Abstract
Objective
This study aimed to evaluate the correlation between the circular RNA VRK serine/threonine kinase 1 (circ‐VRK1) and the clinicopathological features and survival outcomes of breast cancer patients and determine its effects on breast cancer cell proliferation and apoptosis.
Methods
A total of 350 breast cancer tissues and 163 breast cancer adjacent tissues, as controls, were acquired from Specimen House. Circ‐VRK1 expression was measured using qPCR. The correlations between circ‐VRK1 expression and demographic characteristics, tumour features and overall survival were analysed. In vitro, the effects of circ‐VRK1 on breast cancer cell proliferation and apoptosis were measured by upregulating and downregulating circ‐VRK1 expression via plasmid transfection.
Results
Circ‐VRK1 was downregulated in breast cancer tissues compared with adjacent tissues and represented a good value in distinguishing breast cancer tissues from the adjacent tissues. Circ‐VRK1 was associated with smaller tumour size, reduced T stage and lower TNM stage, and circ‐VRK1 was also an independent predictor of better overall survival. According to the in vitro experiments, circ‐VRK1 expression was lower in breast cancer cell lines (including BT474, MDA‐MB‐453 and MDA‐MB‐231) than in a normal breast epithelial cell line (MCF10A), and circ‐VRK1 inhibited cell proliferation but promoted cell apoptosis in MDA‐MB‐231 cells.
Conclusion
Circ‐VRK1 is downregulated in tumour tissues and associated with reduced tumour stage as well as better survival, and it inhibits cell proliferation but promotes cell apoptosis in breast cancer.
Keywords: breast cancer, cell activities, Circ‐VRK1, clinicopathological features, survival
1. INTRODUCTION
Breast cancer, a complex and heterogeneous disease, is responsible for 14% of total cancer‐related deaths and is the leading cause of cancer in women.1 According to the global statistics of 2018, there are estimated 2 088 849 new breast cancer cases and 626 679 breast cancer deaths in 2018, and the incidence and death rate are about 39.2 and 8.6 per 100 000, respectively, in eastern Asia.2 Although early diagnosis, radical surgery, neo‐adjuvant/adjuvant therapy and targeted drug applications have contributed to substantial improvements in the survival rate of breast cancer patients with curative intent, the long‐term mortality rate and high recurrence rate remain urgent clinical problems.3 As a result, for the treatment of breast cancer, it is essential to investigate novel therapeutic targets that can increase the recovery rate and promote long‐term survival.
Circular RNA (circRNA), as a type of the non‐coding RNAs, is identified as by‐product of splicing‐RNA aberration, whose structure is a covalently closed loop without 3′‐end and 5′‐end.4, 5 Additionally, circRNA has been indicated to play a crucial role in the regulation of breast cancer biological processes, including growth, differentiation, metabolism and metastasis.6, 7 For instance, circMYO9B, functioning as miR‐4316 sponges, promotes cell proliferation, migration and invasion and accelerates tumour growth in breast cancer.8 In addition, upregulation of circUBAP2 induces breast cancer progression by sponging miRNA‐661 and increasing the expression of the metastasis‐associated protein MTA1.9 CircRNA VRK serine/threonine kinase 1 (circ‐VRK1), located at chromosome 14 from 97312431 to 97327072, is revealed to inhibit the stemness of breast cancer stem cells, which are cells that are able to self‐renew, differentiate into different cell types and contribute to malignant progression and therapeutic resistance to traditional chemoradiotherapy.10, 11, 12 However, there is currently limited study focusing on the correlation of circ‐VRK1 with tumour characteristics, prognosis and cellular activities in breast cancer. Thus, we conducted this study to evaluate the correlation of circ‐VRK1 with the clinicopathological features and survival in breast cancer patients as well as its effect on breast cancer cell proliferation and apoptosis.
2. MATERIALS AND METHODS
2.1. Participants
A total of 350 breast cancer patients who underwent resections between January 2014 and December 2016 were retrospectively analysed. The eligible criteria were as follows: (a) diagnosed as primary breast cancer by pathological confirmation; (b) age between 18 and 75 years; (c) completed data of tumour feature and survival were able to be retrieved from Medical Record System; (d) frozen tumour tissue was accessible from Specimen House; and (e) without neo‐adjuvant therapies before surgery. This study was approved by the Ethics Committee of our Hospital, and patients or their direct relative families signed the informed consents.
2.2. Sample acquisition and circ‐VRK1 measurement
Tumour tissue samples of 350 included breast cancer patients were obtained from Specimen House; in the meanwhile, 163 adjacent tissue samples from breast cancer patients were acquired from Specimen House as control. Circ‐VRK1 expression in tumour tissues and adjacent tissues was then measured by quantitative polymerase chain reaction (qPCR).
2.3. Data collection and survival calculation
Demographic and tumour features were retrieved from Medical Record System, among which the pathological differentiation was graded as follows: grade 1, well differentiation; grade 2, moderate differentiation; and grade 3, poor differentiation. And the TNM stage was assessed according to the TNM staging system for breast cancer (7th Edition).13 The overall survival (OS) was calculated from the time of resection to the time of death. The median follow‐up duration was 36 months; the last follow‐up date was 2018/6/30.
2.4. Cell sources and culture
Breast cancer cell lines (BT474, MCF7, MDA‐MB‐453 and MDA‐MB‐231) were purchased from Cell Resource Center of Shanghai Institute of Life Sciences, Chinese Academy of Sciences or American Type Culture Collection (ATCC) (Rockefeller). Normal breast epithelial cell line (MCF10A) was purchased from ATCC (Rockefeller). BT474 cell line was cultured in 90% RPMI 1640 sodium (Gibco) with 10% foetal bovine serum (FBS) (Gibco) under 95% air and 5% CO2 at 37°C, MCF7 cell line was cultured in 90% MEM sodium (Gibco) with 10% FBS (Gibco) under 95% air and 5% CO2 at 37°C, and MDA‐MB‐453 and MDA‐MB‐231 cell lines were cultured in 90% L‐15 sodium (Gibco) with 10% FBS (Gibco) under 95% air and 5% CO2 at 37°C. In addition, MCF10A cell line was cultured in MEGM Kit (Sigma) under 95% air and 5% CO2 at 37°C.
2.5. Transfection and measurement of cell proliferation as well as apoptosis
Circ‐VRK1 expression in breast cancer cell lines (BT474, MCF7, MDA‐MB‐453 and MDA‐MB‐231) and normal breast epithelial cell line (MCF10A) was measured by qPCR. Then, control overexpression and shRNA, circ‐VRK1 overexpression and shRNA plasmids were constructed using pGPU6 and pEX‐2 plasmids by Shanghai GenePharma Biotech Company, and transfected into MDA‐MB‐231 cells and accordingly divided into control (+), VRK1 (+), control (−) and VRK1 (−) groups. The MDA‐MB‐231 cells that were not treated with transfection were served as normal group. After transfection, circ‐VRK1 expression was measured at 24 hours by qPCR; cell proliferation was measured at 0, 24, 48 and 72 hours by Counting Kit‐8 (CCK‐8) (Sangon Biotech) according to the instructions of manufacturer; cell apoptosis rate was measured at 24 hours by FITC Annexin V Apoptosis Detection Kit II (BD, USA) according to the instructions of manufacturer.
2.6. Quantitative polymerase chain reaction
Total RNA was firstly extracted from breast cancer tissues, adjacent tissues and harvested cells using TRIzol Reagent (Invitrogen).12, 14 To detect expression of circ‐VRK1, linear RNA was diminished using RNase R (Epicentre). Then, total RNA or RNase R‐treated RNA was reversely transcribed to cDNA using PrimeScript™ RT Reagent Kit (TAKARA). Finally, qPCR was conducted using the QuantiNova SYBR Green PCR Kit (Qiagen). Instrument of ABI 7900HT real‐time PCR system was applied to conduct the PCR. The procedures were carried out as follows: first, 5 minutes at 95°C, 40 cycles of PCR, then followed by standard conditions with 15 seconds of denaturation at 95°C; next, elongation for 1 minute at 60°C. The relative abundance was normalized to GAPDH, and fold change of circ‐VRK1 relative expression was calculated with the 2−△△Ct method.15 The primer sequence of circ‐VRK1 was as follows: forward‐GAACCTGGTGTTGAAGATACGG; reverse‐AATCCTACTTTCCATTCCTTTTTTG. The primer sequence of GAPDH was as follows: forward‐TCCTCACAGTTGCCATGTAGACCC; reverse‐GCGGGCTCAATTTATAGAAACCGGG. The product size and accession number in NCBI of circ‐VRK1 were 195 bp and hsa_circ_0141206, and the product size and accession number in NCBI of GAPDH were 197 bp and NM_002046.7, respectively.
2.7. Statistics
Statistical analysis was performed using SPSS 22.0 software (IBM) and GraphPad 6.01 software (GraphPad Int.). Data were exhibited as mean ± standard deviation, median (1/4‐3/4 quarters) or count (percentage). Comparison among three or above groups was determined by one‐way ANOVA followed by Dunnett's multiple comparison test or the Kruskal‐Wallis H rank sum test; comparison between two groups was determined by t test or Wilcoxon rank sum test; comparison of OS was revealed by Kaplan‐Meier (K‐M) curve and determined by log‐rank test; correlation was determined by the Spearman test; factors predicting OS were determined by univariate and multivariate Cox's proportional hazard regression. P < .05 was considered as significant.
3. RESULTS
3.1. Baseline characteristics
A total of 350 breast cancer patients with the mean age of 52.4 ± 12.4 years were enrolled in this study (Table 1). Regarding pathological grade, there were 76 (21.7%) patients in grade 1, 251 (71.7%) patients in grade 2 and 23 (6.6%) patients in grade 3. The average tumour size of all patients was 3.2 ± 1.6 cm. With respect to T stage, 109 (31.1%) patients were in T1, 209 (59.7%) patients were in T2, and 32 (9.2%) patients were in T3. In regard to N stage, 171 (48.9%), 106 (30.3%), 67 (19.1%) and 6 (1.7%) patients were in the N0, N1, N2 and N3 stage, respectively. Finally, the numbers of patients with TNM stages in I, II and III were 44 (12.6%), 222 (63.4%) and 84 (24.0%), respectively. Other detailed baseline characteristics of breast cancer patients are shown in Table 1.
Table 1.
Baseline characteristics of breast cancer patients
| Characteristics | Patients (N = 350) |
|---|---|
| Age (y) | 52.4 ± 12.4 |
| Pathological grade (n/%) | |
| Grade 1 | 76 (21.7) |
| Grade 2 | 251 (71.7) |
| Grade 3 | 23 (6.6) |
| Tumour size (cm) | 3.2 ± 1.6 |
| T stage (n/%) | |
| T1 | 109 (31.1) |
| T2 | 209 (59.7) |
| T3 | 32 (9.2) |
| N stage (n/%) | |
| N0 | 171 (48.9) |
| N1 | 106 (30.3) |
| N2 | 67 (19.1) |
| N3 | 6 (1.7) |
| M stage (n/%) | |
| M0 | 350 (100.0) |
| TNM stage (n/%) | |
| I | 44 (12.6) |
| II | 222 (63.4) |
| III | 84 (24.0) |
| ER status (n/%) | |
| Negative | 137 (39.1) |
| Positive | 213 (60.9) |
| PR status (n/%) | |
| Negative | 172 (49.1) |
| Positive | 178 (50.9) |
| HER2 status (n/%) | |
| Negative | 230 (65.7) |
| Positive | 120 (34.3) |
Data were presented as mean ± standard deviation or count (%).
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
3.2. Comparison of circ‐VRK1 relative expression in tumour tissue and adjacent tissue in breast cancer
Comparison of circ‐VRK1 relative expression between tumour tissue (n = 350) and adjacent tissue (n = 163) was determined by Wilcoxon rank sum test, which revealed that compared with adjacent tissue (0.851 [0.447‐1.667]), circ‐VRK1 relative expression in tumour tissue (0.339 [0.220‐0.723]) was lower (P < .001) (Figure 1A). The ROC curve demonstrated that circ‐VRK1 was of good value in distinguishing tumour tissue from adjacent tissue with area under curve (AUC) of 0.720 (95% CI: 0.672‐0.768), with the sensitivity and the specificity of the best cut‐off point 61.7% and 79.1%, respectively, and the circ‐VRK1 level at the best cut‐off point was 0.425 (Figure 1B). The best cut‐off point was where the largest sum of sensitivity and specificity occurred.
Figure 1.
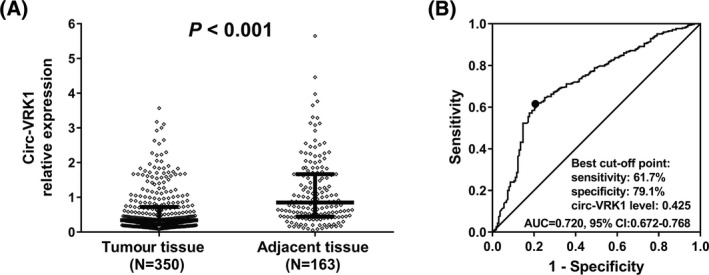
Circ‐VRK1 relative expression in breast cancer patients. Circ‐VRK1 relative expression was decreased in tumour tissue compared with adjacent tissue (A), and it had good value in distinguishing breast tumour tissue from adjacent tissue (B). The Wilcoxon rank sum test was used to compare circ‐VRK1 relative expression between tumour tissue and adjacent tissue, and ROC curve was performed to evaluate the ability of circ‐VRK1 relative expression to discriminate breast tumour tissue from adjacent tissue. P < .05 was considered significant. Circ‐VRK1, circular RNA VRK serine/threonine kinase 1; ROC, receiver operating characteristic.
3.3. Correlation of circ‐VRK1 relative expression with clinicopathological features in breast cancer patients
The difference in circVRK1 expression in breast cancer patients with different clinicopathological features was determined by Kruskal‐Wallis H rank sum test or Wilcoxon rank sum test. Additionally, the correlations of circVRK1 expression with age and tumour size were tested by the Spearman test. Circ‐VRK1 relative expression was elevated in patients with pathology grade 2 compared with those with pathology grade 1 and pathology grade 3 (P = .036) (Figure 2B), and circ‐VRK1 relative expression was also discovered to be negatively associated with tumour size (r = −0.146, P = .006) (Figure 2C), T stage (P < .001) (Figure 2D), N stage (P = .049) (Figure 2E) and TNM stage (P = .016) (Figure 2F). However, there was no correlation of circ‐VRK1 expression with age (P = .374) (Figure 2A), estrogen receptor (ER) (P = .567) (Figure 2G), progesterone receptor (PR) (P = .350) (Figure 2H) or human epidermal growth factor receptor 2 (HER2) (P = .281) (Figure 2I).
Figure 2.
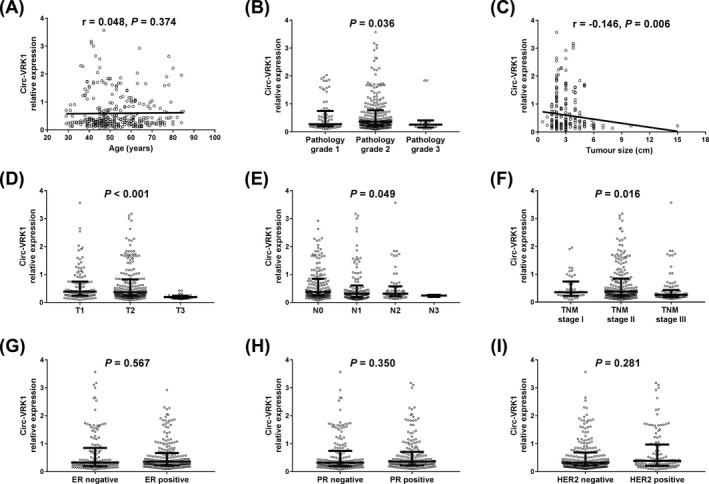
Association of circ‐VRK1 relative expression with clinicopathological features in breast cancer patients. Correlation between circ‐VRK1 and pathology grade was presented (P = .036) (B). Circ‐VRK1 was negatively correlated with tumour size (C), T stage (D), N stage (E) and TNM stage (F). There is no association of circ‐VRK1 with age (A), ER (G), PR (H) or HER2 (I). The Kruskal‐Wallis H rank sum test and the Wilcoxon rank sum test were used to determine the difference in circVRK1 expression in breast cancer patients with different clinicopathological features. And the Spearman test was performed to observe the correlation of circ‐VRK1 with age and tumour size in breast cancer patients. P < .05 was considered significant. Circ‐VRK1, Circular RNA VRK serine/threonine kinase 1; ER, estrogen receptor; HER 2, human epidermal growth factor receptor 2; PR, progesterone receptor; TNM, TNM Classification of Malignant Tumours
3.4. Correlation of circ‐VRK1 relative expression with OS in breast cancer patients
Patients were divided into circ‐VRK1 high‐expression and circ‐VRK1 low‐expression groups according to the median value of circ‐VRK1 expression. Kaplan‐Meier curve was performed to estimate survival, and log‐rank test was used for comparison between the curves. The OS of the circ‐VRK1 high‐expression group was longer (mean OS: 51.3 months, 95% CI: 50.0‐52.7 months) than that of the circ‐VRK1 low‐expression group (mean OS: 46.5 months, 95% CI: 44.1‐44.8 months) (P = .001) (Figure 3).
Figure 3.
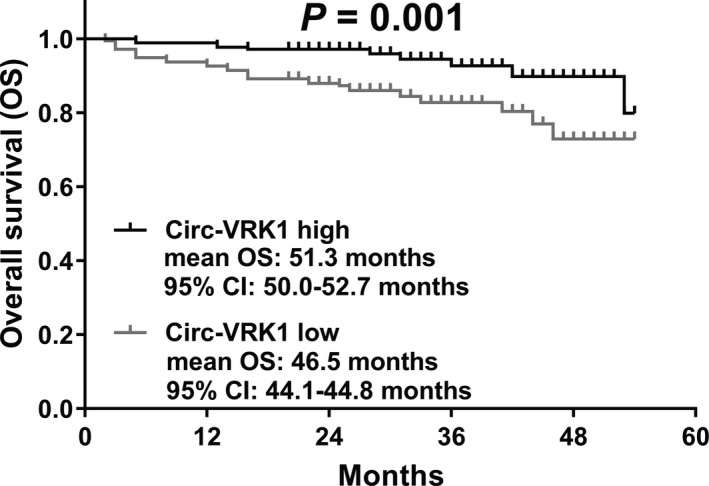
Comparison of OS between circ‐VRK1 high‐expression and low‐expression groups. Compared with the patients with circ‐VRK1 low expression, patients with circ‐VRK1 high expression had better OS. Survival curve was performed by using the Kaplan‐Meier method, and differences between the curves were determined by log‐rank test. P < .05 was considered significant. circ‐VRK1, circular RNA VRK serine/threonine kinase 1; OS, overall survival
3.5. Factors affecting OS in breast cancer patients
Univariate Cox's analysis displayed that circ‐VRK1 (high vs low) was associated with better OS (HR = 0.375, P = .002); however, pathological grade (G3 vs G1/2) (HR = 2.846, P = .011), tumour size (≥3 cm vs <3 cm) (HR = 3.363, P = .001), T stage (T2/T3 vs T1) (HR = 3.603, P = .003), N stage (N1/N2/N3 vs N0) (HR = 3.564, P < .001) and TNM stage (III vs II/I) (HR = 6.496, P < .001) were predictive factors for worse OS for breast cancer patients (Table 2). All factors were further analysed by multivariate Cox's proportional hazard regression analysis, which revealed that circ‐VRK1 (high vs low) was an independent predictor of higher OS (HR = 0.481, P = .031), whereas pathological grade (G3 vs G1/2) (HR = 3.431, P = .016) and TNM stage (III vs II/I) (HR = 5.555, P < .001) independently predicted lower OS (Table 3).
Table 2.
Univariate Cox's proportional hazard regression analysis of factors affecting OS
| Univariate Cox's regression model | ||||
|---|---|---|---|---|
| P value | HR | 95% CI | ||
| Lower | Higher | |||
| Circ‐VRK1 (high vs low) | .002 | 0.375 | 0.201 | 0.700 |
| Age (≥60 y vs <60 y) | .865 | 0.947 | 0.508 | 1.766 |
| Pathological grade (G3 vs G1/2) | .011 | 2.846 | 1.274 | 6.354 |
| Tumour size (≥3 cm vs <3 cm) | .001 | 3.363 | 1.674 | 6.755 |
| T stage (T2/T3 vs T1) | .003 | 3.603 | 1.530 | 8.480 |
| N stage (N1/N2/N3 vs N0) | <.001 | 3.564 | 1.817 | 6.989 |
| TNM stage (III vs II/I) | <.001 | 6.496 | 3.629 | 11.626 |
| ER status (positive vs negative) | .095 | 0.617 | 0.350 | 1.087 |
| PR status (positive vs negative) | .175 | 0.672 | 0.378 | 1.193 |
| HER2 status (positive vs negative) | .414 | 1.273 | 0.713 | 2.273 |
Data were presented as P value, HR (hazards ratio) and 95% CI (confidence interval). P < .05 was considered significant.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; OS, overall survival; PR, progesterone receptor.
Table 3.
Multivariate Cox's proportional hazard regression analysis of factors affecting OS
| Multivariate Cox's regression model | ||||
|---|---|---|---|---|
| P value | HR | 95% CI | ||
| Lower | Higher | |||
| Circ‐VRK1 (high vs low) | .031 | 0.481 | 0.248 | 0.934 |
| Age (≥60 y vs <60 y) | .259 | 1.558 | 0.722 | 3.360 |
| Pathological grade (G3 vs G1/2) | .016 | 3.431 | 1.263 | 9.321 |
| Tumour size (≥3 cm vs <3 cm) | .174 | 2.226 | 0.702 | 7.058 |
| T stage (T2/T3 vs T1) | .672 | 1.358 | 0.329 | 5.599 |
| N stage (N1/N2/N3 vs N0) | .574 | 1.359 | 0.467 | 3.959 |
| TNM stage (III vs II/I) | <.001 | 5.555 | 2.156 | 14.312 |
| ER status (positive vs negative) | .614 | 1.258 | 0.515 | 3.071 |
| PR status (positive vs negative) | .087 | 0.443 | 0.175 | 1.125 |
| HER2 status (positive vs negative) | .151 | 1.623 | 0.838 | 3.145 |
Data were presented as P value, HR (hazards ratio) and 95% CI (confidence interval). P < .05 was considered significant.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; OS, overall survival; PR, progesterone receptor.
3.6. Circ‐VRK1 relative expression in breast cancer cell lines and its effect on cell proliferation and apoptosis
With the purpose of investigating the underlying mechanism of circ‐VRK1 in breast cancer, in vitro experiments were carried out. Circ‐VRK1 relative expression in breast cancer cell lines and normal breast epithelial cell line was detected by qPCR, which observed that circ‐VRK1 was downregulated in BT474 (P < .05), MDA‐MB‐453 (P < .01) and MDA‐MB‐231 (P < .001) cell lines but similar in MCF7 cells (P > .05) compared to normal breast epithelial cell line MCF10A (Figure 4A). The lowest expression of circ‐VRK1 was observed in MDA‐MB‐231 cells, which were chosen to be used in the following experiments. Circ‐VRK1 expression was similar in the normal group compared with the control (+) group (P > .05) and control (−) group (both P > .050), and meanwhile, circ‐VRK1 was upregulated in the VRK1 (+) group compared with the control (+) group (P < .001) and downregulated in the VRK1 (−) group compared with the control (−) group (P < .001), which indicated the successful transfection (Figure 4B). Cell proliferation ability was reduced in the VRK1 (+) group compared with the control (+) group (P < .01), while increased in the VRK1(−) group compared with the control (+) group (P < .05), and similar in the normal group compared with the control (+) group and control (−) group (both P > .050) (Figure 4C). As to cell apoptosis rate, it was elevated in the VRK1 (+) group compared to the control (+) group (P < .01) but lowered in the VRK1 (−) group compared to the control (−) group (P < .05); however, the apoptosis rates were similar in the normal group compared with the control (+) group and control (−) group (both P > .050) (Figure 4D,E). These data suggested that circ‐VRK1 suppressed cell proliferation but promoted cell apoptosis in breast cancer.
Figure 4.
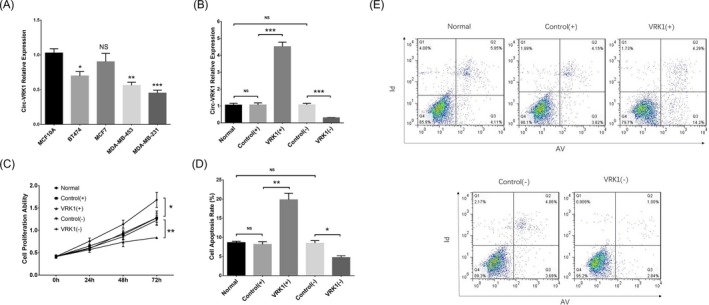
Circ‐VRK1 in cell experiments. Circ‐VRK1 expression was reduced in BT474, MDA‐MB‐453 and MDA‐MB‐231 cell lines but unchanged in MCF7 cell line compared with normal breast epithelial cell line MCF10A (A). Circ‐VRK1 expression was upregulated in the VRK1 (+) group and downregulated in the VRK1 (−) group (B). Cell proliferation was reduced in the VRK (+) group but increased in the VRK (−) group (C). Cell apoptosis rate was increased in the VRK1 (+) group but decreased in the VRK1 (−) group (D, E). Comparison of circ‐VRK1 expression between the normal cell line and each breast cancer cell line was carried out by t test. Comparison of circ‐VRK1 expression, cell proliferation ability and cell apoptosis between the VRK1(+) group and control (+) group as well as the VRK1(−) group and control (−) group was determined by t test. P < .05 was considered significant. *P < .05 and **P < .01. VRK(+), circ‐VRK1 overexpression; control (+), blank overexpression; control (−), blank shRNA; VRK (−), circ‐VRK1 shRNA; circ‐VRK1, circular RNA VRK serine/threonine kinase 1
4. DISCUSSION
In the present study, we found that: (a) circ‐VRK1 expression was downregulated in breast cancer tumour tissue, with its high expression being associated with smaller tumour size, lower T stage and decreased TNM stage; (b) high circ‐VRK1 expression was an independent predictor for better OS in breast cancer patients; (c) in vitro experiments showed that circ‐VRK1 reduced cell proliferation but increased cell apoptosis in breast cancer cells.
Emerging evidences suggest that some circRNAs are dysregulated and are involved in the development and progression in diverse cancers including breast cancer.16, 17, 18 For example, circ‐ZNF609, circ‐MYO9B and circ‐GFRA1 are overexpressed in breast cancer tissues and are associated with larger tumour size, advanced TNM staging, etc in breast cancer patients.8, 19, 20 Previous studies have suggested that some specific circRNAs are dysregulated in tumorous breast tissues and play important roles in the development and progression of breast cancer; however, the majority of circRNAs have not been studied in breast cancer. Circ‐VRK1 has been reported to suppress the expansion and self‐renewal capacity of breast cancer stem cells, but no studies have focused on the clinical implications of circ‐VRK1 expression in breast cancer patients.12 In this study, we compared circ‐VRK1 expression in breast cancer tumour tissues to that in adjacent tissues and found that circ‐VRK1 was downregulated in breast cancer tumour tissues and negatively associated with tumour size, T stage and TNM stage. The first possible explanation for these findings was that circ‐VRK1 might suppress cell proliferation and promote cell apoptosis in breast cancer, thus attenuating the development and progression of breast cancer cells, which might suppress tumour growth and lower disease severity in breast cancer patients. These effects of circ‐VRK1 on cell activities were validated in our subsequent cellular experiments. The second possible explanation was that circ‐VRK1 might be a suppressor for the stemness of breast cancer stem cells, which could reduce drug resistance and metastasis of breast cancer cells; thus, circ‐VRK1 could lead to better treatment response and reduced risk of metastasis, and subsequently contributed to better clinicopathological features in breast cancer patients.12, 21
In addition, a few specific circRNAs are regarded as potential biomarkers in the prognosis of breast cancer by some reports; for example, circ‐ZNF609, circ‐MYO9B and circ‐GFRA1 are shown to be correlated with undesirable outcomes in breast cancer patients and may serve as candidate prognostic biomarkers for breast cancer.8, 20, 22, 23 Regarding circ‐VRK1, although a previous study indicates that circ‐VRK1 reduces drug resistance and influences the prognosis of breast cancer by regulating the stemness of breast cancer stem cells, no study has focused on the effects of circ‐VRK1 on the prognosis in breast cancer patients. In the present study, we investigated the prognostic value of circ‐VRK1 in breast cancer patients, which displayed that high circ‐VRK1 expression was associated with longer OS. The reasons might be that circ‐VRK1 served as an anti‐carcinogenesis gene and was positively associated with disease stage. Thus, patients with higher circ‐VRK1 expression might have less breast cancer severity and better OS in the long term. Additionally, circ‐VRK1 has been reported to suppress the stemness of breast cancer stem cells, which implied that it might attenuate the development of drug resistance or metastasis in breast cancer cells; therefore, patients with circ‐VRK1 high expression would respond more effectively to drug treatments and have a longer survival. However, this explanation needed to be further investigated by study with longer follow‐up to provide more evidence on the prognostic effect of circ‐VRK1. Moreover, the sample size of this study was relatively small, which might influence the statistic power.
Furthermore, a limited number of circRNAs are involved in the biological and molecular mechanisms of breast cancer pathology.24, 25, 26 For example, circ‐Ccnb1 leads to the induction of breast cancer cell death by forming a complex with H2AX and Bclaf1.25 Circ‐Foxo3 is found to suppress tumour growth by upregulating the protein translation of Foxo3.26 In addition, circ_000911, an miR‐449a sponge, inhibits cell proliferation, migration and invasion but promotes cell apoptosis in breast cancer.24 These previous studies suggest that several specific circRNAs are involved in the regulation of molecular processes in different cancers. Regarding circ‐VRK1, a circRNA that was shown to be a tumour suppressor in breast cancer by our clinical analysis, its effects on the cellular activities of breast cancer cells were further explored by in vitro experiments. We discovered that circ‐VRK1 was downregulated in breast cancer cell lines compared with normal breast epithelial cell line, and overexpression of circ‐VRK1 suppressed proliferation but promoted apoptosis in breast cancer cells. These effects might be due to the following: (a) circ‐VRK1 might inactivate the stemness of breast cancer cells, which leads to reduced cell proliferation and increased cell apoptosis,12 and (b) circ‐VRK1 might function as a sponge for several carcinogenic miRNAs (such as miR‐129‐5p and miR‐136‐5p, according to the Tissue‐Specific CircRNA Database [http://gb.whu.edu.cn/TSCD]) and disrupt the protein translation regulated by these miRNAs, which consequently inhibited cell proliferation and promoted cell apoptosis in breast cancer.27, 28 However, this needed to be further verified by functional experiments, which identified specific miRNAs affected by circ‐VRK1 in breast cancer. According to the aforementioned data, we disclosed that circ‐VRK1 represented as an anti‐oncogene by suppressing cell proliferation and promoting cell apoptosis and might serve as a potential therapeutic target for the treatment of breast cancer.
In conclusion, circ‐VRK1 is downregulated in tumour tissues and associated with reduced tumour stage as well as better survival, and it inhibits cell proliferation but promotes cell apoptosis in breast cancer.
ACKNOWLEDGMENTS
None.
Li Y, Li H. Circular RNA VRK1 correlates with favourable prognosis, inhibits cell proliferation but promotes apoptosis in breast cancer. J Clin Lab Anal. 2020;34:e22980 10.1002/jcla.22980
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Clarke M, Collins R, Darby S, et al. Effects of radiotherapy and of differences in the extent of surgery for early breast cancer on local recurrence and 15‐year survival: an overview of the randomised trials. Lancet. 2005;366(9503):2087‐2106. [DOI] [PubMed] [Google Scholar]
- 4. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Z, Xie Q, He D, et al. Circular RNA: new star, new hope in cancer. BMC Cancer. 2018;18(1):834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang JJ, Guo SH, Jia BQ. Down‐regulation of long non‐coding RNA MEG3 serves as an unfavorable risk factor for survival of patients with breast cancer. Eur Rev Med Pharmacol Sci. 2016;20(24):5143‐5147. [PubMed] [Google Scholar]
- 7. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629‐641. [DOI] [PubMed] [Google Scholar]
- 8. Wang N, Gu Y, Li L, et al. Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR‐4316. Arch Biochem Biophys. 2018;653:63‐70. [DOI] [PubMed] [Google Scholar]
- 9. Wang S, Li Q, Wang Y, et al. Upregulation of circ‐UBAP2 predicts poor prognosis and promotes triple‐negative breast cancer progression through the miR‐661/MTA1 pathway. Biochem Biophys Res Commun. 2018;505(4):996‐1002. [DOI] [PubMed] [Google Scholar]
- 10. Dawood S, Austin L, Cristofanilli M. Cancer stem cells: implications for cancer therapy. Oncology (Williston Park). 2014;28(12):1101‐1107, 1110. [PubMed] [Google Scholar]
- 11. Yang F, Xu J, Tang L, Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell Mol Life Sci. 2017;74(6):951‐966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan N, Xu H, Zhang J, et al. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8(56):95704‐95718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greene FL, Balch CM, Fleming ID, et al. AJCC Cancer Staging Handbook: TNM Classification of Malignant Tumors. Berlin: Springer Science & Business Media; 2002. [Google Scholar]
- 14. Tagawa T, Gao S, Koparde VN, et al. Discovery of Kaposi's sarcoma herpesvirus‐encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci U S A. 2018;115(50):12805‐12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods. 2001;25(4):402‐408. [DOI] [PubMed] [Google Scholar]
- 16. Chen Y, Wei S, Wang X, Zhu X, Han S. Progress in research on the role of circular RNAs in lung cancer. World J Surg Oncol. 2018;16(1):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi P, Wan J, Song H, et al. The emerging role of circular RNAs in gastric cancer. Am J Cancer Res. 2018;8(10):1919‐1932. [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Fang L. Advances in circular RNAs and their roles in breast Cancer. J Exp Clin Cancer Res. 2018;37(1):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dimbert B. Epidemiological survey of the oro‐dental health in a school environment in the Belfort area. Rev Odontostomatol (Paris). 1979;8(2):115‐125. [PubMed] [Google Scholar]
- 20. He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR‐34a. J Exp Clin Cancer Res. 2017;36(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bai X, Ni J, Beretov J, Graham P, Li Y Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152‐163. [DOI] [PubMed] [Google Scholar]
- 22. Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR‐145‐5p. Cancer Manag Res. 2018;10:3881‐3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang H, Xiao YI, Wu LI, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA‐000911/miR‐449a pathway in breast carcinogenesis. Int J Oncol. 2018;52(3):743‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fang L, Du WW, Lyu J, et al. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ‐Ccnb1. Cell Death Differ. 2018;25(12):2195‐2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non‐coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35(30):3919‐3931. [DOI] [PubMed] [Google Scholar]
- 27. Li T‐T, Gao X, Gao LI, et al. Role of upregulated miR‐136‐5p in lung adenocarcinoma: a study of 1242 samples utilizing bioinformatics analysis. Pathol Res Pract. 2018;214(5):750‐766. [DOI] [PubMed] [Google Scholar]
- 28. Zhang R‐M, Tang T, Yu H‐M, Yao X‐D. LncRNA DLX6‐AS1/miR‐129‐5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun. 2018;507(1‐4):260‐266. [DOI] [PubMed] [Google Scholar]


