based software for the study of RSA traits. Furthermore, the presented approach can also be useful in learning or describing new traits, and studies on the growth and development in a time-series manner. Unlike above ground traits, at this time root systems do not have a well characterized growth or stages. An understanding of stages and processes is integral to translating root development into mathematical growth models, which can help develop more efficient plants. Compelling visual differences for root shape were uncovered, exhibited by heuristically segmented root images of LG05-4832 and PI 594457A in Fig. 9. Aided by Fourier descriptors, soybean canopy shapes have been previously described [107, 117]. Using a similar approach, we observed that these methods were sufficient to draw root outlines. This system was designed to maximize data acquisition and to reduce environmental differences with minimal errors. Therefore, this pipeline was effective in root trait studies to identify most diverse genotypes from a germplasm set; however further improvements are needed to enable complex organism interaction studies and field grown genotype roots sample assessments. For example, the image based root phenotyping methods still need additional technological refinement and advancement to integrate microorganism-root interaction phenotyping and studies [11, 12, 118, 119].
Fig. 9.
Root shape profiles based on elliptical Fourier transformation (EFT). Example genotypes of a LG05-4832, b EFT derived root outline of LG05-4832 (n = 14), c PI 594457A and, d EFT derived root outline of PI 594457A (n = 14) at 9 days
Controlled environment studies bring advantages of scale and data noise reduction along with cost efficiency gains. Since selection of root traits in row crops is one of the major challenge for breeding programs, we envision a two-step approach: (1) using a germplasm and pre-breeding step in controlled environment screening of root traits similar to this study and previous studies [25, 81, 96, 98] to assemble a smaller collection of accessions or experimental lines for further testing, and (2) field screening for root traits through direct and indirect selection. Since, for most row crop breeding programs, the ultimate goal is increase seed yield, indirect selection for yield traits will need identification of root traits with high heritability and high genetic correlation between root traits and seed yield. While advances in phenotyping and data analytics of above ground traits is gaining exponentially, similar advances in root traits are lacking due to complexity of phenotyping organs below ground and a spatio-temporal scale. Therefore, studies that build on expanding the inference scope of root trait are needed with connectivity with yield performance. While our system shows promise for a time-series root trait data collection, continual work is needed to expand the ability phenotype large number of genotypes in a wider time-series manner and also establishing controlled environment and field grown trait relationship. The novelty of our work is developing an end-to-end phenotyping system, and integration of ML based batch image pre-processing and root trait feature extraction. We envision that approach (1) will help in determining the genetic variation for root traits and thereby influencing selection differential factor of response to selection. This work provides insights on root trait diversity from a large collection of the USDA germplasm bank, and one of the largest such report on soybean root studies.
Conclusions
This report describes innovation through the development of an affordable end-to-end phenotyping system of hundreds of plants, and integration of ML based batch image pre-processing and root trait feature extraction. We have developed a phenotyping pipeline that integrates image capture, image processing and image analysis of growing plant roots in controlled conditions providing a high-throughput, cost effective platform yielding biologically relevant time-series data on root growth and development. The outcome of hardware and software solutions provided a high quantity, cost-effective, efficient, repeatable seedling root phenotyping platform incorporating time series growth capture, and a computer vision based ARIA 2.0 integrated with ML based image preprocessing step. Additionally, we demonstrated the potential of the pipeline to capture RSA trait diversity on three selected soybean genotypes, which can be expanded to larger genotype set. HTP methods together with phenomics and data analytics [120] will give researchers the tools needed to decipher the genetics of RSA trait expression to realize the potential of root driven breeding. Further work is needed to develop methods for 3D reconstruction, as well as methodologies to link and reduce the gap between controlled and field experiment root studies. We envision that approach (1) will help in determining the genetic variation for root traits and thereby influencing selection differential factor of response to selection and prescriptive plant breeding [121] .
Supplementary information
Additional file 1: Video S1. Initiation of germination rolls.
Additional file 2: Video S2. Transplanting soybean seedlings at five days after germination.
Additional file 3: Table S1. A list of imaging system components. Table S2. Validation correlations between 23 RSA traits extracted from ARIA 2.0 using 298 heuristically segmented and CAE segmented root images. Table S3. RSA traits at 6d, 9d and 12d when grouped into country of origin, growth habit type and diversity of genetic background. Table S4. Descriptive statistics for 6 root and shoot traits of 115 maturity group II genotypes of soybean at 6d, 9d and 12d, obtained from BLUP values for each genotype using heuristically segmented images. Table S5. Validation correlation between GiARoot software and ARIA 2.0. Table S6. Minimal root angle diversity among the three genotypes. A Kolmogorov-Smirnov test was used to detect p-value statistical differences in directionality on root branching angle at each of the three time points. Table S7. Correlations between plant dry weight taken at 12d and root traits at 9d for 115 maturity group II genotypes of soybean.
Additional file 4: Video S3. Soybean seedling root imaging.
Additional file 5: Video S4. Color segmentation method of root images using a heuristic approach in MATLAB based on the HSV (hue, saturation, value) color space.
Additional file 6: Figure S1. Root shape profiles derived from elliptical Fourier transformations (EFT) at multiple harmonics (n = 2 to n = 20). Figure S2. PCA plots based on genomic SNP data to further explore the associations between (a) country of origin, (b) growth habit, and (c) genetic diversity (elite, diverse, landrace). Figure S3. Six RSA traits displaying the increase in broad-sense heritability (H2) with each replicate tested (n=14). Figure S4. Validation of primary root length (PRL) using SmartRoot in ImageJ. Figure S5. Correlation between manual and ARIA 2.0 root angles. Correlation resulted in an R2 value of 0.9025. Any root angles calculated by ARIA 2.0 as being less than 10 degrees were considered as being outliers due to a result of very small root segments. Figure S6. Root angles of genotype A (PI 417138; blue), B (PI 643146; red) and C (PI 479718B; green) at 6d (left), 9d (center) and 12d (right) generated from heuristically segmented images. The top row relates to LBA; middle row, LRA; and bottom row is RTA (all expressed as a percentage of total). 0° is the direction of the gravity vector. Figure S7. Overall phenotypic differentiation of three example soybean genotypes: A (PI 417138; blue), B (PI 643146; red) and C (PI 479718B; green) for TRL (total root length), PRL (primary root length), WID (root width), convex area (CVA) LRB (lateral root branching count), VOL (primary root volume), LRA (lateral root branching angle, LED (length distribution, total root length of the upper 1/3 of the root image divided by the total root length in the lower 2/3 of the root image), RHZO (rhizosphere area), WDR (width to depth ratio), Root_weight (dry root weight at 12 days after germination), Shoot_weight (dry shoot weight at 12 days after germination). For every trait each datum is reflective of one root image (therefore there are 14 data points per genotype). The data lines reflect the mean of the data points for respective genotype. Heuristically segmented images were used in this analysis. Figure S8. Dimension reduction analysis. (a) Linear discriminant analysis works as a dimensionality reduction algorithm, is shown using 38 RSA traits to cluster genotype A (PI 417138; blue), B (PI 643146; red) and C (PI 479718B; green) at 6 (triangle), 9 (plus) and 12 (diamond) after germination (n=14). (b) Principal components analysis of the three genotypes at 6, 9 and 12 days after germination. The shaded area enclose 90% of each genotype’s data points (n=14; generated from heuristic segmented images).
Acknowledgements
We sincerely thank the Soybean breeding and Soynomics Research Group members (staff, PDFs, graduate students, undergraduate students) at Iowa State University.
Abbreviations
- ARIA
automatic root imaging analysis
- BLUP
best linear unbiased prediction
- CAE
convolutional auto-encoder
- CNN
convolutional neural network
- CVG
genetic coefficient of variation
- DL
deep learning
- HSD
Tukey’s honest significant difference
- HSV
hue, saturation, value
- HTP
high-throughput phenotyping
- LDA
linear discriminant analysis
- ML
machine learning
- OCR
object character recognition
- PCA
principal components analysis
- RSA
root system architecture
Authors’ contributions
Conceptualization: KF, AKS, BG, AS, SS; experimentation: KF, TZJ; Analysis: KF, TZJ, AKS, BG, VM, KP; project supervision—AKS, BG, AS, SS; writing first draft—KF, AKS; writing and reviewing—all authors. All authors read and approved the final manuscript.
Funding
Authors sincerely appreciate funding support from the R F Baker Center for Plant Breeding at ISU, Iowa Soybean Research Center, Iowa Soybean Association, Monsanto Chair in Soybean Breeding at ISU, Plant Sciences Institute at ISU, and USDA CRIS project (IOW04314).
Availability of data and materials
The ARIA 2.0 code is freely available at the address: https://bitbucket.org/baskargroup/aria2/src/master/. Analysis code is freely available at the address: https://github.com/mighster/ARIA2.0. Raw images and/or segmented masks are available upon request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kevin G. Falk, Email: falk@iastate.edu
Talukder Z. Jubery, Email: znjubery@iastate.edu
Seyed V. Mirnezami, Email: vahid@iastate.edu
Kyle A. Parmley, Email: kparmley@iastate.edu
Soumik Sarkar, Email: soumiks@iastate.edu.
Arti Singh, Email: arti@iastate.edu.
Baskar Ganapathysubramanian, Email: baskarg@iastate.edu.
Asheesh K. Singh, Email: singhak@iastate.edu
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13007-019-0550-5.
References
- 1.Lynch J. Root Architecture and Plant Productivity. Plant Physiol. 1995;109:7–13. doi: 10.1104/pp.109.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.York LM, Galindo-Castañeda T, Schussler JR, Lynch JP. Evolution of US maize (Zea mays L.) root architectural and anatomical phenes over the past 100 years corresponds to increased tolerance of nitrogen stress. J Exp Bot. 2015;66:2347–2358. doi: 10.1093/jxb/erv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manschadi AM, Kaul H-P, Vollmann J, Eitzinger J, Wenzel W. Developing phosphorus-efficient crop varieties—an interdisciplinary research framework. Field Crops Res. 2014;162:87–98. doi: 10.1016/j.fcr.2013.12.016. [DOI] [Google Scholar]
- 4.Comas LH, Becker SR, Cruz VMV, Byrne PF, Dierig DA. Root traits contributing to plant productivity under drought. Front Plant Sci. 2013 doi: 10.3389/fpls.2013.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenta B, Beebe S, Kunert K, Burridge J, Barlow K, Lynch J, et al. Field phenotyping of soybean roots for drought stress tolerance. Agronomy. 2014;4:418–435. doi: 10.3390/agronomy4030418. [DOI] [Google Scholar]
- 6.Monneveux P, Ribaut J-M. Drought adaptation in cereals. Boca Raton: CRC Press; 2006. [Google Scholar]
- 7.Wade LJ, Bartolome V, Mauleon R, Vasant VD, Prabakar SM, Chelliah M, et al. Environmental response and genomic regions correlated with rice root growth and yield under drought in the OryzaSNP panel across multiple study systems. PLoS ONE. 2015;10:e0124127. doi: 10.1371/journal.pone.0124127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.VanToai TT, St Martin SK, Chase K, Boru G, Schnipke V, Schmitthenner AF, et al. Identification of a QTL associated with tolerance of soybean to soil waterlogging. Crop Sci. 2001;41:1247–1252. doi: 10.2135/cropsci2001.4141247x. [DOI] [Google Scholar]
- 9.Guingo E, Hébert Y, Charcosset A. Genetic analysis of root traits in maize. Agronomie. 1998;18:225–235. doi: 10.1051/agro:19980305. [DOI] [Google Scholar]
- 10.Gregory PJ, Bengough AG, Grinev D, Schmidt S, Thomas WB, Wojciechowski T, et al. Root phenomics of crops: opportunities and challenges. Funct Plant Biol. 2009;36:922–929. doi: 10.1071/FP09150. [DOI] [PubMed] [Google Scholar]
- 11.Kuijken RCP, van Eeuwijk FA, Marcelis LFM, Bouwmeester HJ. Root phenotyping: from component trait in the lab to breeding. J Exp Bot. 2015;66:5389–5401. doi: 10.1093/jxb/erv239. [DOI] [PubMed] [Google Scholar]
- 12.Meister R, Rajani MS, Ruzicka D, Schachtman DP. Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 2014;19:779–788. doi: 10.1016/j.tplants.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Ordóñez RA, Castellano MJ, Hatfield JL, Helmers MJ, Licht MA, Liebman M, et al. Maize and soybean root front velocity and maximum depth in Iowa, USA. Field Crops Res. 2018;215:122–131. doi: 10.1016/j.fcr.2017.09.003. [DOI] [Google Scholar]
- 14.Topp CN. Hope in Change: The role of root plasticity in crop yield stability. Plant Physiol. 2016;172:5–6. doi: 10.1104/pp.16.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blum A. Drought resistance—is it really a complex trait? Functional Plant Biol. 2011;38:753–757. doi: 10.1071/FP11101. [DOI] [PubMed] [Google Scholar]
- 16.Bohn M, Novais J, Fonseca R, Tuberosa R, Grift TE. Genetic evaluation of root complexity in maize. Acta Agron Hung. 2006;54:291–303. doi: 10.1556/AAgr.54.2006.3.3. [DOI] [Google Scholar]
- 17.Price AH, Tomos AD. Genetic dissection of root growth in rice (Oryza sativa L.) II: mapping quantitative trait loci using molecular markers. Theor Appl Genet. 1997;95:143–152. doi: 10.1007/s001220050542. [DOI] [Google Scholar]
- 18.Clark RT, Famoso AN, Zhao K, Shaff JE, Craft EJ, Bustamante CD, et al. High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ. 2013;36:454–466. doi: 10.1111/j.1365-3040.2012.02587.x. [DOI] [PubMed] [Google Scholar]
- 19.Kano M, Inukai Y, Kitano H, Yamauchi A. Root plasticity as the key root trait for adaptation to various intensities of drought stress in rice. Plant Soil. 2011;342:117–128. doi: 10.1007/s11104-010-0675-9. [DOI] [Google Scholar]
- 20.Cairns JE, Impa SM, O’Toole JC, Jagadish SVK, Price AH. Influence of the soil physical environment on rice (Oryza sativa L.) response to drought stress and its implications for drought research. Field Crops Res. 2011;121:303–310. doi: 10.1016/j.fcr.2011.01.012. [DOI] [Google Scholar]
- 21.Nagel KA, Kastenholz B, Jahnke S, van Dusschoten D, Aach T, Mühlich M, et al. Temperature responses of roots: impact on growth, root system architecture and implications for phenotyping. Funct Plant Biol. 2009;36:947–959. doi: 10.1071/FP09184. [DOI] [PubMed] [Google Scholar]
- 22.Bengough AG, McKenzie BM, Hallett PD, Valentine TA. Root elongation, water stress, and mechanical impedance: a review of limiting stresses and beneficial root tip traits. J Exp Bot. 2011;62:59–68. doi: 10.1093/jxb/erq350. [DOI] [PubMed] [Google Scholar]
- 23.Hammer GL, Dong Z, McLean G, Doherty A, Messina C, Schussler J, et al. Can changes in canopy and/or root system architecture explain historical maize yield trends in the US Corn Belt? Crop Sci. 2009;49:299–312. doi: 10.2135/cropsci2008.03.0152. [DOI] [Google Scholar]
- 24.Lambers H, Mougel C, Jaillard B, Hinsinger P. Plant-microbe-soil interactions in the rhizosphere: an evolutionary perspective. Plant Soil. 2009;321:83–115. doi: 10.1007/s11104-009-0042-x. [DOI] [Google Scholar]
- 25.Adu MO, Chatot A, Wiesel L, Bennett MJ, Broadley MR, White PJ, et al. A scanner system for high-resolution quantification of variation in root growth dynamics of Brassica rapa genotypes. J Exp Bot. 2014;65:2039–2048. doi: 10.1093/jxb/eru048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer-Pascuzzi AS, Symonova O, Mileyko Y, Hao Y, Belcher H, Harer J, et al. Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 2010;152:1148–1157. doi: 10.1104/pp.109.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagel KA, Putz A, Gilmer F, Heinz K, Fischbach A, Pfeifer J, et al. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct Plant Biol. 2012;39:891. doi: 10.1071/FP12023. [DOI] [PubMed] [Google Scholar]
- 28.Robinson H, Kelly A, Fox G, Franckowiak J, Borrell A, Hickey L. Root architectural traits and yield: exploring the relationship in barley breeding trials. Euphytica. 2018;214:151. doi: 10.1007/s10681-018-2219-y. [DOI] [Google Scholar]
- 29.Bucksch A, Burridge J, York LM, Das A, Nord E, Weitz JS, et al. Image-based high-throughput field phenotyping of crop roots. Plant Physiol. 2014;166:470–486. doi: 10.1104/pp.114.243519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burridge J, Jochua CN, Bucksch A, Lynch JP. Legume shovelomics: high—throughput phenotyping of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata subsp, unguiculata) root architecture in the field. Field Crops Res. 2016;192:21–32. doi: 10.1016/j.fcr.2016.04.008. [DOI] [Google Scholar]
- 31.Colombi T, Kirchgessner N, Marié CAL, York LM, Lynch JP, Hund A. Next generation shovelomics: set up a tent and REST. Plant Soil. 2015;388:1–20. doi: 10.1007/s11104-015-2379-7. [DOI] [Google Scholar]
- 32.Trachsel S, Kaeppler SM, Brown KM, Lynch JP. Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil. 2011;341:75–87. doi: 10.1007/s11104-010-0623-8. [DOI] [Google Scholar]
- 33.Forde BG. Is it good noise? The role of developmental instability in the shaping of a root system. J Exp Bot. 2009;60:3989–4002. doi: 10.1093/jxb/erp265. [DOI] [PubMed] [Google Scholar]
- 34.Ito K, Tanakamaru K, Morita S, Abe J, Inanaga S. Lateral root development, including responses to soil drying, of maize (Zea mays) and wheat (Triticum aestivum) seminal roots. Physiol Plant. 2006;127:260–267. doi: 10.1111/j.1399-3054.2006.00657.x. [DOI] [Google Scholar]
- 35.Kato Y, Kamoshita A, Yamagishi J, Imoto H, Abe J. Growth of rice (Oryza Sativa L.) cultivars under upland conditions with different levels of water supply Root system development, soil moisture change and plant water status. Plant Prod Sci. 2007;10:3–13. doi: 10.1626/pps.10.3. [DOI] [Google Scholar]
- 36.Lynch JP. Roots of the second green revolution. Aust J Bot. 2007;55:493. doi: 10.1071/BT06118. [DOI] [Google Scholar]
- 37.Herder GD, Van Isterdael G, Beeckman T, De Smet I. The roots of a new green revolution. Trends Plant Sci. 2010;15:600–607. doi: 10.1016/j.tplants.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, et al. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature. 2012;488:535–539. doi: 10.1038/nature11346. [DOI] [PubMed] [Google Scholar]
- 39.Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, et al. Control of root system architecture by deeper rooting 1 increases rice yield under drought conditions. Nat Genet. 2013;45:1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- 40.Burton AL, Brown KM, Lynch JP. Phenotypic diversity of root anatomical and architectural traits in Zea species. Crop Sci. 2013;53:1042–1055. doi: 10.2135/cropsci2012.07.0440. [DOI] [Google Scholar]
- 41.Abdel-Haleem H, Lee G-J, Boerma RH. Identification of QTL for increased fibrous roots in soybean. Theor Appl Genet. 2011;122:935–946. doi: 10.1007/s00122-010-1500-9. [DOI] [PubMed] [Google Scholar]
- 42.Gregory PJ. Plant roots: growth, activity and interactions with the soil. Hoboken: Wiley; 2008. [Google Scholar]
- 43.O’Toole JC, Bland WL. Genotypic variation in crop plant root systems. In: Brady NC, editor. Advances in agronomy. Cambridge: Academic Press; 1987. pp. 91–145. [Google Scholar]
- 44.Zhao J, Fu J, Liao H, He Y, Nian H, Hu Y, et al. Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm. ChinSciBull. 2004;49:1611–1620. [Google Scholar]
- 45.Lynch J, van Beem JJ. Growth and architecture of seedling roots of common bean genotypes. Crop Sci. 1993;33:1253–1257. doi: 10.2135/cropsci1993.0011183X003300060028x. [DOI] [Google Scholar]
- 46.Uga Y, Ebana K, Abe J, Morita S, Okuno K, Yano M. Variation in root morphology and anatomy among accessions of cultivated rice (Oryza sativa L.) with different genetic backgrounds. Breed Sci. 2009;59:87–93. doi: 10.1270/jsbbs.59.87. [DOI] [Google Scholar]
- 47.Nakhforoosh A, Grausgruber H, Kaul H-P, Bodner G. Wheat root diversity and root functional characterization. Plant Soil. 2014;380:211–229. doi: 10.1007/s11104-014-2082-0. [DOI] [Google Scholar]
- 48.Singh RJ, Hymowitz T. Soybean genetic resources and crop improvement. Genome. 1999;42:605–616. doi: 10.1139/g99-039. [DOI] [Google Scholar]
- 49.de Dorlodot S, Forster B, Pagès L, Price A, Tuberosa R, Draye X. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 2007;12:474–481. doi: 10.1016/j.tplants.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Hoisington D, Khairallah M, Reeves T, Ribaut J-M, Skovmand B, Taba S, et al. Plant genetic resources: What can they contribute toward increased crop productivity? PNAS. 1999;96:5937–5943. doi: 10.1073/pnas.96.11.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gizlice Z, Carter TE, Burton JW. Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci. 1994;34:1143–1151. doi: 10.2135/cropsci1994.0011183X003400050001x. [DOI] [Google Scholar]
- 52.Naik HS, Zhang J, Lofquist A, Assefa T, Sarkar S, Ackerman D, et al. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods. 2017;13:23. doi: 10.1186/s13007-017-0173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Naik HS, Assefa T, Sarkar S, Reddy RVC, Singh A, et al. Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci Rep. 2017;7:44048. doi: 10.1038/srep44048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao T, Emadi H, Saha H, et al. A novel multirobot system for plant phenotyping. Robotics. 2018;7(4):61. doi: 10.3390/robotics7040061. [DOI] [Google Scholar]
- 55.Tattaris M, Reynolds MP, Chapman SC. A direct comparison of remote sensing approaches for high-throughput phenotyping in plant breeding. Front Plant Sci. 2016 doi: 10.3389/fpls.2016.01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh A, Ganapathysubramanian B, Singh AK, Sarkar S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016;21:110–124. doi: 10.1016/j.tplants.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 57.Singh AK, Ganapathysubramanian B, Sarkar S, Singh A. Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci. 2018 doi: 10.1016/j.tplants.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 58.Camargo AV, Mackay I, Mott R, Han J, Doonan JH, Askew K, et al. Functional mapping of quantitative trait loci (QTLs) associated with plant performance in a wheat MAGIC mapping population. Front Plant Sci. 2018 doi: 10.3389/fpls.2018.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tardieu F, Cabrera-Bosquet L, Pridmore T, Bennett M. Plant Phenomics, from sensors to knowledge. Curr Biol. 2017;27:770–783. doi: 10.1016/j.cub.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 60.Burgess AJ, Retkute R, Pound MP, Foulkes J, Preston SP, Jensen OE, et al. High-resolution three-dimensional structural data quantify the impact of photoinhibition on long-term carbon gain in wheat canopies in the field. Plant Physiol. 2015;169:1192–1204. doi: 10.1104/pp.15.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fahlgren N, Gehan MA, Baxter I. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr Opin Plant Biol. 2015;24:93–99. doi: 10.1016/j.pbi.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Hartmann A, Czauderna T, Hoffmann R, Stein N, Schreiber F. HTPheno: an image analysis pipeline for high-throughput plant phenotyping. BMC Bioinformatics. 2011;12:148. doi: 10.1186/1471-2105-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korhonen L, Heikkinen J. Automated analysis of in situ canopy images for the estimation of forest canopy cover. For Sci. 2009;55:323–334. [Google Scholar]
- 64.Mielewczik M, Friedli M, Kirchgessner N, Walter A. Diel leaf growth of soybean: a novel method to analyze two-dimensional leaf expansion in high temporal resolution based on a marker tracking approach (Martrack Leaf) Plant Methods. 2013;9:30. doi: 10.1186/1746-4811-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Virlet N, Sabermanesh K, Sadeghi-Tehran P, Hawkesford MJ. Field scanalyzer: an automated robotic field phenotyping platform for detailed crop monitoring. Funct Plant Biol. 2017;44:143–153. doi: 10.1071/FP16163. [DOI] [PubMed] [Google Scholar]
- 66.Lobet G, Pagès L, Draye X. A Novel Image-analysis toolbox enabling quantitative analysis of root system architecture1[W][OA] Plant Physiol. 2011;157:29–39. doi: 10.1104/pp.111.179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Atkinson JA, Lobet G, Noll M, Meyer PE, Griffiths M, Wells DM. Combining semi-automated image analysis techniques with machine learning algorithms to accelerate large-scale genetic studies. Gigascience. 2017;6:1–7. doi: 10.1093/gigascience/gix084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mairhofer S, Zappala S, Tracy SR, Sturrock C, Bennett M, Mooney SJ, et al. RooTrak: automated recovery of three-dimensional plant root architecture in soil from X-ray microcomputed tomography images using visual tracking. Plant Physiol. 2012;158:561–569. doi: 10.1104/pp.111.186221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Topp CN, Bray AL, Ellis NA, Liu Z. How can we harness quantitative genetic variation in crop root systems for agricultural improvement?: quantifying root architecture for crops. J Integr Plant Biol. 2016;58:213–225. doi: 10.1111/jipb.12470. [DOI] [PubMed] [Google Scholar]
- 70.Lobet G, Draye X, Périlleux C. An online database for plant image analysis software tools. Plant Methods. 2013;9:38. doi: 10.1186/1746-4811-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wasson A, Bischof L, Zwart A, Watt M. A portable fluorescence spectroscopy imaging system for automated root phenotyping in soil cores in the field. J Exp Bot. 2016;67:1033–1043. doi: 10.1093/jxb/erv570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gray SB, Strellner RS, Puthuval KK, Ng C, Shulman RE, Siebers MH, et al. Minirhizotron imaging reveals that nodulation of field-grown soybean is enhanced by free-air CO2 enrichment only when combined with drought stress. Funct Plant Biol. 2013;40:137. doi: 10.1071/FP12044. [DOI] [PubMed] [Google Scholar]
- 73.Mooney SJ, Pridmore TP, Helliwell J, Bennett MJ. Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil. 2012;352:1–22. doi: 10.1007/s11104-011-1039-9. [DOI] [Google Scholar]
- 74.Mairhofer S, Zappala S, Tracy S, Sturrock C, Bennett MJ, Mooney SJ, et al. Recovering complete plant root system architectures from soil via X-ray μ-computed tomography. Plant Methods. 2013;9:8. doi: 10.1186/1746-4811-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jahnke S, Menzel MI, Dusschoten DV, Roeb GW, Bühler J, Minwuyelet S, et al. Combined MRI–PET dissects dynamic changes in plant structures and functions. Plant J. 2009;59:634–644. doi: 10.1111/j.1365-313X.2009.03888.x. [DOI] [PubMed] [Google Scholar]
- 76.Clark RT, MacCurdy RB, Jung JK, Shaff JE, McCouch SR, Aneshansley DJ, et al. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 2011;156:455–465. doi: 10.1104/pp.110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee C-R, Zurek PR, Symonova O, et al. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci. 2013;110:1695–1704. doi: 10.1073/pnas.1304354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu J, Ingram PA, Benfey PN, Elich T. From lab to field, new approaches to phenotyping root system architecture. Curr Opin Plant Biol. 2011;14:310–317. doi: 10.1016/j.pbi.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 79.Delory BM, Li M, Topp CN, Lobet G. archiDART v.30: A new data analysis pipeline allowing the topological analysis of plant root systems. F1000Res. 2018 doi: 10.12688/f1000research.13541.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pace J, Lee N, Naik HS, Ganapathysubramanian B, Lübberstedt T. Analysis of maize (Zea mays L.) seedling roots with the high-throughput image analysis tool ARIA (automatic root image analysis) PLoS ONE. 2014;9:e108255. doi: 10.1371/journal.pone.0108255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Le Bot J, Serra V, Fabre J, Draye X, Adamowicz S, Pagès L. DART: a software to analyse root system architecture and development from captured images. Plant Soil. 2010;326:261–273. doi: 10.1007/s11104-009-0005-2. [DOI] [Google Scholar]
- 82.Das A, Schneider H, Burridge J, Ascanio AKM, Wojciechowski T, Topp CN, et al. Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods. 2015;11:51. doi: 10.1186/s13007-015-0093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, et al. EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J. 2009;57:945–956. doi: 10.1111/j.1365-313X.2008.03739.x. [DOI] [PubMed] [Google Scholar]
- 84.Galkovskyi T, Mileyko Y, Bucksch A, Moore B, Symonova O, Price CA, et al. GiA roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol. 2012;12:116. doi: 10.1186/1471-2229-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rellán-Álvarez R, Lobet G, Lindner H, Pradier P-L, Sebastian J, Yee M-C, et al. GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. Elife. 2015;4:e07597. doi: 10.7554/eLife.07597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu J, Wu Q, Pagès L, Yuan Y, Zhang X, Du M, et al. RhizoChamber-Monitor: a robotic platform and software enabling characterization of root growth. Plant Methods. 2018;14:44. doi: 10.1186/s13007-018-0316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pound MP, French AP, Atkinson JA, Wells DM, Bennett MJ, Pridmore T. RootNav: navigating images of complex root architectures. Plant Physiol. 2013;162:1802–1814. doi: 10.1104/pp.113.221531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leitner D, Felderer B, Vontobel P, Schnepf A. Recovering root system traits using image analysis exemplified by two-dimensional neutron radiography images of lupine. Plant Physiol. 2014;164:24–35. doi: 10.1104/pp.113.227892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.French A, Ubeda-Tomas S, Holman TJ, Bennett MJ, Pridmore T. High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol. 2009;150:1784–1795. doi: 10.1104/pp.109.140558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akintayo A, Tylka GL, Singh AK, Ganapathysubramanian B, Singh A, Sarkar S. A deep learning framework to discern and count microscopic nematode eggs. Sci Rep. 2018;8:9145. doi: 10.1038/s41598-018-27272-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kumar P, Huang C, Cai J, Miklavcic SJ. Root phenotyping by root tip detection and classification through statistical learning. Plant Soil. 2014;380:193–209. doi: 10.1007/s11104-014-2071-3. [DOI] [Google Scholar]
- 92.Lobet G, Koevoets IT, Noll M, Meyer PE, Tocquin P, Pagès L, et al. Using a structural root system model to evaluate and improve the accuracy of root image analysis pipelines. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oliveira MF, Nelson RL, Geraldi IO, Cruz CD, de Toledo JFF. Establishing a soybean germplasm core collection. Field Crops Res. 2010;119:277–289. doi: 10.1016/j.fcr.2010.07.021. [DOI] [Google Scholar]
- 94.Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, et al. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE. 2013;8:e54985. doi: 10.1371/journal.pone.0054985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hund A, Trachsel S, Stamp P. Growth of axile and lateral roots of maize: I development of a phenotying platform. Plant Soil. 2009;325:335–349. doi: 10.1007/s11104-009-9984-2. [DOI] [Google Scholar]
- 96.Gioia T, Galinski A, Lenz H, Müller C, Lentz J, Heinz K, et al. GrowScreen-PaGe, a non-invasive, high-throughput phenotyping system based on germination paper to quantify crop phenotypic diversity and plasticity of root traits under varying nutrient supply. Funct Plant Biol. 2017;44:76–93. doi: 10.1071/FP16128. [DOI] [PubMed] [Google Scholar]
- 97.Dupuy LX, Wright G, Thompson JA, Taylor A, Dekeyser S, White CP, et al. Accelerating root system phenotyping of seedlings through a computer-assisted processing pipeline. Plant Methods. 2017;13:57. doi: 10.1186/s13007-017-0207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Atkinson JA, Wingen LU, Griffiths M, Pound MP, Gaju O, Foulkes MJ, et al. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J Exp Bot. 2015;66:2283–2292. doi: 10.1093/jxb/erv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abdel-Ghani AH, Kumar B, Reyes-Matamoros J, Gonzalez-Portilla PJ, Jansen C, Martin JPS, et al. Genotypic variation and relationships between seedling and adult plant traits in maize (Zea mays L.) inbred lines grown under contrasting nitrogen levels. Euphytica. 2013;189:123–133. doi: 10.1007/s10681-012-0759-0. [DOI] [Google Scholar]
- 100.de Sousa SM, Clark RT, Mendes FF, de Oliveira AC, de Vasconcelos MJV, Parentoni SN, et al. A role for root morphology and related candidate genes in P acquisition efficiency in maize. Funct Plant Biol. 2012;39:925–935. doi: 10.1071/FP12022. [DOI] [PubMed] [Google Scholar]
- 101.Smith JR, Mengistu A, Nelson RL, Paris RL. Identification of soybean accessions with high germinability in high-temperature environments. Crop Sci. 2008;48:2279. doi: 10.2135/cropsci2008.01.0026. [DOI] [Google Scholar]
- 102.Germination Rolls. Vimeo. https://vimeo.com/288177394. Accessed 17 Apr 2019.
- 103.Transplanting. Vimeo. https://vimeo.com/288837774. Accessed 17 Apr 2019.
- 104.Smart Shooter Photography Software. https://kuvacode.com/. Accessed 2 Apr 2019.
- 105.Singh Soybean Root Phenomics: Seedling Roots. Vimeo. https://vimeo.com/264556231. Accessed 17 Apr 2019.
- 106.Fitter A. Characteristics and functions of root systems. In: Waisel Y, Eshel A, Beeckman T, Kafkafi U, editors. Plant roots: the hidden half. Boca Raton: CRC Press; 1991. [Google Scholar]
- 107.Jubery TZ, Shook J, Parmley K, Zhang J, Naik HS, Higgins R, et al. Deploying Fourier coefficients to unravel soybean canopy diversity. Front Plant Sci. 2017 doi: 10.3389/fpls.2016.02066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fiorani F, Schurr U. Future scenarios for plant phenotyping. Annu Rev Plant Biol. 2013;64:267–291. doi: 10.1146/annurev-arplant-050312-120137. [DOI] [PubMed] [Google Scholar]
- 109.Pound MP, Burgess AJ, Wilson MH, Atkinson JA, Griffiths M, Jackson AS, et al. Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. 2016. https://biorxiv.org/lookup/doi/10.1101/053033. Accessed 17 May 2016.
- 110.Kato Y, Abe J, Kamoshita A, Yamagishi J. Genotypic variation in root growth angle in rice (Oryza sativa L.) and its association with deep root development in upland fields with different water regimes. Plant Soil. 2006;287:117–129. doi: 10.1007/s11104-006-9008-4. [DOI] [Google Scholar]
- 111.Kashiwagi J, Krishnamurthy L, Purushothaman R, Upadhyaya HD, Gaur PM, Gowda CLL, et al. Scope for improvement of yield under drought through the root traits in chickpea (Cicer arietinum L.) Field Crops Res. 2015;170:47–54. doi: 10.1016/j.fcr.2014.10.003. [DOI] [Google Scholar]
- 112.Singh V, van Oosterom JE, Jordan DR, Hunt CH, Hammer GL. Genetic variability and control of nodal root angle in sorghum. Crop Sci. 2011;51:2011–2020. doi: 10.2135/cropsci2011.01.0038. [DOI] [Google Scholar]
- 113.Lynch JP. Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol. 2011;156:1041–1049. doi: 10.1104/pp.111.175414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Walk TC, Jaramillo R, Lynch JP. Architectural tradeoffs between adventitious and basal roots for phosphorus acquisition. Plant Soil. 2006;279:347–366. doi: 10.1007/s11104-005-0389-6. [DOI] [Google Scholar]
- 115.Yu L-X, Ray JD, O’Toole JC, Nguyen HT. Use of wax-petrolatum layers for screening rice root penetration. Crop Sci. 1995;35:684–687. doi: 10.2135/cropsci1995.0011183X003500030005x. [DOI] [Google Scholar]
- 116.Zheng H, Babu RC, Pathan MS, Ali L, Huang N, Courtois B, et al. Quantitative trait loci for root-penetration ability and root thickness in rice: comparison of genetic backgrounds. Genome. 2000;43:53–61. doi: 10.1139/g99-065. [DOI] [PubMed] [Google Scholar]
- 117.Kuhl FP, Giardina CR. Elliptic Fourier features of a closed contour. Comput Graph Image Process. 1982;18:236–258. doi: 10.1016/0146-664X(82)90034-X. [DOI] [Google Scholar]
- 118.Singh AK, Hamel C, DePauw RM, Knox RE. Genetic variability in arbuscular mycorrhizal fungi compatibility supports the selection of durum wheat genotypes for enhancing soil ecological services and cropping systems in Canada. Can J Microbiol. 2012;58:293–302. doi: 10.1139/w11-140. [DOI] [PubMed] [Google Scholar]
- 119.Ellouze W, Hamel C, Singh AK, Mishra V, DePauw RM, Knox RE. Abundance of the arbuscular mycorrhizal fungal taxa associated with the roots and rhizosphere soil of different durum wheat cultivars in the Canadian prairies. Can J Microbiol. 2018;64:527–536. doi: 10.1139/cjm-2017-0637. [DOI] [PubMed] [Google Scholar]
- 120.Parmley K, Nagasubramanian K, Sarkar S, Ganapathysubramanian B, Singh AK. Development of optimized phenomic predictors for efficient plant breeding decisions using phenomic-assisted selection in soybean. Plant Phenomics. 2019;2019:1–15. doi: 10.34133/2019/5809404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Parmley KA, Higgins RH, Ganapathysubramanian B, Sarkar S, Singh AK. Machine learning approach for prescriptive plant breeding. Sci Rep. 2019;9(1):17132. doi: 10.1038/s41598-019-53451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Video S1. Initiation of germination rolls.
Additional file 2: Video S2. Transplanting soybean seedlings at five days after germination.
Additional file 3: Table S1. A list of imaging system components. Table S2. Validation correlations between 23 RSA traits extracted from ARIA 2.0 using 298 heuristically segmented and CAE segmented root images. Table S3. RSA traits at 6d, 9d and 12d when grouped into country of origin, growth habit type and diversity of genetic background. Table S4. Descriptive statistics for 6 root and shoot traits of 115 maturity group II genotypes of soybean at 6d, 9d and 12d, obtained from BLUP values for each genotype using heuristically segmented images. Table S5. Validation correlation between GiARoot software and ARIA 2.0. Table S6. Minimal root angle diversity among the three genotypes. A Kolmogorov-Smirnov test was used to detect p-value statistical differences in directionality on root branching angle at each of the three time points. Table S7. Correlations between plant dry weight taken at 12d and root traits at 9d for 115 maturity group II genotypes of soybean.
Additional file 4: Video S3. Soybean seedling root imaging.
Additional file 5: Video S4. Color segmentation method of root images using a heuristic approach in MATLAB based on the HSV (hue, saturation, value) color space.
Additional file 6: Figure S1. Root shape profiles derived from elliptical Fourier transformations (EFT) at multiple harmonics (n = 2 to n = 20). Figure S2. PCA plots based on genomic SNP data to further explore the associations between (a) country of origin, (b) growth habit, and (c) genetic diversity (elite, diverse, landrace). Figure S3. Six RSA traits displaying the increase in broad-sense heritability (H2) with each replicate tested (n=14). Figure S4. Validation of primary root length (PRL) using SmartRoot in ImageJ. Figure S5. Correlation between manual and ARIA 2.0 root angles. Correlation resulted in an R2 value of 0.9025. Any root angles calculated by ARIA 2.0 as being less than 10 degrees were considered as being outliers due to a result of very small root segments. Figure S6. Root angles of genotype A (PI 417138; blue), B (PI 643146; red) and C (PI 479718B; green) at 6d (left), 9d (center) and 12d (right) generated from heuristically segmented images. The top row relates to LBA; middle row, LRA; and bottom row is RTA (all expressed as a percentage of total). 0° is the direction of the gravity vector. Figure S7. Overall phenotypic differentiation of three example soybean genotypes: A (PI 417138; blue), B (PI 643146; red) and C (PI 479718B; green) for TRL (total root length), PRL (primary root length), WID (root width), convex area (CVA) LRB (lateral root branching count), VOL (primary root volume), LRA (lateral root branching angle, LED (length distribution, total root length of the upper 1/3 of the root image divided by the total root length in the lower 2/3 of the root image), RHZO (rhizosphere area), WDR (width to depth ratio), Root_weight (dry root weight at 12 days after germination), Shoot_weight (dry shoot weight at 12 days after germination). For every trait each datum is reflective of one root image (therefore there are 14 data points per genotype). The data lines reflect the mean of the data points for respective genotype. Heuristically segmented images were used in this analysis. Figure S8. Dimension reduction analysis. (a) Linear discriminant analysis works as a dimensionality reduction algorithm, is shown using 38 RSA traits to cluster genotype A (PI 417138; blue), B (PI 643146; red) and C (PI 479718B; green) at 6 (triangle), 9 (plus) and 12 (diamond) after germination (n=14). (b) Principal components analysis of the three genotypes at 6, 9 and 12 days after germination. The shaded area enclose 90% of each genotype’s data points (n=14; generated from heuristic segmented images).
Data Availability Statement
The ARIA 2.0 code is freely available at the address: https://bitbucket.org/baskargroup/aria2/src/master/. Analysis code is freely available at the address: https://github.com/mighster/ARIA2.0. Raw images and/or segmented masks are available upon request.