Abstract
Pharmacogenomics (PGx) decision support and return of results is an active area of precision medicine. One challenge of implementing PGx is extracting genomic variants and assigning haplotypes in order to apply prescribing recommendations and information from the Clinical Pharmacogenetics Implementation Consortium (CPIC), the US Food and Drug Administration (FDA), the Pharmacogenomics Knowledgebase (PharmGKB), etc. Pharmacogenomics Clinical Annotation Tool (PharmCAT) (i) extracts variants specified in guidelines from a genetic data set derived from sequencing or genotyping technologies, (ii) infers haplotypes and diplotypes, and (iii) generates a report containing genotype/diplotype‐based annotations and guideline recommendations. We describe PharmCAT and a pilot validation project comparing results for 1000 Genomes Project sequences of Coriell samples with corresponding Genetic Testing Reference Materials Coordination Program (GeT‐RM) sample characterization. PharmCAT was highly concordant with the GeT‐RM data. PharmCAT is available in GitHub to evaluate, test, and report results back to the community. As precision medicine becomes more prevalent, our ability to consistently, accurately, and clearly define and report PGx annotations and prescribing recommendations is critical.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ The scientific pharmacogenomics community is well aware of the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines and the importance of using them to make prescribing recommendations. However, the ability to consistently infer the haplotypes/diplotypes upon which the guidelines are based is a current challenge in the field.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ This study addresses the need to implement an algorithm and software for taking genotyping or sequencing data as a variant call format file input, combined with the CPIC allele definitions and guideline recommendations, and generate a report for each individual's genetic variation.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ This study adds the knowledge that using a reproducible and robust tool, genetic data from a variant call format file, can be consistently annotated with the appropriate CPIC guidelines and prescribing recommendations.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The Pharmacogenomics Clinical Annotation Tool will enable the clinical pharmacology and translational science community to gain confidence and the capability to return pharmacogenomics results to individual participants consistently.
Precision medicine continues to develop at the forefront of health care, and pharmacogenomic implementation programs are one major focus. As previously described, it remains a challenge to accurately assign diplotypes for pharmacogenomics (PGx) genes based on extracted genomic variants from genotype or sequence data files.1 Many important PGx genes encode metabolizing enzymes such as cytochrome P450 (CYP) enzymes and thiopurine methyltransferase. The sequences of these genes are known to have substantial variation in the population, and variant sequences are referred to as “star” alleles, e.g., CYP2D6*2. In some cases, a star allele is defined by a single genetic variant, such as a single‐nucleotide polymorphism (SNP), but in other cases the star allele is a name for a combination of variants across the gene, i.e., a haplotype. In these cases, the phenotype depends on the haplotypes present on both chromosomes, referred to as the diplotype. Without phasing information, haplotype and diplotype assignment can be quite difficult if not impossible in some instances. However, in many instances, educated assumptions can be made to enable prediction. The ability to assign haplotypes and diplotypes, and corresponding annotations and published prescribing recommendations, such as those from Clinical Pharmacogenetics Implementation Consortium (CPIC) or Dutch Pharmacogenetics Working Group (DPWG) guidelines, US Food and Drug Administration (FDA)–approved drug labels, or other sources, is essential to move PGx implementation programs into reality.
One of the greatest challenges in this process is the lack of standardization of star allele (haplotype) definitions across different labs and testing platforms. Currently, the process of PGx haplotype assignment varies from one lab to another.2, 3, 4, 5 These differences are largely driven by the lack of uniformity in which DNA positions are used to determine haplotype assignment and what is assayed by each lab. Many labs assay for “tag” SNPs, which are used to represent a haplotype rather than assaying multiple variant positions. However, some of these SNPs appear in more than one known haplotype. In these cases, the tag SNP alone does not provide enough information to distinguish between multiple (albeit sometimes rare) haplotypes. Assuming that a tag SNP can only be present in one particular haplotype can lead to incorrect haplotype assignment. If the PGx community can better standardize the assignment of haplotypes from sequence and genotype data, then haplotype assignment will not be dependent on the particular lab selected, which would be extremely advantageous for the field.6
Here we discuss the development of PharmCAT—the Pharmacogenomics Clinical Annotation Tool. PharmCAT extracts PGx variants of interest from a genetic data set derived from sequencing or genotyping technologies represented as a variant call format (VCF) file, infers the haplotypes and diplotypes, and generates an interpretation report. The herein described version of PharmCAT includes prescribing recommendations from published CPIC guidelines and extracts variants included in CPIC's Supplemental Materials. Future versions will include other variants, annotations, and guidelines, such as those listed previously.1 The PharmCAT report can be used to inform prescribing decisions based on the given genetic variants. PharmCAT provides researchers and healthcare systems implementing PGx with an approach to more consistently and transparently interpret genomic results and link those results to annotations and prescribing recommendations.1
Two previously published tools (Stargazer and Astrolabe) predict CYP2D6 star alleles from sequence data. Stargazer uses binary alignment map (BAM) files as input, performs variant and copy number variation calling and annotation, and outputs CYP2D6 diplotypes.7 The latest version of Stargazer also interprets CYP2A6/CYP2A7 diplotypes, according to the Stargazer website (https://stargazer.gs.washington.edu/stargazerweb/).8 Astrolabe (formerly known as Constellation) takes BAM and VCF files as input, uses a probabilistic scoring system, and outputs CYP2D6 diplotypes.9 PharmCAT differs from both of these tools as it allows the use of either sequence data or genotypes (such as from a genotyping chip) in the form of a VCF file and outputs a report containing diplotype assignment, annotations, and clinical guideline recommendations. The initial version of PharmCAT interprets 11 genes from published CPIC guidelines (see below for a list of genes), not including CYP2D6. CYP2D6 is known for its highly variable sequence and copy number variation, making it especially difficult to determine diplotypes. Instead of reinventing advanced algorithms for this particular gene, PharmCAT accepts CYP2D6 diplotypes derived independently and provides CYP2D6‐based annotations and recommendations in the output report. The Astrolabe team is part of the PharmCAT consortium and CYP2D6 calls from Astrolabe can be passed to PharmCAT.
In this paper, we describe in detail the PharmCAT software tool. PharmCAT is available in GitHub under the Mozilla Public License (MPL 2.0) for the scientific and clinical community to review, test, and improve. It adheres to the FAIR (findable, accessible, interoperable, and reusable) guiding principles.10
Results
To verify that the Named Allele Matcher component of PharmCAT (see Methods for description) generates accurate genotypes, PharmCAT‐derived genotypes were compared with those provided by the Centers for Disease Control and Prevention (CDC)‐based Genetic Testing Reference Material Coordination Program (GeT‐RM). GeT‐RM characterized 137 genomic samples from Coriell cell lines for 28 genes in a targeted genotype approach by selected laboratories. Sample selection, distribution, laboratories, and assay selection are described in detail by Pratt et al.3 Ten of these genes are included in the initial PharmCAT release: CYP2C9, CYP2C19, CYP3A5, CYP4F2, DPYD, SLCO1B1, TPMT, UGT1A1, VKORC1, and IFNL3 (CYP2D6 is not presented here). Integrated call sets derived from low‐coverage and whole‐exome sequence data with additional imputed genotypes are publicly available for 59 of the 137 GeT‐RM‐characterized Coriell samples as part of the 1000 Genomes Phase 3 data release (see Methods for access to the data). Table 1 summarizes the concordance between the GeT‐RM and PharmCAT results (a per sample comparison by gene is available in Table S1 ). For eight genes, results were considered concordant when the PharmCAT result and the GeT‐RM result matched. The results for CYP2C19, CYP2C9, CYP3A5, IFNL3, and VKORC1 were concordant for all 59 samples, and for TPMT for 58 out of the 59 samples. For SLCO1B1, only five samples had comparable star allele genotypes. Of the 59 publicly available samples in 1000 Genomes, GeT‐RM reported CYP4F2 and UGT1A1 genotypes for only 45 of them. Of those 45, concordance was found for 36 CYP4F2 genotypes.
Table 1.
Summary of concordance between GeT‐RM and PharmCAT results
| Gene | Number of samplesa | Number of concordant samplesb | % Concordance |
|---|---|---|---|
| CYP2C19 c | 59 | 59 | 100 |
| CYP2C9 | 59 | 59 | 100 |
| CYP3A5 | 59 | 59 | 100 |
| CYP4F2 | 45 | 36 | 80.0 |
| DPYD d | 59 | 56 | 94.9 |
| IFNL3 e | 59 | 59 | 100 |
| SLCO1B1 d | 59 | 5 | 8.5 |
| TPMT | 59 | 58 | 98.3 |
| UGT1A1 d | 45 | 24 | 53.3 |
| VKORC1 | 59 | 59 | 100 |
CYP, cytochrome P450; GeT‐RM, Genetic Testing Reference Materials Coordination Program.
aNumber of GeT‐RM characterized Coriell samples with available 1000 Genome VCF files (1000 Genomes Phase 3 data release). bNumber of samples with agreement between GeT‐RM consensus genotypes and PharmCAT‐derived genotypes using 1000 Genomes VCF files. c CYP2C19*1/*4 is considered concordant to CYP2C19*1/*4A based on presence of rs28399504; c.1A>G. dDetails of how concordance was calculated are described in the Supplementary Material . eGeT‐RM IFNL3 genotypes retrieved from the non consensus table (only one testing company provided IFNL3 genotypes).
For UGT1A1, concordance was determined slightly differently. CPIC uses seven commonly tested alleles from the UDP‐Glucoronosyltransferase (UGT) Alleles Nomenclature website (http://prime.vetmed.wsu.edu/resources/udp‐glucuronsyltransferase‐homepage).11 These star allele definitions are based on a single variant each; however, it is known that combinations of UGT1A1 variants on one chromosome are possible. PharmCAT reports UGT1A1 results as a string of all identified variants per allele in phased data (the 1000 Genomes samples in our study were phased). The GeT‐RM results do not include UGT1A1*80, which is in high linkage disequilibrium (though not 100%) with UGT1A1*28. Therefore, *80 was disregarded in the PharmCAT results for the comparison with the GeT‐RM results (see supplementary for more information). For example, the GeT‐RM result of *60/(*28 + *60) for NA11832 was considered concordant with the PharmCAT result of *60/*28 + *60 + *80. For UGT1A1, results were concordant for 24 samples.
Concordance for DPYD was also determined in a different manner. While a star allele nomenclature exists containing some variants for this gene, CPIC recommendations for fluoropyrimidine dosing are instead based on combinations of individual DPYD variants.12 The PharmCAT DPYD definition table contains the variants used in the CPIC recommendations. However, GeT‐RM reported DPYD star alleles. To allow for comparison, the star allele results from GeT‐RM were mapped to the variants included in the CPIC guideline by using the DPYD allele functionality table found on the PharmGKB website (https://www.pharmgkb.org/page/dpydRefMaterials).13 Further details are described in the supplement . The results were concordant for 56 samples.
Discussion
The PharmCAT performance was evaluated using 59 sequences from 1000 Genomes for Coriell samples which were characterized through the GeT‐RM project.
Discordant results
The discordance between PharmCAT results and the GeT‐RM genotypes can be divided into a few categories.
First, variants that exist in known haplotypes were detected in novel combinations in the sample VCF files. PharmCAT detects only variants already existing in the allele definitions (not novel variation), and the combination of variants found is compared with the existing definitions to assign a haplotype. However, combinations of known variants were detected that are not currently found in the existing definitions from the CYP4F2 and SLCO1B1 nomenclatures. PharmCAT relies on the allele definitions included in the CPIC guidelines derived from these sources. For example, manual inspection of the VCF file for one Coriell sample showed the sample to be homozygous for the variant found in CYP4F2*3, and heterozygous for the variant found in CYP4F2*2, meaning that both the *3 variant AND the *2 variant were present on the same chromosome. This sample was reported as CYP4F2*3/*3 by GeT‐RM. There is no match for this sample because there is no existing star allele definition in which both the *3 and *2 variants co‐exist on the same chromosome (https://www.pharmvar.org/gene/CYP4F2).14 Therefore, in this case, the genotype is reported by PharmCAT as ‘not called.’ Seven samples had discordant results for CYP4F2 due to this issue (see Supplementary Materials ). This outcome could be improved, expanding the allele definitions contained in the nomenclature resources.
In the same vein, PharmCAT was unable to match SLCO1B1 star alleles for 54 of the 59 samples. A comparison of the variants in the sample VCF files to the allele definitions showed that some variants do not exclusively exist in the defined star allele combinations. Unlike CYP4F2, the star allele definitions for SLCO1B1 were extracted from several articles.15 For example, one variant is in both the SLCO1B1*18 and *19 definitions but it is also present in combination with other variants. While this common variant was present in many of the samples, the other variants included in the *18 and *19 definitions were not, so PharmCAT could not match the samples to the *18 or *19 definitions. Yet the samples were clearly not *1, so PharmCAT could not give a diplotype result. As described in the Methods section and Supplementary Materials , PharmCAT reports the rs4149056 genotype if star alleles for SLCO1B1 cannot be determined. This SNP is associated with decreased function and increased risk for myopathy with statin use regardless of whether it is found alone or in combination with other variants, and CPIC recommendations exist for this variant genotype. A comparison of the phenotype for the GeT‐RM diplotypes and the phenotype for the PharmCAT rs4149056 genotypes resulted in phenotype concordance for 48 more samples. This means that the GeT‐RM results and the PharmCAT results translate to the same SLCO1B1 allele function, phenotype and CPIC prescribing recommendation for simvastatin for these samples.
The panels used for the GeT‐RM targeted genotyping do not include as many star alleles or variants as are included in PharmCAT with the allele definitions. PharmCAT identified decreased function variants in DPYD in three samples and the TPMT*16 allele in one sample, none of which were interrogated variants in GeT‐RM. However, in one case, a sample was discordant because PharmCAT reported UGT1A1*1/*1 while GeT‐RM reported UGT1A1*1/(*7) because *7 is not considered in the CPIC guideline and therefore not in the PharmCAT allele definition table. With increasing knowledge in the PGx field, the number of variants included in genotyping applications may increase, possibly reducing the gap between array‐based testing and sequencing data. Until then, results from targeted genotyping vs. sequencing may differ depending on the gene.
In some cases, GeT‐RM reported a variant that we could not find in the 1000 Genomes VCF files. Two samples for CYP4F2 and eight samples for UGT1A1 were discordant due to this issue. Further details are described in the Supplementary Materials .
An additional 12 UGT1A1 results were discordant because 1000 Genome VCF files with phased data were run on PharmCAT. Eleven samples were reported by GeT‐RM as UGT1A1*60/*28 and by PharmCAT as UGT1A1*1/*28 + *60 + *80, probably because GeT‐RM did not have phased data while PharmCAT did. Even though the genotype results differed, both the GeT‐RM and PharmCAT genotypes result in the same phenotype and recommendation for these samples. For one sample, GeT‐RM reported UGT1A1*27/(*28 + *60) and PharmCAT reported UGT1A1*1/*27 + *28 + *60 + *80, which did result in a different metabolizer phenotype.
A table summarizing the reasons for discordance by gene is included in the Supplementary Materials .
Coverage
The available samples from GeT‐RM contained all the possible variants or star alleles included in the PharmCAT allele definitions for CYP4F2, IFNL3, and VKORC1. Table S2 summarizes the star alleles and variants for CYP2C19, CYP2C9, CYP3A5, TPMT, and UGT1A1 that were validated with the samples. Table S2 further highlights the star alleles and variants that were not present in the Coriell samples tested with PharmCAT. DPYD is not included in Table S2 because of the reporting difference between PharmCAT, which uses variants based on the CPIC guidelines, and the GeT‐RM, which uses star alleles. Though in silico examples of alleles and variants in the PharmCAT allele definitions have been tested, testing on real DNA samples is needed for further validation.
CFTR variants covered in the CPIC guideline are included with PharmCAT but were not part of this validation set. GeT‐RM provides reference material for CFTR independent from the 137 genomic samples but the sequences are not part of the available 1000 Genome data set for testing with PharmCAT.
PharmCAT could be validated for SLCO1B1 rs4149056 detection. However, it should be noted that preference is given by PharmCAT to report star alleles if determinable, based on the CPIC guidelines.
Seeking community input and testing
With this initial beta release of PharmCAT, the pharmacogenomics community is asked to support the continuing evaluation of this freely available tool by running VCF samples, documenting issues and successfully identifying variants. GitHub can be used to communicate feedback (https://github.com/PharmGKB/PharmCAT/issues).16 Prior to release, we validated PharmCAT with in silico test VCF files for variant positions. Only a limited number of combinations have been tested on “real world” samples given the availability of publicly accessible samples with orthogonal genotypes and the lack of sample diversity. Issues with allele definitions or variant representation in the VCF files are possible. Of special interest are VCF representations of multi‐base insertion/deletions, and variants with overlapping definitions. For example, the CFTR allele definition table includes three deletions in close proximity. While our constructed in silico test VCF files allowed the call of these variants, they might not reflect how these variants are represented in other VCF files.
Limitations and future directions
As with any initial version of a tool, PharmCAT has limitations. PharmCAT assumes the sample VCF file has already undergone extensive quality control. PharmCAT produces an output report containing diplotypes and recommendations based on the genetic data provided. If the genetic data that are entered into PharmCAT are of low quality, inaccurate diplotypes may result, potentially leading to inaccurate recommendations. Also, PharmCAT uses allele definitions based on GRCh38 requiring users to provide samples in the same build. However, tools exist to transfer GRCh37 to GRCh38.
PharmCAT only considers variants contained in the allele definition files; novel variants and variants not in existing allele definitions are not considered. The initial release of PharmCAT contains genes and variants found in CPIC guidelines and provides CPIC recommendations in the output. We intend to expand the annotations in the report output to include other annotations such as DPWG guidelines, FDA‐approved drug labels and PharmGKB information. We also intend to expand the genes included in PharmCAT as applicable to add information from these other sources and new CPIC guidelines. In the initial release, PharmCAT processes one VCF file and generates one output report at a time. In a future release, we intend to accommodate batch file processing.
Methods
Tool description
PharmCAT takes input data (A), processes it (B), and provides a report output (C). Each of these components is described below.
(A) Input data. PharmCAT requires several types of input, including a user‐provided VCF file and multiple files packaged with the tool.
User‐provided normalized VCF file. PharmCAT processes a single VCF file. Key VCF requirements are alignment to the GRCh38 and verification that the variant representation format matches that in the allele definition files (see below). The VCF file requirements, including variant standardization, are described in detail in the documentation found in GitHub on the PharmCAT “VCF Requirements” webpage (https://github.com/PharmGKB/PharmCAT/wiki/VCF‐Requirements).17 The user is responsible for providing an appropriate VCF file based on the requirements. VCF preprocessing documentation is available in GitHub on the PharmCAT “Preparing VCF Files” webpage (https://github.com/PharmGKB/PharmCAT/wiki/Preparing‐VCF‐Files).18 It is important to note that QUAL and FILTER columns are not read or interpreted by PharmCAT. The user is responsible for determining quality criteria in their own data set and removing data not meeting that criteria before submitting to PharmCAT.
User‐provided CYP2D6 diplotype (optional). PharmCAT accepts Astrolabe output containing a CYP2D6 diplotype as an input file and includes relevant CYP2D6 clinical guideline recommendations in the report output. The Astrolabe software, distributed by Children's Mercy Hospital (https://www.childrensmercy.org/Health_Care_Professionals/Research/Pediatric_Genomic_Medicine/Software_Tools/),19 must be acquired separately and run independently from PharmCAT.
Allele definitions. Allele definitions are provided in standardized tables by gene and can be accessed on the PharmCAT website (http://www.pharmcat.org).20 The first release of PharmCAT includes allele definitions for the following genes included in CPIC guidelines: CYP2C19, CYP2C9, CYP3A5, CYP4F2, CFTR, DPYD, IFNL3, SLCO1B1, TPMT, UGT1A1, VKORC1. The allele definitions are based on the CPIC/PharmGKB tables found on the PharmGKB website (https://www.pharmgkb.org/page/pgxGeneRef)21 with some alterations (for more information, see the PharmCAT website). Further description about the allele definitions are included in the Supplementary Materials .
Curated guideline recommendations and additional information. PharmGKB routinely curates CPIC guidelines. As part of the curation process, allele‐based and diplotype‐based information provided by the guidelines is entered into the PharmGKB database. Gene diplotypes are combined into phenotype groups based on star allele function. Phenotype groups are annotated with guideline‐specific information such as implications for specific drugs, prescribing recommendations and strength of recommendation (all from the CPIC guidelines and CPIC's Supplementary Materials ). These annotations are retrieved from the PharmGKB database, packaged with each PharmCAT version, and integrated into the output report (see section B2).
(B) Processes. The PharmCAT tool consists of two main processes (Figure 1 ). The Named Allele Matcher matches variant information from the user‐supplied VCF file to the gene allele definitions to generate a diplotype call for each gene. The Reporter takes the output of the Matcher and generates a report containing drug prescribing recommendations.
Figure 1.
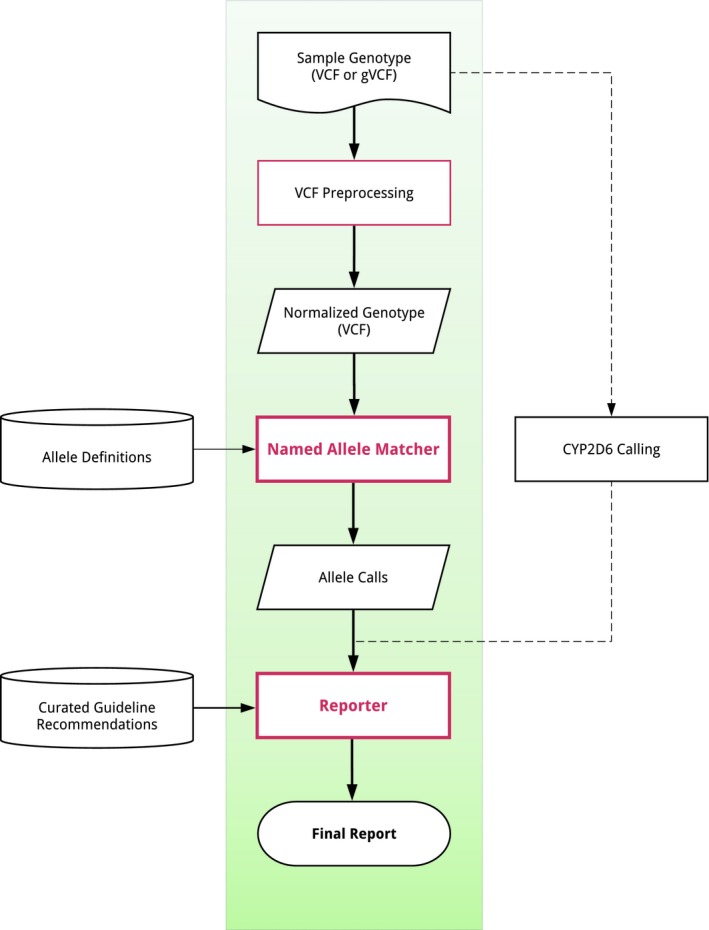
Overview of the PharmCAT tool. Sample VCF file is provided by the user. Allele definitions and recommendations (extracted from PGx guidelines) are combined with additional curated information such as notes, caveats, and warnings. The NamedAlleleMatcher and Reporter are core components of PharmCAT. CYP2D6 calls are done externally and passed to the Reporter for the Final Report output. CYP, cytochrome P450. VCF, variant call format. [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
-
1
Named Allele Matcher. The basic Named Allele Matcher process for unphased data is summarized by the following steps:
Star allele definitions from (A3) above are read into the matcher. By default, any nonreference star allele that does not contain a base call for a given position (i.e., blank spots in the definition table) will default to the reference row's base call.
The sample data in VCF file are read into the matcher. Any calls reported in the VCF at positions that are not used in the allele definition tables are ignored by the matcher.
Per gene, all possible genotype combinations for the variants in the VCF file are generated, representing potential diplotypes. Both alleles in each potential diplotype are compared with the allele definitions provided. If matches to defined alleles are obtained, the potential diplotypes are considered valid. Valid diplotypes are scored and only the top‐scoring one is returned by the Named Allele Matcher.
Further information about the variant matching and the scoring algorithm is available in the PharmCAT GitHub documentation (https://github.com/PharmGKB/PharmCAT/wiki/NamedAlleleMatcher‐101)22 (https://github.com/PharmGKB/PharmCAT/wiki/NamedAlleleMatcher‐201).23
-
2
Reporter. The Reporter collects and combines different data sources and creates an HTML document with sections for recommendations, gene information, and disclaimers/warnings. The input into the Reporter are the output from the Named Allele Matcher, curated guideline recommendations (see section A4) and curated information provided by PharmCAT consisting primarily of caveats, disclaimers, and genomic positions of interest. The CYP2D6 diplotype from Astrolabe is optional input.
The Reporter has a basic algorithm for getting output from the Named Allele Matcher and storing it in an internal representation, which can then be displayed in multiple places on the final report. For genes without star allele representation, genotypes are used instead of diplotypes. As the Reporter and output report contain both genotypes and diplotypes, the term “genotype” is used generically in to refer to both. In addition to genotypes, the reporter records which positions from the allele definition files had calls in the VCF file. Also, calls from the aforementioned genomic positions of interest in the curated information input are recorded (e.g., rs4149056 in SLCO1B1).
The Reporter has a general algorithm for most genes that reports the output of the Named Allele Matcher. However, some genes require a “custom” algorithm for determining genotype based on logic specific to that gene. Basically, the algorithm for processing Named Allele Matcher output can be overridden when necessary. In the initial version of PharmCAT, this is done for the SLCO1B1 and UGT1A1 genes. Due to reasons listed in the Discussion section and Supplementary Materials , SLCO1B1 and UGT1A1 genotypes are frequently impossible to assign based on the allele definition files. Therefore, rule‐based systems were developed for both genes in the Reporter tool. For SLCO1B1, if the Named Allele Matcher is not able to find a diplotype match based on the allele definition file, the rs4149056 genotype is used if present in the VCF file since the CPIC guideline for simvastatin includes recommendations for the rs4149056 genotype alone, as well as the star alleles. The rule‐based system for UGT1A1 is more complicated due to the reasons found in the Results section and is described in detail in the Supplementary Materials .
Once genotypes are finalized for all genes, the Reporter compares them to the curated clinical guideline recommendations discussed in section A4. For each gene, the Reporter determines what phenotype group contains the given genotype, and then marks it as a match for display in the final report. Some guidelines provide information based on multiple genes, so the Reporter constructs the matching genotype string in a particular way to compare against genotype strings specified in the guideline annotation groups. Each matched group has multiple annotations that are displayed to the end user like “phenotype,” “allele functionality,” and “recommendations.”
The Reporter uses a data model called Message Annotation to integrate additional information for further explanatory or cautionary text in particular places in the final report. These messages are predefined (see the PharmCAT website for more information) and triggered by certain conditions like a particular genotype result or if a particular sequence location is not specified in the VCF file. Each Message Annotation has associated logic to determine whether to display it in the final report and if so, where. The final report is an HTML document by default (described in the next section), but PharmCAT can be configured to output the supporting data model as a JSON document.
(C) Output report. The output of PharmCAT is an HTML‐formatted report. This format was chosen because of the complexity of the output, which requires extensive formatting and layout in order to be human‐readable and because of the vast number of existing tools for authoring and viewing HTML. Excerpts from the report for the 1000 Genomes VCF file for Coriell sample NA12717 is shown in Figure 2 (an example full report is provided in Supplementary Materials ). The PharmCAT report is divided in to four sections, (i) Genotype Summary, (ii) CPIC Recommendation, (iii) Allele Call Details, and (iv) Disclaimers, which are described in the Supplementary Materials .
Figure 2.
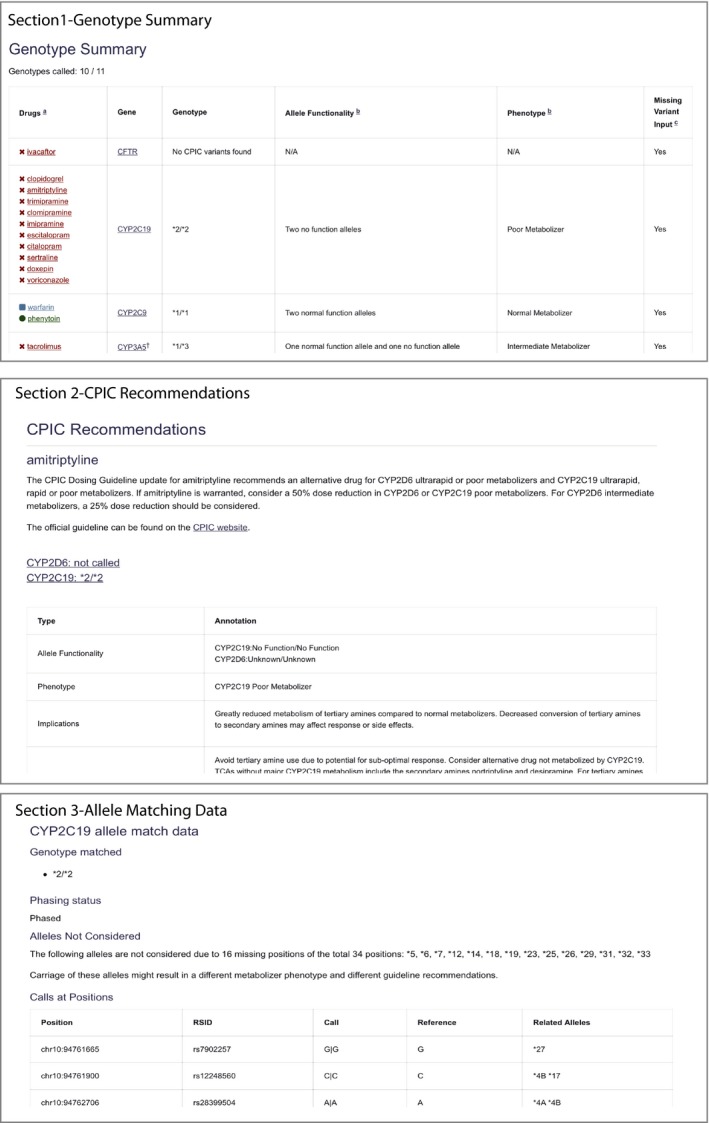
PharmCAT report sections for the 1000 Genomes VCF file for Coriell sample NA12717. The PharmCAT report consists of four parts: (i) genotype summary table, (ii) Clinical Pharmacogenetics Implementation Consortium (CPIC) recommendations section by drug in alphabetical order, (iii) gene information about the interrogated variants, and (iv) disclaimer. In the summary table, the drugs are colored to indicate whether CPIC recommends a prescribing change based on the given genotype. The last column in the table indicates star alleles that could not be considered for the genotype assignment due to missing variant information in the VCF file. It is important to note missing information because it could result in changes to the phenotype and/or CPIC recommendation. PharmCAT, pharmacogenomics clinical annotation tool. [Colour figure can be viewed at https://www.wileyonlinelibrary.com]
Validation sample description
We compared PharmCAT output with the GeT‐RM results for CYP2C9, CYP2C19, CYP3A5, CYP4F2, DPYD, SLCO1B1, TPMT, UGT1A1, VKORC1, and IFNL3. Details about the commercial platforms/assays and sample selection for the GeT‐RM data are described in Pratt et al. 3
For 59 of the 137 GeT‐RM characterized Coriell samples, integrated call sets derived from low‐coverage and whole‐exome sequence data with additional imputed genotypes are publicly available as part of the 1000 Genomes Phase 3 data release. The GRCh38 liftover VCF files from the 05‐02‐2013 release were downloaded from the 1000 Genomes FTP server (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/GRCh38_positions/).24 These publicly available genomic data were reformatted according to the VCF requirements listed above (A1). To ensure reproducibility, we have made stable copies of the VCFs used for this analysis, which are available for download (https://purl.stanford.edu/rd572fp2219).25 These files were run with PharmCAT v0.5.1 (https://github.com/PharmGKB/PharmCAT/releases/tag/v0.5.1).26 We note that, although GeT‐RM provides reference material for CFTR independent from the 137 genomic samples, we were not able to access VCF files as part of the 1000 Genome data set and therefore were unable to use these samples for PharmCAT comparison.
Funding
This work is supported by the NIH/NIGMS grant GM61374 (TEK, RBA), NIH grant U01 HL065962 (MDR), and National Science Foundation Graduate Research Fellowship (AL).
Conflict of Interest
M.D.R. is on the scientific advisory board for Cipherome. R.B.A. is a stockholder in Personalis Inc., 23andMe, and Youscript. The other authors declared no competing interests for this work.
Author Contributions
K.S., M.W‐C., M.D.R., and T.E.K. wrote the manuscript; K.S., M.W‐C., R.M.W., M.W., M.D.R., and T.E.K. designed the research; K.S., M.W‐C., R.M.W., M.W., A.L., and A.V. performed the research; K.S., M.W‐C., R.B.A., and L.C. analyzed the data.
Supporting information
Table S1. The GeT‐RM and PharmCAT results per sample, by gene.
Table S2. A summary of the variants/alleles from the allele definitions that are validated with the GeT‐RM samples and those that require validation.
Supplementary Materials
Supplement Example Report.pdf
References
- 1. Klein, T.E. & Ritchie, M.D. PharmCAT: a pharmacogenomics clinical annotation tool. Clin. Pharmacol. Ther. 104, 19–22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kalman, L.V. et al Pharmacogenetic allele nomenclature: international workgroup recommendations for test result reporting. Clin. Pharmacol. Ther. 99, 172–185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pratt, V.M. et al Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: a GeT‐RM collaborative project. J. Mol. Diagn. 18, 109–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Relling, M.V. & Evans, W.E. Pharmacogenomics in the clinic. Nature 526, 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Driest, S.L. et al Association of arrhythmia‐related genetic variants with phenotypes documented in electronic medical records. JAMA 315, 47–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bush, W.S. et al Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin. Pharmacol. Ther. 100, 160–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee, S.B. et al Stargazer: a software tool for calling star alleles from next‐generation sequencing data using CYP2D6 as a model. Genet. Med., 21, 361–372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stargazer . <https://stargazer.gs.washington.edu/stargazerweb/>. Accessed August 5, 2019.
- 9. Twist, G.P. et al Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole‐genome sequences. NPJ Genom. Med. 1, 15007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilkinson, M.D. et al The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. UGT Nomenclature Committee . <https://prime.vetmed.wsu.edu/resources/udp-glucuronsyltransferase-homepage>. Accessed August 5, 2019.
- 12. Amstutz, U. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin. Pharmacol. Ther. 103, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gene-specific information tables for DPYD . <https://www.pharmgkb.org/page/dpydRefMaterials>. Accessed August 5, 2019.
- 14.PharmVar CYP4F2. <https://www.pharmvar.org/gene/CYP4F2>. Accessed August 5, 2019.
- 15. Wilke, R.A. et al The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin‐induced myopathy. Clin. Pharmacol. Ther. 92, 112–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. PharmCAT issues . <https://github.com/PharmGKB/PharmCAT/issues>. Accessed August 5, 2019.
- 17. VCF requirements . <https://github.com/PharmGKB/PharmCAT/wiki/VCF-Requirements>. Accessed August 5, 2019.
- 18. Preparing VCF files . <https://github.com/PharmGKB/PharmCAT/wiki/Preparing-VCF-Files>. Accessed August 5, 2019.
- 19. Astrolabe . <https://www.childrensmercy.org/Health_Care_Professionals/Research/Pediatric_Genomic_Medicine/Software_Tools/>. Accessed August 26, 2019.
- 20. Pharmacogenomics Clinical Annotation Tool . <http://www.pharmcat.org>. Accessed August 5, 2019.
- 21. PGx Gene-specific information tables . <https://www.pharmgkb.org/page/pgxGeneRef>. Accessed August 5, 2019.
- 22. NamedAlleleMatcher 101 . <https://github.com/PharmGKB/PharmCAT/wiki/NamedAlleleMatcher-101>. Accessed August 5, 2019.
- 23. NamedAlleleMatcher 201 . <https://github.com/PharmGKB/PharmCAT/wiki/NamedAlleleMatcher-201>. Accessed August 5, 2019.
- 24. Index of /vol1/ftp/releast/20130502/supporting/GRCh38_positions/ . <ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/GRCh38_positions/> Accessed August 5, 2019.
- 25. Pre-validated GeT-RM Coriell Samples for use with PharmCAT and the code used to generate them . <https://purl.stanford.edu/rd572fp2219>. Accessed August 5, 2019.
- 26. v0.5.1 . <https://github.com/PharmGKB/PharmCAT/releases/tag/v0.5.1>. Accessed August 5, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The GeT‐RM and PharmCAT results per sample, by gene.
Table S2. A summary of the variants/alleles from the allele definitions that are validated with the GeT‐RM samples and those that require validation.
Supplementary Materials
Supplement Example Report.pdf


