Abstract
Genetic epidemiology requires an appropriate approach to measure genetic variation within the population. The aim of this study was to evaluate the characteristics and genotyping results of DNA extracted from 2 human DNA sources, selected for their rapid and noninvasive sampling, and the use of simple and standardized protocols that are essential for large-scale epidemiologic studies. Saliva and urine samples were collected at the same day from 20 subjects aged 9–10 yr. Genomic DNA was extracted using commercial kits. Quantitative and qualitative evaluation was done by assessing the yield, the purity, and integrity of the extracted DNA. As a proof-of-concept, genotyping was performed targeting CC16 A38G and uteroglobin-related protein 1 (UGRP1)-112G/A. Saliva was found to provide the highest yield and concentration of total DNA extracted. Salivary DNA showed higher purity and a significantly less degraded state compared to urinary DNA. Consequently, the salivary DNA gave better genotyping results than urinary DNA. Therefore, if the choice exists, saliva is the preferred noninvasive matrix for genotyping purposes in large-scale genetic epidemiologic studies. Only in particular cases using urine could nevertheless be considered useful, although specific limitations need to be taken into account.
Keywords: biomarkers, genotype, real-time PCR, genetic epidemiology, children
INTRODUCTION
Genetic epidemiology is the study of the role of genes and their interaction with environmental factors in the occurrence of disease in human populations.1 One of the genetic factors playing an important role in genetic epidemiology are single nucleotide polymorphisms (SNPs).2, 3 These simple and stable changes occur at an average frequency of ∼1 in 1.3 and 1.9 kb overall.4–6 Most SNPs have no effect on health or development, but some of them may be implicated in disease incidence and susceptibility. These can be observed between individuals in a population and could influence promoter activity (gene expression), mRNA conformation (stability), and translational efficiency. Therefore, they may play a role in the susceptibility of an individual to many common diseases, medicinal drug metabolism, and genome evolution. They could potentially also play a direct role with or without other factors in the phenotypic expression of diseases or traits.7–10 SNPs can be the marker of choice for many applications of genetic epidemiology because of the high genotyping efficiency, data quality, genome-wide coverage, and analytical simplicity.11
The measurement of the genetic variations, i.e., genotyping, is a crucial factor to take into consideration for the development of public health genomics, where genome-based knowledge is used to benefit public health, by implementing it into public health policy and services for the benefit of the population health. For instance, in the United Kingdom, public health genomics led to an increased personalized treatment, e.g., in cancer, ameliorating the public health care. Moreover, genomic knowledge will enable subpopulations at risk to be defined, i.e., precision public health. Subsequently, by focusing intensive lifestyle interventions on and motivating lifestyle modifications in those with increased genomic risks (because of specific SNPs), the cost-effectiveness of health care might be improved.12, 13 Also in Belgium, such an approach is envisioned in the future.14
To perform genotyping assays in epidemiologic studies, which are often done on a healthy, nonhospitalized population, a straightforward and noninvasive workflow is needed. However, traditionally, blood or buffy coats remain the commonly used sources of human nucleic acids for genotyping, implying several limitations such as the need of a professional training in phlebotomy, equipment, and infrastructure, thereby hampering sampling in a more epidemiologic setting. Furthermore, blood sampling is not easily accepted as it can cause anxiety and/or physical discomfort, particularly in vulnerable populations. Contrarily, collecting urine and saliva is an extremely noninvasive process. Urine is already a highly desirable biospecimen for large population-based studies because it can be collected easily and recurrently by noninvasive techniques, in relatively large volumes. Furthermore, it contains about 400 epithelial cells (i.e., squamous, renal tubular, and transitional urothelial) and macrophages per milliliter of human urine, which makes urine-derived DNA-based detection possible.15–24 Another very desirable body fluid for diagnosis and monitoring is saliva because it is easily accessible noninvasively; its collection is relatively inexpensive, safe, and easy; and can be performed without the help of health care workers.25 It is composed of ∼75% epithelial cells (∼430,000 cells/ml26) and ∼25% leukocytes (2–136,000 cells/ml27, 28) and is a good source of high-quality DNA for the use of genomic applications.14, 29–31 Therefore, urine and saliva could be the ideal sources for genotyping assays in epidemiologic studies, especially for the use in children or people that will not comply with blood collections. It has been shown that simple, self-administered sample collection methods, which are possible when collecting urine and saliva, significantly increase the participation rates in epidemiologic studies.32
The goal of this study was to evaluate the noninvasive biofluids urine and saliva by extracting genomic DNA (gDNA) of children with the aim to measure genetic variations (genotyping) using a real-time PCR assay, which could be applied in future large-scale epidemiologic studies. To deliver a proof-of-concept for genotyping using the extracted DNA from urine and saliva, we measured 2 SNPs that have already been reported to be implicated in susceptibility for respiratory diseases. Firstly, we investigated the SNP rs3741240, a polymorphism located in the CC16 gene, coding for club cell protein, an anti-inflammatory protein secreted by the club cells in the deep lung33 and associated with decreased lung function and with increased susceptibility to asthma and other lung diseases.34–36 Secondly, we investigated the SNP rs1368408, located in the promoter of the secretoglobin family 3A member 2 gene [also called uteroglobin-related protein 1 (UGRP1) gene], which plays an important role in secreting lung surfactant protein and for which it has been shown to result in susceptibility for asthma.37–39 Based on the obtained results, we could recommend which source of DNA to use for which type of epidemiologic studies. Although each of the biofluids has been already previously used for genotyping22, 40 this is the first time this type of comparison of biofluids from children for genotyping has been made.
MATERIALS AND METHODS
Population, urine, and saliva samples
The urine and saliva samples were collected from 20 children (9–10 yr old, 8 girls, 12 boys), recruited in the framework of a feasibility study concerning the search for biomarkers linked to respiratory diseases and air pollution (ethical committee approval registration number B403201734310).
The urine and saliva samples were collected anonymously on March 9th, 2018, in a school in the neighborhood of Leuven, Belgium, between 9:45 AM and 10:25 AM. Although the first urine is more concentrated in cells, this is not always practical in epidemiologic studies. Thus, the subsequent, presumably “second-morning” urine was collected using a 125-ml sample vessel (VWR, Leuven, Belgium), immediately divided in aliquots, and stored at −80°C. No preservative was added to avoid potential impact on downstream analyses. The saliva was collected using the Oragene DNA tubes (DNA Genotek, Kanata, ON, Canada). This allowed the saliva to be stored at room temperature for a prolonged time (for up to 5 yr, according to the manufacturer), maintaining the integrity of the DNA as it contains reagents to preserve the high MW DNA by inhibiting degradation and preventing bacterial growth.41
Ethics
The Ethics Committee of the Faculty of Medicine of the Catholic University of Louvain approved the study protocol, which complied with all applicable requirements of international regulations (registration number B403201734310). All human subjects participating in the study have given the requisite informed consent.
DNA extraction from the urine and saliva samples
Total urinary DNA was extracted from the 20 samples 4 mo after their collection. The urine aliquots were thawed overnight at 4°C and extracted the following day using the Gentra Puregene Blood kit (Qiagen, Hilden, Germany), following the protocol for body fluids.42 The protocol was adapted for extraction out of 3 ml of urine, which is the maximum volume specified in the protocol provided by the manufacturer and which is also the maximum of volume possible to be easily extracted for many samples simultaneously. During the purification procedure, cells were lysed with an anionic detergent in the presence of a DNA stabilizer, which limits the activity of DNases found intracellularly and elsewhere in the environment and which was removed again during precipitation. RNA was removed by treatment with an RNA digesting enzyme. Other contaminants and enzyme inhibitors such as proteins and divalent cations were removed by salt precipitation. The purified DNA was dissolved in 50 µl of water and stored at −20°C.
DNA from 2 ml saliva of each of the 20 children was extracted 4 mo after its collection using the prepIT-L2P protocol for manual purification of DNA from whole sample (DNA Genotek). This is an alcohol precipitation–based method including a heat-treatment and removal of impurities and inhibitors. The protocol that was performed, did not contain an RNA removal step. DNA was recovered by precipitation with alcohol, dissolved in 200 µl Tris-EDTA buffer, and stored at −20°C.
Yield and purity of total DNA extracted
The DNA concentration (ng/μl), the yield (ng/ml biofluid), and the purity (A260/A280) were first determined by spectrophotometry using the Nanodrop ND-2000 (Thermo Fisher Scientific, Waltham, MA, USA).
The quantity and concentration of the extracted DNA was also investigated using a Qubit dsDNA BR and HS Assay Kit with the Qubit 4.0 fluorometer (Thermo Fischer Scientific).
DNA integrity of the extracted DNA
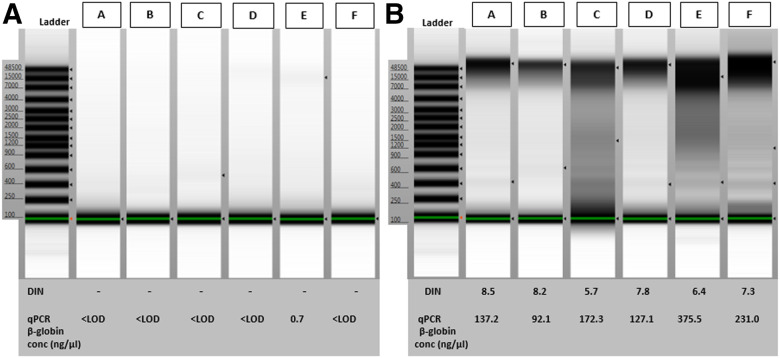
The DNA integrity of the gDNA was analyzed using the Agilent 4200 TapeStation Automated Electrophoresis System (Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer’s instruction. One microliter of the DNA sample was measured using a gDNA screen tape (Agilent Technologies), which has a sizing range of 200–>60,000 bp and a sizing precision of 200–15,000 bp. The results were analyzed with the TapeStation Analysis Software, A.02.02, including the generation of a DNA Integrity Number (DIN), providing an indication of integrity (distribution to a scale of DIN 1–10; a high DIN indicates highly intact gDNA, and a low DIN a strongly degraded gDNA sample). A selection of 6 higher and lower concentrated samples (i.e., A, B, C, D, E, F), based on the Nanodrop measurements, was analyzed as representatives for possible best- and worst-case scenarios.
To estimate the amount of nondegraded DNA, the yield of intact amplifiable human DNA was determined by analyzing 10 ng of total DNA (based on Nanodrop results) of each urine and saliva sample by using an SYBR Green real-time qPCR assay, which amplifies a 164-bp fragment of the β-globin gene (Accession Number EF450778), as described previously.23
Genotyping assays: CC16 SNP rs3741240 and UGRP1 SNP rs1368408
As a proof-of-concept, the extracted DNA was used in a genotyping assay targeting 2 SNPs. Fifty nanograms of total DNA (based on the measurements with the Nanodrop) extracted from the urine and saliva samples was used for 2 allelic discrimination assays performed on the StepOnePlusReal-Time PCR system (Thermo Fisher Scientific). Two commercially available kits Taqman SNP Genotyping assays (rs3741240 ID_C__25473445_10 and rs1368408 ID_C__7515585_20; Thermo Fisher Scientific) were used following the manufacturer’s instructions to analyze the SNP CC16 A38G and the SNP UGRP1-112G/A, respectively. The mutated allele corresponds with the VIC dye (allele1) and the wild-type dye corresponds to the fluorescein amidites (FAM) dye (allele 2). A negative template control was included for each assay. A positive control was not added. The assay was performed in duplicate. If one of the 2 measurements resulted in “undetermined,” the result of that sample was recorded as “undetermined.” The “undetermined” samples were genotyped again by increasing the input DNA to 200 ng (based on the Nanodrop measurements).
In order to validate the genotyping results, Sanger sequencing was performed on the PCR fragments of CC16 and URGP1 of salivary DNA samples (primers: CC16 forward: 5′-TAATACGACTCACTATAGGGTTACCTATCCCACCAAGCCAATGC-3′, reverse: 5′-GGAAACAGCTATGACCATGTGGGCAGCTCACTCCTTCTTCTG-3′; UGRP1: forward: 5′-TAATACGACTCACTATAGGGAGAAGGATTCGTTGGGCTCTTTGC-3′; reverse: 5′-GGAAACAGCTATGACCATGAGCCAGTGGTTCCACTTGCCATAC-3′). The sequences were determined using the ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA, Life Technologies, Carlsbad, CA, USA) and analyzed with the Sequence Scanner V.1.0 software (Applied Biosystems, Life Technologies).
Because no information was available on the fragment size of the amplified real-time PCR fragments for the commercial assays, a capillary electrophoresis was performed on the obtained real-time PCR fragments. This was done using an Agilent 4200 TapeStation Automated Electrophoresis System (Agilent Technologies) with a D1000 screen tape for a sizing of 35–1000 bp, according to the manufacturer’s instruction. The results were analyzed with the TapeStation Analysis Software, A.02.02. The investigation was done on 1 µl of each real-time PCR product (CC16 SNP A38G, UGRP1 SNP-112G/A to be compared with β-globin qPCR product) of 2 samples (sample A and D, representing each gender, representative of all samples because this concerns an allelic discrimination assay).
Statistical analysis
Results were presented as median with interquartile range or mean with sd for the continuous variables (concentration, yield, and purity). The comparison of yield and concentration between both biofluids and between Nanodrop (including the purity) and Qubit was done using the nonparametric Wilcoxon signed rank-sum test. The comparison between boys and girls was done using the nonparametric Wilcoxon rank-sum test. All P values were 2-sided with a level of statistical significance set at <0.05. Statistical analyses were made with the JMPs 14.1 software from SAS Institute (Cary, NC, USA).
RESULTS
Total yield
The median DNA concentration (ng/µl DNA) and median total DNA yield (ng/ml biofluid) are summarized per gender in Table 1. Both with UV spectroscopy (Nanodrop) as with fluorescence spectroscopy (Qubit), it was found that under the conditions used, saliva contains the highest concentration and yield of total DNA. Clearly, for both genders, the concentrations measured by fluorescence spectroscopy are significantly lower than those measured by UV spectroscopy, with a more pronounced difference in the urinary DNA values compared to salivary DNA. We observed no significant differences in yield between gender, except for the low urinary yields, measured with Qubit.
TABLE 1.
DNA concentration, yield and purity of urinary and salivary DNA
| Population |
Urine |
Saliva |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Nanodrop |
Qubit |
Nanodrop |
Qubit |
|||||||
| DNA conc (ng/µl) | Total yield (ng DNA/ml biofluid) | A260/A280 | DNA conc (ng/µl) | Total yield (ng/ml biofluid) | DNA conc (ng/µl) | Total yield (ng DNA/ml biofluid) | A260/A280 | DNA conc (ng/µl) | Total yield (ng/ml biofluid) | |
| Girls (n = 8) | 37.6 [34.3–40.5] | 626 [571–675] | 1.57 ± 0.22a | 0.43 [0.26–0.58]b | 7.20 [4.33–9.66]b | 290 [284–502] | 22,100 [18,800–41,700] | 1.87 ± 0.04a | 111 [85–160] | 9690 [6700–12,500] |
| Boys (n = 12) | 34.5 [30.4–38.2] | 575 [506–637] | 1.62 ± 0.08c | 0.11 [0.10–0.13]b | 1.83 [1.60–2.16]b | 376 [264–502] | 33,500 [25,900–38,300] | 1.85 ± 0.05c | 123 [110–176] | 12,100 [8700–15,900] |
Measurements were performed with Nanodrop and Qubit. Values are represented as median (interquartile range) for concentration and yield and mean ± sd for the A260/A280 ratio.
Significant differences (P < 0.001) were observed for the mean ratio of A260/A280 between urine and saliva for girls when using the Wilcoxon signed rank-sum test.
When using the Wilcoxon rank-sum test, significant differences (P < 0.001) of urinary yield and concentration were observed between boys and girls, when measured by Qubit.
Significant differences (P < 0.001) were also observed for the mean ratio of A260/A280 between urine and saliva for boys when using the Wilcoxon signed rank-sum test.
DNA purity
The purity was assessed based on the ratio A260/A280 as measured by UV spectrometry, which is summarized for urine and saliva in Table 1, including a summary per gender. The results indicate that the DNA extracted from urine has a lower mean ratio of A260/A280 compared to the ratio of saliva (P < 0.001). This could indicate that the urinary DNA has a lower purity than the salivary DNA. However, a low A260:A280 ratio may also be caused by the very low concentration (<10 ng/μl) of nucleic acid in the urine samples.43 No significant differences are observed between genders.
DNA integrity
The DNA integrity of the samples was evaluated as described in the Materials and Methods sections. In Fig. 1, the TapeStation results on a selection of 6 higher and lower concentrated samples (i.e., A, B, C, D, E, F), show that the extracted urinary gDNA was too dilute and too degraded to give any interpretable results. In comparison, the gDNA extracted from saliva contained a high proportion of long DNA fragments for most samples, with a band above the 48,500 bp ladder band and an average DIN of 7.3 (±sd 1.10), with sample C being more degraded, although no impact on genotyping results was observed (see “Genotyping Assays”). A higher volume (15 µl) of the urinary DNA samples was also loaded on a classic agarose gel. However, due to the low total concentrations and the degraded state of the urinary DNA, no distinct bands were visible (unpublished results).
FIGURE 1.
Integrity of DNA extracted from urine and saliva. TapeStation results and concentrations (ng/μl) obtained from β-globin qPCR of DNA extracted form urine (A) and saliva (B) on a selection of 6 samples (boys: A, B, C, E; girls: D, F). The corresponding yields (nanograms per microliter biofluid) are for urine (A): sample A, B, C, D, F: undetermined; sample E: 11.1 ng/ml; and for saliva (B): sample A: 8495 ng/ml, sample B: 6244 ng/ml, sample C: 12,577 ng/ml, sample D: 10,008 ng/ml, sample E: 33,527 ng/ml, sample F; 17,977 ng/ml.
A real-time qPCR assay using the human-specific β-globin primers was used to determine more specifically the amount of human-intact amplifiable DNA in the total DNA extracted from urine and saliva. The yield of human-intact amplifiable DNA extracted from urine was close to the detection limit (0.13 ng/µl corresponding to 10 copies/well) and could only be determined for 3 out of the 20 samples (i.e., 1.7–10–11.1 ng of human-intact amplifiable DNA per milliliter of urine). This was in contrast with the human-intact amplifiable DNA extracted from saliva, ranging from 6244 to 84,408 ng/ml saliva. No difference between genders was observed. As an example, the individual qPCR results obtained for the 6 individuals A–F are summarized in Fig. 1. These were in line with the observed Tapestation results.
Genotyping assays
To evaluate if this experimental setup could be applied for future genetic epidemiologic studies on a larger scale, the total DNA extracted from urine and saliva was genotyped for rs3741240 and rs1368408. For this, the amount of input DNA was determined based on the Nanodrop concentration values because these will be in practice the least time- and cost-consuming measurements to be done in large-scale studies. Table 2 summarizes results from the genotyping assays based on the allelic discrimination plots that discriminate between the homozygote and the heterozygote genotypes as well as the undetermined or nonamplified samples. When using 50 ng of input DNA (based on Nanodrop measurement), a higher genotype call rate (GCR) is observed for salivary samples (100%) compared to urinary DNA samples (GCR 60%). The GCR was also dependent on which assay that was used, i.e., better results for CC16 than for UGRP1 were obtained (75 and 60% success rate with 50 ng of input DNA, respectively). The increase of input of urinary DNA of the undetermined samples from 50 to 200 ng (based on the Nanodrop measurements) resulted in a successful genotyping of 4 out of 5 undetermined samples for the SNP CC16 and successful genotyping of 6 out of 8 undetermined samples for the UGRP1 SNP (unpublished results). This improved the GCR, using urinary DNA from 60 to 90% for the UGRP1 SNP assay and from 75 to 95% for the CC16 SNP assay. Finally, the Sanger sequencing results tested with the salivary DNA support and confirm the outcomes generated for each genotyping assay (see Supplemental Table S1 in supplemental data). The allele frequencies (based on the results of salivary DNA) are 45% wild type and 55% heterozygous for CC16 SNP A38G and 70% wild type and 30% heterozygous for UGRP1 SNP-112G/A with no homozygous mutants genotyped in either of the assays.
TABLE 2.
Genotyping results for the CC16 SNP A38G and the UGRP1 SNP-112G/A
| Characteristic |
CC16 SNP A38G |
UGRP1 SNP-112G/A |
||
|---|---|---|---|---|
| Saliva | Urine | Saliva | Urine | |
| Total number of samples | 20 | 20 | 20 | 20 |
| Homozygous wild type (%) | 9 (45%) | 8 | 14 (70%) | 9 |
| Heterozygous (%) | 11 (55%) | 7 | 6 (30%) | 3 |
| Homozygous mutant (%) | 0 (0) | 0 | 0 (0) | 0 |
| Determined (%) | ||||
| 50 ng DNA input | 20 (100%)a | 15 (75%) | 20 (100%)a | 12 (60%) |
| 200 ng DNA input (b) | 19 (95%) | 18 (90%) | ||
Both genotyping assays were performed twice with the DNA extracted from saliva (n = 20) and urine (n = 20) 0.50 ng of DNA input (based on Nanodrop) was used.
All salivary samples yielded a 100% GCR, including for the samples A, B, C, D, E, F, randomly selected for the DNA integrity verificatio, [with C not impacted by the lower DIN value (see Fig. 1)]. This is in contrast with the selection of the 6 urinary DNA samples for which samples C and F resulted in undetermined CC16 SNP genotype and sample F in undetermined UGRP1 SNP genotype.
The failed urinary samples were repeated with 200 ng DNA input. As all saliva samples could be successfully genotyped, the percentage of each obtained genotype based on the total amount of samples tested, was calculated based on the saliva values (indicated between brackets).
Because no information was given on the amplicon size, we determined the fragment size of the qPCR products of sample A and D using capillary electrophoresis (Tapestation) (unpublished results). The amplicons from the genotyping assays gave a band around 80 and 140 bp for the CC16 and the UGRP1 genotyping assay, respectively. The β-globin qPCR showed a band around 160 bp, as expected.23
DISCUSSION
Urine and saliva were investigated as possible DNA sources for genotyping assays suited for larger-scale epidemiologic studies. Salivary and urinary sampling present major advantages compared to the blood-dependent methods. They were selected in this study to investigate the effectiveness and efficiency of DNA genotyping assays. Both biofluids are noninvasive, easily collected, processed, and assayed, particularly in vulnerable strata of the population, such as children. Both sources contain DNA and could therefore potentially be used for downstream genotyping assays. However, some differences at several levels exist, which should be taken into account depending on the type of study to be conducted.
Under the conditions tested and taking into account the chosen storage conditions and processing methods, the extraction of total DNA from saliva resulted in significantly higher yields, higher concentrations, better quality, and purer and less degraded DNA compared to the total DNA extracted from urine. The yield was quantified using 2 distinct quantification methods, i.e., UV and fluorescence spectroscopy. The latter provides the amount of intact double stranded DNA (dsDNA) with minimal interference of proteins, RNA, single stranded DNA (ssDNA), or other contaminants that can influence the UV absorbance, with a DNA yield decreasing with increasing levels of fragmentation and denaturation of DNA. The former measures maximal absorbance of nucleic acids without distinguishing between dsDNA, ssDNA, RNA, and nucleotides.44 Thus, as expected, our results showed that the UV method gave significantly higher concentrations, both for urine and saliva-derived DNA. The presence of contamination in the sample, i.e., protein (indicated by the A260:A280 ratio, especially in case of urinary DNA) as well as RNA, ssDNA, and other contaminants could influence the UV absorption ultimately leading to an overestimation of the DNA concentration compared to fluorescence spectroscopy. Indeed, the protocol for urinary DNA extraction contains an optional RNA removal step, which was not performed during the extraction of salivary DNA, and which could lead to an overestimation of the concentration of the total salivary DNA, measured with Nanodrop. Additionally, the presence of RNA in the samples also has an impact on the difference in purity between the DNA extracted from both biofluids, which could lead to an overestimation of the purity (A260:A280) of the salivary DNA. It should also be noted that DNA quantification of urinary DNA by Qubit in our results could potentially be underestimated. Optimal Qubit quantification of frozen DNA samples requires a sufficient amount of salt in the DNA solution. In this case, the urinary DNA was dissolved in water. This could potentially lead to a destabilization of dsDNA, hindering the binding of the fluorescent dye and thus to an underestimation of the DNA concentration.45
Other factors could also play a role in the measured yield and quality of the recovered urinary DNA. First, the timing of urine collection has been described to have an impact on the yield. It has been shown that the first urine results in the highest yields because it contains more cells,46 although it should be noted that urine taken at other moments also gives acceptable yield and could be used in routine.23 Due to practical reasons, a presumably second-morning urine sample was collected in this field study, with lower expected urine yield, but still enough to perform the majority of the genotyping assays. Additionally, the second-morning urine could, at a technical level, be a better choice in this case because the first morning urine contains more degraded DNA.23 Secondly, the gender could also impact the obtained DNA concentration and yield. It is known that female urine contains more cells and therefore a higher amount of DNA than male urine. However, this was mainly previously described in adults with a fully developed urogenital tract.19, 23 In our results, using urine samples collected from 9- to 10-yr-old children, this was only confirmed for the very low yield of urinary DNA measured with the Qubit. Thirdly, urine is generally not considered as an ideal source of DNA due to the low concentration of nucleated cells present. As mentioned previously, saliva contains a significantly higher number of cells compared to urine. This explains the substantial difference in yield obtained between the salivary and the urinary DNA. However, the obtained concentrations should be compared with caution because different starting volumes, elution volumes, and buffers were used for both extraction protocols of saliva and urine. Nevertheless, with a larger starting biofluid volume and a lower elution volume, it was expected that urinary DNA would be more concentrated. In our case, this was not observed, confirming once more that the initial DNA present in urine is much lower than in saliva. Fourthly, the yield interpretation is influenced by the amount of degraded DNA present. The highly degraded state of the urinary DNA in our study is a result of the presence of urea, uric acid, creatinine, mineral salt, urokinase, and bacteria.47 This results in a quantification of urinary, fragmented DNA that is underestimated by fluorescence spectroscopy because this method underestimates with 70% the concentration of dsDNA with a size <23 kb.48 This is in contrast with the Nanodrop, which measures total DNA, including fragmented DNA. Furthermore, it is known that urinary and salivary DNA contain coextracted microbial nucleotides present in these biofluids, which could lead to an overestimation of the actual human DNA concentration measured by both of these nonspecific spectrophotometric or fluorometric methods.23, 26, 40, 49 The use of a real-time qPCR assay using the human-specific β-globin primers, overcomes these overestimations by determining more specifically the amount of human-intact amplifiable DNA in the total DNA extracted from urine or saliva. Our results showed that the DNA extracted from saliva contains a significant proportion of human intact DNA fragments of at least 164 bp long in contrast with the urinary DNA for which this human proportion could not be determined because it was below the detection limit for most of the samples. This result was rather unexpected because there should be a certain amount of human DNA present in the urine, confirmed by the successful genotyping assays, targeting human genes. Perhaps the presence of contaminants, suggested by the lower A260/A280 values, although this could also be due to the low DNA concentration,43 could play a role in obtaining good results in the qPCR quantification assay for urinary DNA. Removing them would possibly improve the outcome of the quantification assay but has not been done here because this would include an additional time-consuming step, which is not ideal in a large-scale epidemiologic study. Moreover, the ultimate goal is the genotyping of the samples, for which a straightforward concentration measurement to determine input DNA (such as based on Nanodrop measurements) is preferred to be used. Additionally, there is a significant difference in size of the amplicon between the different assays, namely around 80 and 20 bp between, respectively, the CC16 SNP genotyping assay and the β-globin qPCR and between UGRP1 SNP genotyping assay and the β-globin qPCR. A possible explanation for the less successful quantification qPCR assay could be that the fragment to be amplified in the β-globin qPCR is too large for the degraded human DNA present in urine, resulting in an unsuccessful outcome of the β-globin qPCR and potentially in an underestimation of the human DNA present in urinary DNA. This would imply that qPCR assays, including human DNA concentration measurements and genotyping assays, should be carefully selected when working with degraded urine-derived DNA. To further elucidate this, a more thorough investigation should be done by using different primers to target different fragment sizes of 1 gene.
In our study, always a 100% GCR was obtained for salivary DNA, which was not the case for urinary DNA. This makes salivary DNA definitely the preferred biofluid, if the choice exists. When starting with the same input DNA, based on the Nanodrop measurements, we could observe a lower GCR for urine, which is due to the lower DNA quality and the difficulty to estimate concentration of the extracted DNA, compared with salivary DNA. Furthermore, there is a difference in GCR between assays. We could observe better results when using the CC16 SNP genotyping assay, compared to the UGRP1 assay. As mentioned above, 1 possibility might be the difference in amplicon size. The impact of the size of the qPCR product and the efficiency of the assay has been previously observed, i.e., when developing a qPCR method, a difference in size of a qPCR product can impact the amplification efficiency.50 However, when increasing the input DNA up to “200 ng” (based on Nanodrop measurements, and hence not reflecting the “true” quantity for reasons explained above), most of the undetermined samples were successfully genotyped for the CC16 SNP as well as for the UGRP1 SNP, reducing the difference of GCR between the assays. Based on these results, we could suggest using a higher amount of starting material (based on Nanodrop measurements) when using urinary DNA compared to salivary DNA. Increasing the input of DNA, based on Nanodrop, to at least “200 ng” solves a part of the undetermined cases. However, it also requires significantly more material of DNA, limiting the number of assays that could be done with the DNA extracted from urine in 1 extraction run. This is in contrast with salivary DNA, where a lower DNA input of “50 ng” (based on Nanodrop measurements) is sufficient to generate successful data. The genotyping results from the salivary DNA indicate a same trend as the expected allele frequencies described by the 1000 genome Project (45% wild type, 44% heterozygous, 11% homozygous mutants for CC16 SNP A38G, and 78% wild type and 21% heterozygous, 0.014% homozygous mutant for UGRP1 SNP-112G/A).51 The slight deviation can be due to the differences in heterogeneity of ethnicity of the population compared to the 1000 Genome Project and to the relatively small sample size of our study.
The higher yielding saliva has the advantage that only 1 extraction run will allow the performance of a multitude of genotyping assays. This is not the case for urine because the yielded concentration and quantity are relatively low, limiting the number of genotyping assays to be done out of 1 extraction. Although urine is available in large volumes, the extraction of urine is limited to a maximum volume to be extracted at once, depending on the extraction kit used. This has as an additional consequence that the remaining volume of the collected urine should be stored properly if no aliquots were made immediately after sampling. Storing the urine samples at room temperature or 4°C induces bacterial growth and quick degradation of human DNA over time, impeding the potential utility of urine.47 Freezing the urine improves the recovery to some extent, although the choice of which temperature, the storage period, and the related storage costs are important parameters to consider.52 In contrast, although using a more costly sampling device, with the appropriate collection conditions for saliva in this study, the extracted salivary DNA is very stable, even at room temperature, yielding perfect results for the 2 different genotyping assays. This has been confirmed by previous studies, which show that the Oragene saliva sample is of such high quality that it can be used as an alternative to blood DNA in epidemiologic studies.14, 40, 41, 53 The storage of saliva at room temperature with this preservative, whereas urine was stored around 4 mo period without a preservative (though at −80°C) until DNA extraction, might have favored a better outcome for salivary DNA. To circumvent some of the drawbacks of urine, immediate DNA extraction or alternative or additional handling, not interfering with genotyping, could be considered.16, 52, 54, 55 Also, the choice of which commercial extraction kit to use, i.e., its specific efficiency, could have an impact on the quality or yield of the extracted DNA from urine.23 Nevertheless, immediate extraction is not realistic in large epidemiologic studies and one of the main requirements for these types of studies is a quick and easy sampling, thus preferably limiting the number of additional steps.
Based on the above, we can conclude that, if the choice exists, saliva is obviously the preferred matrix for genotyping purposes in large-scale epidemiologic studies, where the noninvasive character, the rapidity of the sampling, and the use of simple and standardized protocols are essential. Despite the logistical convenience of urine, the results are not satisfactory, with a lower DNA quality and quantity and variable GCR depending on the selected target. Furthermore, the cost-effectiveness could be challenged due to increasing costs related to higher storage costs and possible repeated sample processing linked to limited accuracy. However, using urine might be eventually considered when investigating retrospective studies of banked urine where no other source is available and when there is no possibility to recontact the participants to collect saliva. However, when using urine for genotyping, the limitations specified in this study should be taken into account. In summary, although urine has other properties of interest, saliva is the best-suited noninvasive source of high-quality DNA for molecular assays in genetic epidemiologic studies.
Supplementary Material
This article includes supplemental data. Please visit http://jbt.abrf.org/ to obtain this information.
ACKNOWLEDGMENTS
The authors thank the colleagues from Transversal Activities in Applied Genomics (TAG) at Sciensano for their help in the preparation of the field study. They thank the Sequencing platform of TAG for running the sequencing reactions. Finally, they thank the participating school and the CLB for their enthusiastic collaboration during the field study. This research is funded by the Belgian Federal Science Policy Office (BELSPO, BR/165/PI/ PMOLLUGENIX-V2) through the Project PMOLLUGENIX-V2 and by Sciensano PJ/RP through research project Respikid. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data nor in writing the manuscript. The authors declare no conflicts of interest.
REFERENCES
- 1.Last JM, Spasoff RA, Harris SS, Thuriaux MC, eds. A Dictionary of Epidemiology, 4th ed. New York, NY: Oxford University Press, 2001. [Google Scholar]
- 2.Collins A, Lonjou C, Morton NE. Genetic epidemiology of single-nucleotide polymorphisms. Proc Natl Acad Sci USA. 1999;96:15173–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schork NJ, Fallin D, Lanchbury JS. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet. 2000;58:250–264. [DOI] [PubMed] [Google Scholar]
- 4.Sachidanandam R, Weissman D, Schmidt SC, et al.International SNP Map Working Group . A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. [DOI] [PubMed] [Google Scholar]
- 5.Cargill M, Altshuler D, Ireland J, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238; erratum: 23, 373. [DOI] [PubMed] [Google Scholar]
- 6.Halushka MK, Fan JB, Bentley K, et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. [DOI] [PubMed] [Google Scholar]
- 7.Martin N, Boomsma D, Machin G. A twin-pronged attack on complex traits. Nat Genet. 1997;17:387–392. [DOI] [PubMed] [Google Scholar]
- 8.LeVan TD, Bloom JW, Bailey TJ, et al. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol. 2001;167:5838–5844. [DOI] [PubMed] [Google Scholar]
- 9.Shastry BS. SNPs: impact on gene function and phenotype. In Komar AA. (ed): Methods in Molecular Biology, 2nd ed Cleveland, OH: Humana Press, c/o Springer Science + Business Media, 2009:3–22. [DOI] [PubMed] [Google Scholar]
- 10.Chanock S. Candidate genes and single nucleotide polymorphisms (SNPs) in the study of human disease. Dis Markers. 2001;17:89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin PA, Luikart G, Wayne RK. SNPs in ecology, evolution and conservation. Trends Ecol Evol. 2004;19:208–216. [Google Scholar]
- 12.The Human Genomics Strategy Group Building on our inheritance: genomic technology in healthcare. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/213705/dh_132382.pdf. Published January 2012. Accessed September 15, 2019.
- 13.Burton H, Jackson C, Abubakar I. The impact of genomics on public health practice. Br Med Bull. 2014;112:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van den Eynden J, Descamps T, Delporte E, et al. The genetic structure of the Belgian population. Hum Genomics. 2018;12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannas A, Kalunga G, Green C, et al.TB trDNA consortium . Implications of storing urinary DNA from different populations for molecular analyses. PLoS One. 2009;4:e6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Zhao S, Zhu R, Zhang G, Li C. UTI preventing DNA degradation of storing urinary samples for genotyping. Forensic Sci International Genet Suppl Ser. 2011;3:e3–e4. [Google Scholar]
- 17.Aoki K, Tanaka H, Ueki M. DNA typing for personal identification of urine after long-term preservation for testing in doping control. Drug Test Anal. 2017;9:1116–1123. [DOI] [PubMed] [Google Scholar]
- 18.Chiarella P, Carbonari D, Capone P, et al. Susceptibility biomarker detection in urine exfoliate DNA. Biomark Med. 2017;11:957–966. [DOI] [PubMed] [Google Scholar]
- 19.Vu NT, Chaturvedi AK, Canfield DV. Genotyping for DQA1 and PM loci in urine using PCR-based amplification: effects of sample volume, storage temperature, preservatives, and aging on DNA extraction and typing. Forensic Sci Int. 1999;102:23–34. [DOI] [PubMed] [Google Scholar]
- 20.Botezatu I, Serdyuk O, Potapova G, et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem. 2000;46:1078–1084. [PubMed] [Google Scholar]
- 21.Mundt LA, Shanahan K. Graff’s Textbook of Routine Urinalysis and Body Fluids. Philadelphia, PA: Wolters Kluwer / Lippincott Williams & Wilkins, 2011. [Google Scholar]
- 22.Haufroid V, Clippe A, Knoops B, Bernard A, Lison D. Genotyping in urine: an interesting tool for epidemiological studies. Clin Chem. 1998;44:2210–2211. [PubMed] [Google Scholar]
- 23.El Bali L, Diman A, Bernard A, Roosens NHC, De Keersmaecker SCJ. Comparative study of seven commercial kits for human DNA extraction from urine samples suitable for DNA biomarker-based public health studies. J Biomol Tech. 2014;25:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan Z, Zhou Z, Chen H, et al. PCR-ready human DNA extraction from urine samples using magnetic nanoparticles. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;881–882:63–68. [DOI] [PubMed] [Google Scholar]
- 25.Tabak LA. A revolution in biomedical assessment: the development of salivary diagnostics. J Dent Educ. 2001;65:1335–1339. [PubMed] [Google Scholar]
- 26.Dawes C. Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch Oral Biol. 2003;48:329–336. [DOI] [PubMed] [Google Scholar]
- 27.Thiede C, Prange-Krex G, Freiberg-Richter J, Bornhäuser M, Ehninger G. Buccal swabs but not mouthwash samples can be used to obtain pretransplant DNA fingerprints from recipients of allogeneic bone marrow transplants. Bone Marrow Transplant. 2000;25:575–577. [DOI] [PubMed] [Google Scholar]
- 28.Endler G, Greinix H, Winkler K, Mitterbauer G, Mannhalter C. Genetic fingerprinting in mouthwashes of patients after allogeneic bone marrow transplantation. Bone Marrow Transplant. 1999;24:95–98. [DOI] [PubMed] [Google Scholar]
- 29.Vriens A, Nawrot TS, Saenen ND, et al. Recent exposure to ultrafine particles in school children alters miR-222 expression in the extracellular fraction of saliva. Environ Health. 2016;15:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wren ME, Shirtcliff EA, Drury SS. Not all biofluids are created equal: chewing over salivary diagnostics and the epigenome. Clin Ther. 2015;37:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koni AC, Scott RA, Wang G, et al.IDEFICS Consortium . DNA yield and quality of saliva samples and suitability for large-scale epidemiological studies in children. Int J Obes. 2011; 35(Suppl 1):S113–S118. [DOI] [PubMed] [Google Scholar]
- 32.Hansen TVO, Simonsen MK, Nielsen FC, Hundrup YA. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol Biomarkers Prev. 2007;16:2072–2076. [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee AB, Zhang Z, Chilton BS. Uteroglobin: a steroid-inducible immunomodulatory protein that founded the Secretoglobin superfamily. Endocr Rev. 2007;28:707–725. [DOI] [PubMed] [Google Scholar]
- 34.Hong K, Kim H-K, Kang M-J, et al. CC10 A38G Polymorphism (rs3741240) is associated with asthma susceptibility and bronchial hyperresponsiveness mediated by the eosinophilic inflammation in Korean children. J Allergy Clin Immunol. 2013;131:AB205. [Google Scholar]
- 35.Zhao G, Lin X, Zhou M, Zhao J. Association between CC10 +38A/G polymorphism and asthma risk: a meta-analysis. Pak J Med Sci. 2013;29:1439–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Candelaria PV, Backer V, Laing IA, et al. Association between asthma-related phenotypes and the CC16 A38G polymorphism in an unselected population of young adult Danes. Immunogenetics. 2005;57:25–32. [DOI] [PubMed] [Google Scholar]
- 37.Niimi T, Munakata M, Keck-Waggoner CL, et al. A polymorphism in the human UGRP1 gene promoter that regulates transcription is associated with an increased risk of asthma. Am J Hum Genet. 2002;70:718–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie H, Wu M, Shen B, Niu Y, Huo Y, Cheng Y. Association between the -112G/A polymorphism of uteroglobulin-related protein 1 gene and asthma risk: a meta-analysis. Exp Ther Med. 2014;7:721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SK, Seok H, Park HJ, et al. Association between secretoglobin family 3A member 2 (SCGB3A2) gene polymorphisms and asthma in a Korean population. Med Sci Monit. 2017;23:1880–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abraham JE, Maranian MJ, Spiteri I, et al. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics. 2012;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunes AP, Oliveira IO, Santos BR, et al. Quality of DNA extracted from saliva samples collected with the OrageneTM DNA self-collection kit. BMC Med Res Methodol. 2012;12:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiagen Sample and Assay Technologies. In: Gentra Puregene Handbook, 3rd ed Valencia, CA, USA: Qiagen, 2009:32–33. [Google Scholar]
- 43.Thermo Fisher Scientific Assessment of Nucleic Acid Purity. Technical Bulletin, NanoDrop Spectrophotometers. Wilmington, DE, USA: Thermo Fisher Scientific, 2015:1–2. [Google Scholar]
- 44.O’Neill M, McPartlin J, Arthure K, Riedel S, McMillan ND. Comparison of the TLDA with the Nanodrop and the reference Qubit system. J Phys Conf Ser. 2011;307:012047. [Google Scholar]
- 45.Nakayama Y, Yamaguchi H, Einaga N, Esumi M. Pitfalls of DNA quantification using DNA-binding fluorescent dyes and suggested solutions. PLoS One. 2016;11:e0150528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kavaler E, Landman J, Chang Y, Droller MJ, Liu BC. Detecting human bladder carcinoma cells in voided urine samples by assaying for the presence of telomerase activity. Cancer. 1998;82:708–714. [DOI] [PubMed] [Google Scholar]
- 47.Khan G, Kangro HO, Coates PJ, Heath RB. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J Clin Pathol. 1991;44:360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Georgiou CD, Papapostolou I. Assay for the quantification of intact/fragmented genomic DNA. Anal Biochem. 2006;358:247–256. [DOI] [PubMed] [Google Scholar]
- 49.Garbieri TF, Brozoski DT, Dionísio TJ, Santos CF, Neves LT. Human DNA extraction from whole saliva that was fresh or stored for 3, 6 or 12 months using five different protocols. J Appl Oral Sci. 2017;25:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbau-Piednoir E, Lievens A, Mbongolo-Mbella G, et al. SYBR Green qPCR screening methods for the presence of “35S promoter” and “NOS terminator” elements in food and feed products. Eur Food Res Technol. 2010;230:383–393. [Google Scholar]
- 51.Abecasis GR, Altshuler D, Auton A, et al. ; The 1000 Genomes Project Consortium . A map of human genome variation from population scale sequencing. Nature. 2011;467:1061–1073; erratum: 473, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hilhorst M, Theunissen R, van Rie H, van Paassen P, Tervaert JWC. DNA extraction from long-term stored urine. BMC Nephrol. 2013;14:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rylander-Rudqvist T, Håkansson N, Tybring G, Wolk A. Quality and quantity of saliva DNA obtained from the self-administrated oragene method--a pilot study on the cohort of Swedish men. Cancer Epidemiol Biomarkers Prev. 2006;15:1742–1745. [DOI] [PubMed] [Google Scholar]
- 54.Yokota M, Tatsumi N, Tsuda I, Takubo T, Hiyoshi M. DNA extraction from human urinary sediment. J Clin Lab Anal. 1998;12:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nozawa N, Koyano S, Yamamoto Y, Inami Y, Kurane I, Inoue N. Real-time PCR assay using specimens on filter disks as a template for detection of cytomegalovirus in urine. J Clin Microbiol. 2007;45:1305–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.