Abstract
Background
Small nuclear ribonucleoproteins (snRNPs) complexes of protein and noncoding RNA accumulate in the cell nucleus and catalyze pre-mRNA splicing to form the spliceosome. This study aimed to investigate the role of the spliceosome, splicing factor 3b subunit 1 (SF3B1), in AGS and MKN28 human gastric cancer cells in vitro, including gene knockdown with small interfering RNA (siRNA), and the use of the selective mRNA splicing inhibitor of SF3B1, pladienolide B.
Material/Methods
In AGS and MKN28 human gastric cancer cells, SF3B1expression was inhibited with siRNA and pladienolide B. Following SF3B1 inhibition, the Cell Counting Kit-8 (CCK-8) assay measured cell proliferation, and flow cytometry was used to investigate cell apoptosis and cell cycle arrest. The downstream HOXA10 and AKT pathways were studied by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot. The presence of alternative splicing, or differential splicing, of single-gene coding for multiple proteins, was analyzed using The Cancer Genome Atlas (TCGA) SpliceSeq.
Results
Inhibition of SF3B1 reduced the proliferation rate of AGS and MKN28 human gastric cancer cells by inducing apoptosis and G2/M phase arrest. SF3B1 knockdown resulted in reduced homeobox A10 (HOXA10) mRNA expression and expression of long noncoding RNA (lncRNA) isoforms of HOXA10 (exons 1 and 3) and HOXA10 (exons 2 and 3). SF3B1 inhibition increased PTEN levels and reduced AKT protein phosphorylation.
Conclusions
In AGS and MKN28 human gastric cancer cells in vitro, inhibition of SF3B1 reduced cell proliferation, induced apoptosis, and resulted in cell cycle arrest by regulating HOXA10 splicing.
MeSH Keywords: Alternative Splicing; Genes, Homeobox; snRNP Core Proteins; Stomach Neoplasms
Background
Worldwide, gastric cancer is one of the most common malignant tumors and is the third most common cause of cancer-related mortality [1]. The treatment of gastric cancer has improved with a multidisciplinary approach that includes surgery as the main treatment option, combined with radiotherapy and chemotherapy. However, many patients still suffer from recurrence and metastasis following initial treatment. With the rapid development of research on gastric cancer, gene therapy is becoming a possibility, as the abnormal expression of several oncogenes and tumor suppressor genes have been identified in the occurrence and development of gastric cancer [2]. However, research continues to identify the biological and molecular mechanisms for the development and progression of gastric cancer, and new treatments continue to be investigated.
Pre-mRNA splicing is a universal mechanism in eukaryotic cells where mRNA isoforms are generated from limited genomes by alternative splicing, or differential splicing, of single-gene coding for multiple proteins, which results in the synthesis of alternative proteins from increased transcript diversity [3]. Pre-mRNA splicing has a vital role in mRNA modification, especially for specific genes involved in tumor occurrence, and dysregulation of the splicing modulators is involved in the progression of several types of cancer [4].
Small nuclear ribonucleoproteins (snRNPs) participate in splicing RNA precursors in RNA alternative splicing. The spliceosome, splicing factor 3b subunit 1 (SF3B1), encodes the subunit 1 of splicing factor 3b, a component of the U2 snRNP complex. SF3B1 is located on chromosome 2q33.1 and results in alternative splicing events. Dysregulation of SF3B1 is associated with the progression of multiple malignant diseases, including breast cancer [5,6], head and neck cancer [7], and leukemia [8]. Importantly, snRNPs are attractive targets for anticancer treatment, and the agents targeting them has attracted considerable attention. Currently, several anticancer modulators of SF3B1 have been identified [9–11], including pladienolide B, which directly binds to SF3B1, to inhibit the splicing of tumor cells [12,13]. However, SF3B1 and the effects of its inhibitor, pladienolide B, have been rarely studied in human cancer cells, and their effects gastric cancer remain unknown.
Therefore, this study aimed to investigate the role of the spliceosome, SF3B1, in AGS and MKN28 human gastric cancer cells in vitro, including gene knockdown with small interfering RNA (siRNA) and the use of a specific SF3B1 inhibitor, pladienolide B.
Material and Methods
Cell culture and cell transfection
The AGS and MKN28 human gastric cancer cells were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA). All cells were cultured in a 37°C humidified incubator with 5% CO2. The spliceosome, splicing factor 3b subunit 1 (SF3B1), was investigated. The cells were transfected with small interfering RNA (siRNA)-SF3B1 or siRNA-NC. The siRNA-SF3B1 oligonucleotide sequence was 5′-UAUUGCAACAUUCUACAUGGU-3′, and the passenger sequence was 5′-CAUGUAGAAUGUUGCAAUAUU-3′. Cell transfection was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions.
Cell Counting Kit-8 (CCK-8) assay for cell proliferation
The cell growth rate was assessed using the Cell Counting Kit-8 (CCK-8) assay. After treatment, cells were aliquoted into a 96-well plate (100 μL/well) at a concentration of 1×105/mL and incubated for between 1–6 days. During detection, 20 μL of CCK-8 solution (KeyGen Biotech Co. Ltd., Nanjing, China) was added into each well, and plates were incubated for a further 2 h. The absorbance at 450 nm was measured with a microplate spectrophotometer reader.
Flow cytometry for evaluation of the cell cycle and cell apoptosis
For the cell cycle experiment, cells were harvested and fixed with 70% ethanol. The fixed cells were centrifuged and washed thrice with phosphate-buffered saline (PBS) and then stained with propidium iodide (PI) solution (KeyGen Biotech Co. Ltd., Nanjing, China). For cell apoptosis assay, the cells were collected and washed twice with PBS. Annexin V-fluorescein isothiocyanate (FITC) (KeyGen Biotech Co. Ltd., Nanjing, China) and PI were added to the cell suspension, according to the manufacturer’s instructions. Cells in the fourth quadrant of the flow cytometry cytogram were regarded as apoptotic cells. All the samples were analyzed using a BD flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Each experiment was performed in triplicate.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
The qRT-PCR for mRNA was performed using a one-step RT-PCR detecting kit (Takara, Minato-ku, Tokyo, Japan) on an HT7500 System (Applied Biosystems, Foster City, CA, USA). The primer sequences were:
SF3B1, forward: 5′-GTGGGCCTCGATTCTACAGG-3′;
SF3B1, reverse: 5′-GATGTCACGTATCCAGCAAATCT-3′;
HOXA10 exon 1, forward: 5′-TGCATATTTGGAATGCGCCG-3′;
HOXA10 exon 1, reverse: 5′-GAGAGACGGAAACCAGCTCC-3′;
HOXA10 exon 2, forward: 5′-CTCGCCCATAGACCTGTGG-3′;
HOXA10 exon 2, reverse: 5′-GTTCTGCGCGAAAGAGCAC-3′;
HOXA10 exon 3, forward: 5′-TGGCTCACGGCAAAGAGTG-3′;
HOXA10 exon 3, reverse: 5′-GCTGCGGCTAATCTCTAGGC-3′;
GADPH, forward: 5′-GAAGGTGAAGGTCGGAGTC-3′;
GADPH, reverse: 5′-GAAGATGGTGATGGGATTTC-3′.
GADPH served as an endogenous control to normalize the expression of SF3B1 and HOXA10 mRNA. The comparative cycle threshold (2−ΔΔCT) method was used to calculate the relative abundance of RNA compared with GADPH expression, and the fold difference relative to GADPH was determined.
Western blot
Cells were harvested, and total protein was extracted and then measured by the Bradford protein assay (Bio-Rad, Hercules, CA, USA). Protein from each sample was separated on 12% Bis-Tris polyacrylamide gels by electrophoresis and blotted onto nitrocellulose membranes (GE Healthcare Life Sciences, Logan, UT, USA). Antibodies against SF3B1, HOXA10, PTEN, AKT, and p-AKT (Abcam, Cambridge, MA, USA) were used, and horseradish peroxidase-linked anti-rabbit or anti-mouse IgG secondary antibodies (Beyotime, Shanghai, China) were used. The primary antibodies were diluted using the following concentrations: HOXA10, AKT, and p-AKT (1: 1000), SF3B1 (1: 5000), and PTEN (1: 10000). Blots were immunostained with the primary antibodies at 4°C overnight, and with the secondary antibody at room temperature for 1 h. Proteins were visualized using an electrochemiluminescence (ECL) microplate reader and Western blot detection reagents (GE Healthcare Life Sciences, Logan, UT, USA). The results were normalized against GADPH levels.
Analysis of alternative splicing events
Data on alternative splicing, or differential splicing, of single-gene coding for multiple proteins were analyzed using The Cancer Genome Atlas (TCGA) SpliceSeq (https://bioinformatics.mdanderson.org/TCGASpliceSeq/). Alternative splicing events were quantified by the percent spliced in index (PSI), with values ranging from 0–100%. The alternative splicing events were screened with a PSI rate of >2.5 times in gastric cancer and nontumor samples in the database. The RNA expression profiles of the SF3B1 and HOXA10 genes were analyzed in the cBioPortal for Cancer Genomics (http://www.cbioportal.org/) using the data of the TCGA gastric cancer cohort.
Statistical analysis
Data were expressed as the mean±standard deviation (SD) of at least three independent experiments. Student’s t-test determined differences between the groups. Data analysis was performed using SPSS version 20 software (IBM, Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
Inhibition of splicing factor 3b subunit 1 (SF3B1), reduced the viability of AGS and MKN28 human gastric cancer cells in vitro
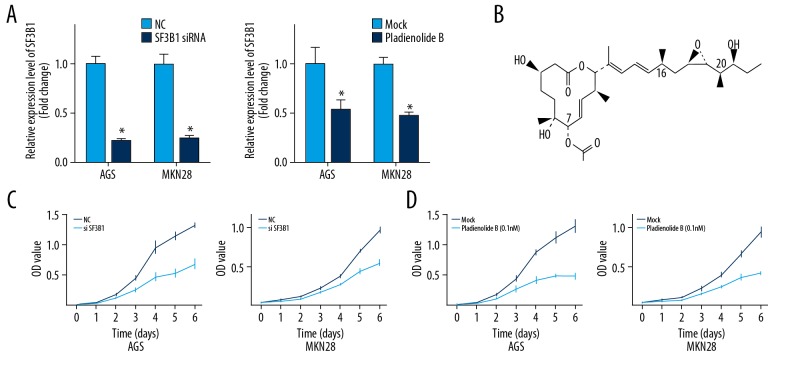
The knockdown of the SF3B1 gene was performed with small interfering RNA (siRNA) (Figure 1A). The Cell Counting Kit-8 (CCK-8) assay showed that the SF3B1 knockdown resulted in growth arrest (Figure 1C). The effect of inhibition of SF3B1 with pladienolide B, which specifically binds to SF3B1, was evaluated in gastric cancer cells (Figure 1B). Pretreatment with pladienolide B resulted in a significant reduction in SF3B1 expression in the gastric cancer cells (Figure 1A), and the growth rate of the cells was significantly lower than that of the control group (Figure 1D).
Figure 1.
AGS and MKN28 human gastric cancer cell viability was significantly inhibited by the use of splicing factor 3b subunit 1 (SF3B1) small interfering RNA (siRNA) and pladienolide B. (A) The effect of siRNA and pladienolide B on SF3B1 expression was shown by quantitative analysis. (B) The chemical structure of pladienolide B, a selective mRNA splicing inhibitor of SF3B1. (C) The cell viability AGS and MKN28 human gastric cancer cells with SF3B1 siRNA and (D) pladienolide B evaluated by the Cell Counting Kit-8 (CCK-8) assay. The experiments were performed in triplicate. The mean±SD represents the data. * p<0.05 (vs. the control group).
SF3B1 dysfunction induces apoptosis and cell cycle arrest
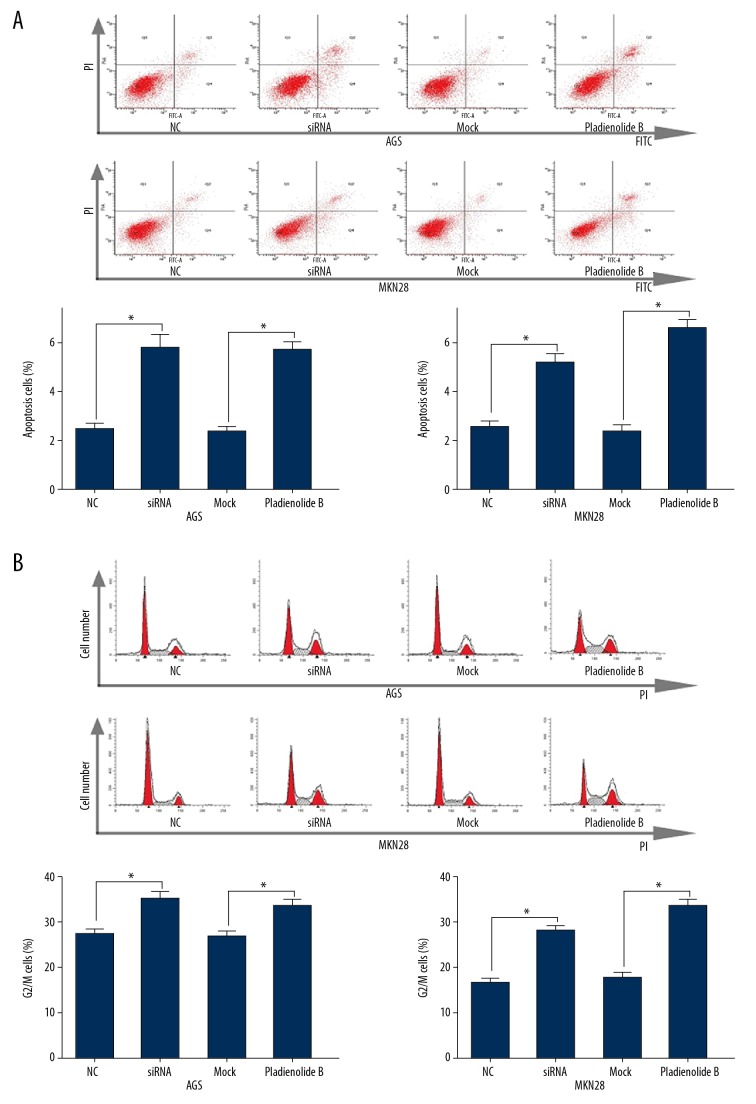
Following knockdown and inhibition of SF3B1 in the AGS and MKN28 human gastric cancer cells, flow cytometry was used to detect cell apoptosis and components of the cell cycle. The number of apoptotic cells was significantly increased (Figure 2A), and there was G2/M phase arrest after SF3B1 siRNA or pladienolide B treatment (Figure 2B).
Figure 2.
Inhibition of splicing factor 3b subunit 1 (SF3B1) by small interfering RNA (siRNA) and pladienolide B induced apoptosis and cell cycle arrest in AGS and MKN28 human gastric cancer cells. (A) Apoptosis of AGS and MKN28 human gastric cancer cells and the percentage of cells undergoing apoptosis evaluated by flow cytometry. (B) The cell cycle assay and percentage of cells in the G2/M phase. The experiments were performed in triplicate. The mean±SD represents the data. * p<0.05 (vs. the control group).
SF3B1 knockdown resulted in reduced homeobox A10 (HOXA10) mRNA expression and expression of long noncoding RNA (lncRNA) isoforms of HOXA10 (exons 1 and 3) and HOXA10 (exons 2 and 3)
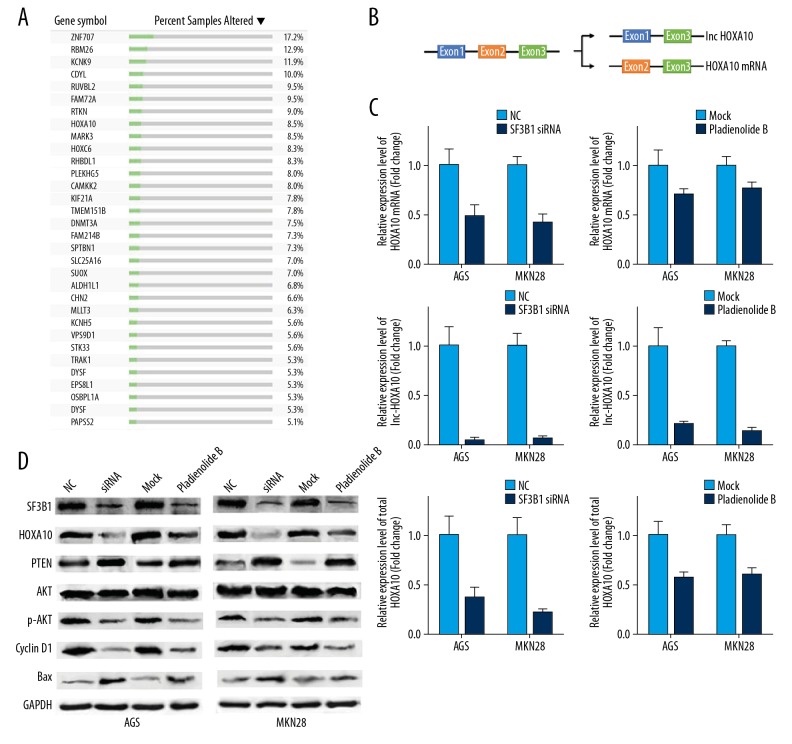
The presence of alternative splicing, or differential splicing, of single-gene coding for multiple proteins, was analyzed using The Cancer Genome Atlas (TCGA) SpliceSeq. Ninety-seven events that included 89 genes were screened (Table 1). Among them, 32 genes were abnormally expressed in more than 5% TCGA gastric cancer tumor tissues (Figure 3A). HOXA10 is a critical gene in cancer progression and acts as an independent factor for the survival of gastric cancer [14,15]. HOXA10 was identified as a potential effector oncogene of alternative splicing.
Table 1.
Genes with a percent spliced in index (PSI) rate of >2.5 times the alternative splicing events differently in gastric cancer samples.
| Splice type | Exon | Gene symbol | PSI (tumor tissue) | PSI (normal tissue) | Ratio of PSI change |
|---|---|---|---|---|---|
| Upregulation | |||||
| ES | 5 | MAGI2 | 39.1 | 2.7 | 14.48148 |
| AP | 1 | HOXC4 | 22.3 | 2.0 | 11.15 |
| AT | 5 | CLEC3A | 46.7 | 5.4 | 8.648148 |
| ES | 2 | CKMT2 | 42.2 | 5.9 | 7.152542 |
| RI | 3.2 | CLU | 32.7 | 5.7 | 5.736842 |
| AP | 1 | ALDH1L1 | 47.8 | 8.7 | 5.494253 |
| AP | 4.1 | TGFBR3 | 26.9 | 5.0 | 5.38 |
| AP | 5 | KCNN2 | 86.5 | 17.1 | 5.05848 |
| AP | 5 | COLEC11 | 42.6 | 8.9 | 4.786517 |
| ES | 2 | ZMAT1 | 54.5 | 12.6 | 4.325397 |
| AP | 2 | ISLR | 47.6 | 11.1 | 4.288288 |
| AP | 2 | ARHGAP28 | 29.0 | 6.9 | 4.202899 |
| AP | 8.1 | EPS8L1 | 45.6 | 11.0 | 4.145455 |
| AP | 10 | SH3KBP1 | 37.0 | 9.1 | 4.065934 |
| AP | 1 | PKIB | 34.8 | 8.6 | 4.046512 |
| RI | 4.2 | FAM214B | 27.0 | 6.8 | 3.970588 |
| AP | 2 | ANKMY1 | 27.8 | 7.1 | 3.915493 |
| AT | 13 | CNGB1 | 28.3 | 7.4 | 3.824324 |
| ES | 4.2: 5.1: 5.2 | ZNF707 | 27.1 | 7.1 | 3.816901 |
| AP | 1 | APOLD1 | 47.3 | 12.4 | 3.814516 |
| ES | 13 | PLCD4 | 62 | 16.3 | 3.803681 |
| AD | 1.2: 1.3: 1.4: 1.5: 1.6 | RUVBL2 | 29.5 | 7.8 | 3.782051 |
| AP | 7 | LDLRAD4 | 27.5 | 7.4 | 3.716216 |
| ES | 2.1: 2.2 | SLC25A16 | 36.6 | 10.0 | 3.66 |
| ES | 16 | SORBS1 | 46.3 | 13.0 | 3.561538 |
| AP | 5 | CDYL | 32.4 | 9.3 | 3.483871 |
| AP | 8 | CASQ1 | 71.0 | 20.6 | 3.446602 |
| ES | 6 | COL6A3 | 51.4 | 15.0 | 3.426667 |
| AP | 1 | HOXC6 | 36.0 | 10.9 | 3.302752 |
| AP | 2 | IL20RB | 35.3 | 10.9 | 3.238532 |
| AT | 4.2 | ARHGEF4 | 39.2 | 12.3 | 3.186992 |
| AP | 14 | AK5 | 42.2 | 13.3 | 3.172932 |
| AP | 1 | CCL14 | 58.9 | 18.9 | 3.116402 |
| AP | 2 | LRRC32 | 32.0 | 10.4 | 3.076923 |
| AP | 8 | PLEKHG5 | 41.5 | 13.8 | 3.007246 |
| AP | 6 | HDAC9 | 55.0 | 18.8 | 2.925532 |
| AP | 2.1 | VPS9D1 | 39.6 | 13.6 | 2.911765 |
| AP | 1 | HOXA10 | 50.2 | 17.5 | 2.868571 |
| AP | 5.1 | SUOX | 30.8 | 10.8 | 2.851852 |
| ES | 5 | PDLIM7 | 33.8 | 11.9 | 2.840336 |
| AP | 1.1 | OSBPL1A | 35.9 | 12.7 | 2.826772 |
| ES | 12: 13: 14: 15: 16: 19 | TNC | 48.2 | 17.1 | 2.818713 |
| AP | 7.1 | ANGEL1 | 39.6 | 14.2 | 2.788732 |
| AP | 5.1 | STK33 | 32.6 | 12.1 | 2.694215 |
| AT | 10.2 | MYPN | 35.2 | 13.2 | 2.666667 |
| AP | 1 | WNT2B | 43.6 | 16.4 | 2.658537 |
| AP | 2.1 | RHBDL1 | 49.5 | 18.7 | 2.647059 |
| AP | 4 | 4-Sep | 34.7 | 13.2 | 2.628788 |
| ES | 9 | ZNF667 | 38.4 | 14.7 | 2.612245 |
| AT | 8 | PPP1R1A | 35.7 | 13.7 | 2.605839 |
| ES | 3: 4: 5: 6: 7: 8: 9: 10: 11: 12: 13: 14: 15: 16: 17 | COL12A1 | 89.8 | 35.0 | 2.565714 |
| ES | 2: 3.1: 3.2 | SHROOM1 | 37.7 | 14.7 | 2.564626 |
| AP | 2 | SMAD6 | 34.5 | 13.5 | 2.555556 |
| AP | 2 | DYSF | 49.9 | 19.6 | 2.545918 |
| RI | 3.4: 3.5 | SH2B1 | 39.5 | 15.6 | 2.532051 |
| AP | 8 | DNMT3A | 35.4 | 14.1 | 2.510638 |
| Down-regulation | |||||
| AP | 2 | NRG2 | 1.8 | 22.8 | 12.66667 |
| AP | 4 | LSP1 | 4.0 | 32.5 | 8.125 |
| AP | 1 | TRAK1 | 4.5 | 28.0 | 6.222222 |
| AT | 36 | LAMB4 | 4.4 | 25.2 | 5.727273 |
| AP | 2 | KCNH5 | 6.2 | 34.2 | 5.516129 |
| AP | 7 | MLLT3 | 6.5 | 30.3 | 4.661538 |
| ES | 29: 30: 32 | KIF21A | 6.2 | 28.3 | 4.564516 |
| ES | 19 | DYSF | 11.4 | 51.7 | 4.535088 |
| ES | 24 | KIF21A | 8.6 | 35.4 | 4.116279 |
| ES | 19: 20: 21: 22 | MICAL3 | 7.1 | 27.1 | 3.816901 |
| AP | 12.1 | CHN2 | 14.9 | 55.0 | 3.691275 |
| ES | 23: 24 | TNS1 | 11.3 | 40.9 | 3.619469 |
| ES | 13: 14 | TBC1D1 | 7.9 | 28.1 | 3.556962 |
| AP | 4 | GAS7 | 10.5 | 37.3 | 3.552381 |
| ES | 16 | SVIL | 12.6 | 43.3 | 3.436508 |
| AP | 2 | RTKN | 8.7 | 29.8 | 3.425287 |
| AP | 1 | ASTN2 | 16.7 | 55.8 | 3.341317 |
| AP | 1 | SPTBN1 | 12.1 | 39.7 | 3.280992 |
| AP | 1 | ARHGEF9 | 9.0 | 29.4 | 3.266667 |
| ES | 11 | FBLN2 | 22.6 | 73.3 | 3.243363 |
| AP | 1 | RARB | 10.8 | 34.7 | 3.212963 |
| AP | 1 | PDE8B | 19.9 | 63.9 | 3.211055 |
| ES | 9 | PAPSS2 | 10.4 | 32.6 | 3.134615 |
| AT | 7.3 | TRIM73 | 12.3 | 38.4 | 3.121951 |
| AT | 3 | TMEM151B | 17.6 | 54.0 | 3.068182 |
| ES | 19 | VCL | 18.0 | 51.7 | 2.872222 |
| AT | 2 | FAM72A | 12.0 | 34.2 | 2.85 |
| AP | 2 | PLEKHG5 | 13.2 | 37.3 | 2.825758 |
| AP | 3.1 | COL18A1 | 16.0 | 44.9 | 2.80625 |
| AT | 20 | GRIK2 | 13.1 | 36.3 | 2.770992 |
| AP | 1 | NTM | 15.4 | 42.3 | 2.746753 |
| ES | 21 | SVIL | 23.0 | 63.1 | 2.743478 |
| AP | 1 | CASQ1 | 29.0 | 79.4 | 2.737931 |
| AP | 1 | FAM72A | 12.5 | 33.9 | 2.712 |
| ES | 14 | RBM26 | 12.7 | 34.3 | 2.700787 |
| AP | 1 | HDAC9 | 17.7 | 46.9 | 2.649718 |
| AP | 3 | ALDH1L1 | 32.3 | 84.2 | 2.606811 |
| AT | 2.2 | KCNK9 | 20.4 | 53.0 | 2.598039 |
| AT | 7.2 | ADRA1A | 13.7 | 35.5 | 2.591241 |
| ES | 17 | CAMKK2 | 13.0 | 33.5 | 2.576923 |
| ES | 17: 18 | MARK3 | 13.5 | 34.3 | 2.540741 |
Figure 3.
Inhibition of splicing factor 3b subunit 1 (SF3B1) by small interfering RNA (siRNA) and pladienolide B and the expression of homeobox A10 (HOXA10) and the AKT pathway in AGS and MKN28 human gastric cancer cells. (A) List of genes in Table 1 that showed altered expressed in more than 5% of samples using data from The Cancer Genome Atlas (TCGA) gastric cancer cohort in cBioPortal. (B) HOXA10 gene structure and alternative transcripts. (C) The effect of siRNA and pladienolide B on HOXA10 mRNA/lnc-HOXA10/total HOXA10 expression shown by quantitative analysis. (D) Representative Western blot images of relative protein expression in AGS and MKN28 human gastric cancer cells. The experiments were performed in triplicate. The mean±SD represents the data. * p<0.05 (vs. the control group).
The HOXA10 gene possesses three exons, long noncoding (lnc)-HOXA10 (exons 1 and 3) and HOXA10 (exons 2 and 3) isoforms (Figure 3B). After SF3B1 inhibition, HOXA10 pre-mRNA formed HOXA10 mRNA. Two specific primers were constructed to detect the mRNA levels of HOXA10 mRNA (exon 2) and lnc-HOXA10 (exon 1). The levels of HOXA10 mRNA and lnc-HOXA10 were both reduced in AGS and MKN28 human gastric cancer cells after SF3B1 siRNA or pladienolide B treatment compared with the control group. Also, levels of lnc-HOXA10 were reduced much more than for HOXA10 mRNA. We further designed the primer targeting the total amount of HOXA10 RNA (exon 3) and found that the total HOXA10 RNA also significantly decreased (Figure 3C).
The levels of HOXA10 proteins were studied. Compared with the control group, a significant decrease in the HOXA10 protein level was found (Figure 3D). Therefore, expression of the HOXA10 gene was regulated by SF3B1, resulting in the manipulation of the downstream target genes of the HOXA10 protein. To determine whether SF3B1 promoted cell growth via the PTEN/AKT pathway in AGS and MKN28 human gastric cancer cells, the expression of these mediators was analyzed. SF3B1 inhibition by siRNA or pladienolide B induced a significant increase in the PTEN protein level and a significant decrease in the p-AKT and protein level. Key regulators in the AKT pathway, including the apoptosis marker, Bax, and the cell cycle marker, cyclin D1, were also increased (Figure 3D). These findings showed that SF3B1 affected the growth of AGS and MKN28 human gastric cancer cells in vitro via the regulation of PTEN by HOXA10.
Discussion
Alternative splicing, or differential splicing, of single-gene coding for multiple proteins, is a common cell regulatory mechanism that results in the diversity of RNA translation [16]. Small nuclear ribonucleoprotein (snRNP) complexes of protein and noncoding RNA accumulate in the cell nucleus. The snRNPs catalyze pre-mRNA splicing to form the spliceosome. Spliceosomes, which are aggregated by multiple nucleoproteins and have the functions of identifying 5′ splicing sites and 3′ splicing sites of RNA exons, are multi-component complexes formed during RNA splicing [17,18]. The spliceosome, splicing factor 3b subunit 1 (SF3B1), has previously been shown to play an important role in tumorigenesis and tumor progression [5,6]. However, the mechanism of SF3B1 in cancer and its downstream pathways remain unclear.
In the present study, SF3B1 gene knockdown with small interfering RNA (siRNA), and the use of the selective mRNA splicing inhibitor of SF3B1, pladienolide B were used to inhibit gene expression and to explore gene function. Pladienolide B, a natural product targeting SF3B1, exhibited antitumor activity in AGS and MKN28 human gastric cancer cells in vitro. Pladienolide B has potential as a chemotherapeutic drug and as a tool to study the role of SF3B1 in alternative splicing and cancer development. Also, in this study, apoptosis and the cell cycle were studied, and the effect of SF3B1 on cell proliferation was investigated. When SF3B1 was inhibited, the cell cycle in the AGS and MKN28 cells was arrested, and apoptosis increased. This finding was consistent with the results of previous studies on human malignant cell lines [19].
The knockdown of SF3B1 described in this study resulted in HOXA10 alternative splicing. The HOXA10 pre-mRNA can be transcribed into one coding isoform (HOXA10 mRNA) and one noncoding isoform (lnc-HOXA10) [20]. In this study, when SF3B1 was downregulated, the HOXA10 mRNA and lnc-HOXA10 decreased. The homeobox (HOX) gene, which is present in most eukaryotic cells, is an evolutionarily conserved family of polygenes [20]. HOX gene-encoded proteins are a class of transcription factors containing homologous heteromorphic domains and have an important role during cancer development in vertebrates [21]. The HOXA10 gene is an important member of the homeobox gene family. Recently, studies in oncology have shown that HOXA10 gene is involved in the development of human malignant tumors, including colorectal cancer [22], ovarian cancer [23], and medulloblastoma [24]. In gastric cancer, HOXA10 expression has previously been shown to be increased in gastric cancer tissues [14], and this phenomenon is related to the biological characteristics of the tumor [15]. Cell proliferation and cell migration are enhanced in gastric cancer cells that overexpressed HOXA10 but were inhibited in cells with silenced HOXA10 [25]. Also, lnc-HOXA10 can increase HOXA10 transcription by recruiting SNF2L [26]. Therefore, when the lnc-HOXA10 level was significantly reduced, HOXA10 expression will be significantly inhibited, and a loop amplification effect will be formed. This phenomenon eventually decreases HOXA10 mRNA levels.
The HOXA10 protein downregulation found in this study was consistent with previously reported findings of PTEN upregulation in cancer cells [22,27]. PTEN can reduce AKT activation and prevent downstream signal transduction events regulated by AKT. In the present study, to determine whether the AKT pathway was a component of the SF3B1 downstream pathway regulating the proliferation of AGS and MKN28 human gastric cancer cells, the levels of AKT, p-AKT, and PTEN protein were compared in all groups. The AKT pathway contains several genes that are involved in the normal growth and renewal of eukaryote cells and is critical for the development and metastasis of many cancers. AKT can increase the expression level of cyclin D1 and other factors, and can directly activate the CDK1 protein, so that cells can pass through cell cycle checkpoints to accelerate cell proliferation. Also, AKT regulates the function of members of the Bcl-2 family, inhibits the formation of dimers of bcl-2, and promotes the inactivation of the pro-apoptotic factor, Bax, to inhibit cell apoptosis [28,29]. Inhibition of the AKT pathway is an important approach to anticancer therapy [30,31]. The findings from this study showed that SF3B1 inhibition increased the levels of PTEN and reduced AKT protein phosphorylation in AGS and MKN28 human gastric cancer cells in vitro.
Conclusions
This study aimed to investigate the role of the spliceosome, splicing factor 3b subunit 1 (SF3B1), in AGS and MKN28 human gastric cancer cells in vitro, and used small interfering RNA (siRNA) and pladienolide B, a selective inhibitor of splicing factor 3b that inhibits mRNA splicing, to inhibit SF3B1. The findings showed that inhibition of SF3B1 resulted in cycle arrest and induced cell apoptosis by modulating the alternative splicing of HOXA10. PTEN/AKT/PI3K may be a downstream signaling pathway for SF3B1 in gastric cancer cells. SF3B1 and its inhibitor, pladienolide B, require further studies on their potential role in gastric cancer.
Footnotes
Source of support: This study was supported by a grant from the Natural Science Foundation of Jiangsu, China (No. BK20180215)
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Pereira A, Moreira F, Vinasco-Sandoval T, et al. miRNome reveals new insights into the molecular biology of field cancerization in gastric cancer. Front Genet. 2019;10:592. doi: 10.3389/fgene.2019.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Marabti E, Younis I. The cancer spliceome: Reprograming of alternative splicing in cancer. Front Mol Biosci. 2018;5:80. doi: 10.3389/fmolb.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maguire SL, Leonidou A, Wai P, et al. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J Pathol. 2015;235(4):571–80. doi: 10.1002/path.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu X, Tian M, Gu J, et al. SF3B1 mutation is a poor prognostic indicator in luminal B and progesterone receptor-negative breast cancer patients. Oncotarget. 2017;8(70):115018–27. doi: 10.18632/oncotarget.22983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y, Trivedi S, Ferris RL, Koide K. Regulation of HPV16 E6 and MCL1 by SF3B1 inhibitor in head and neck cancer cells. Sci Rep. 2014;4:6098. doi: 10.1038/srep06098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaida D, Schneider-Poetsch T, Yoshida M. Splicing in oncogenesis and tumor suppression. Cancer Sci. 2012;103(9):1611–16. doi: 10.1111/j.1349-7006.2012.02356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagisetti C, Palacios G, Goronga T, et al. Optimization of antitumor modulators of pre-mRNA splicing. J Med Chem. 2013;56(24):10033–44. doi: 10.1021/jm401370h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamano T, Kubo S, Yano A, et al. Splicing modulator FR901464 is a potential agent for colorectal cancer in combination therapy. Oncotarget. 2019;10(3):352–67. doi: 10.18632/oncotarget.26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato M, Muguruma N, Nakagawa T, et al. High antitumor activity of pladienolide B and its derivative in gastric cancer. Cancer Sci. 2014;105(1):110–16. doi: 10.1111/cas.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Di C, Yan J, et al. Inhibition of SF3b1 by pladienolide B evokes cycle arrest, apoptosis induction and p73 splicing in human cervical carcinoma cells. Artif Cells Nanomed Biotechnol. 2019;47(1):1273–80. doi: 10.1080/21691401.2019.1596922. [DOI] [PubMed] [Google Scholar]
- 14.Sentani K, Oue N, Naito Y, et al. Upregulation of HOXA10 in gastric cancer with the intestinal mucin phenotype: Reduction during tumor progression and favorable prognosis. Carcinogenesis. 2012;33(5):1081–88. doi: 10.1093/carcin/bgs121. [DOI] [PubMed] [Google Scholar]
- 15.Lim JY, Yoon SO, Seol SY, et al. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;19(41):7078–88. doi: 10.3748/wjg.v19.i41.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeVaux RS, Kuentzel M, Herschkowitz J, Chittur SV. Determination of Alternate Splicing Events Using Transcriptome Arrays. Methods Mol Biol. 2017;1507:245–59. doi: 10.1007/978-1-4939-6518-2_18. [DOI] [PubMed] [Google Scholar]
- 17.Huang W, Huang Y, Xu J, Liao JL. How does the spliceosome catalyze intron lariat formation? Insights from quantum mechanics/molecular mechanics free-energy simulations. J Phys Chem B. 2019;123(28):6049–55. doi: 10.1021/acs.jpcb.9b04377. [DOI] [PubMed] [Google Scholar]
- 18.Charenton C, Wilkinson ME, Nagai K. Mechanism of 5′ splice site transfer for human spliceosome activation. Science. 2019;364(6438):362–67. doi: 10.1126/science.aax3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorge J, Petronilho S, Alves R, et al. Apoptosis induction and cell cycle arrest of pladienolide B in erythroleukemia cell lines. Invest New Drugs. :2019. doi: 10.1007/s10637-019-00796-2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Ramos-Womack M, Han C, et al. Changes throughout a genetic network mask the contribution of Hox gene evolution. Curr Biol. 2019;29(13):2157–66. doi: 10.1016/j.cub.2019.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Huang Q, Wei GH. The role of HOX transcription factors in cancer predisposition and progression. Cancers (Basel) 2019;11(4) doi: 10.3390/cancers11040528. pii: E528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Y, Sun S, Jiao N, et al. Upregulation of HOXA10 protein expression predicts poor prognosis for colorectal cancer. Genet Test Mol Biomarkers. 2018;22(6):390–97. doi: 10.1089/gtmb.2017.0240. [DOI] [PubMed] [Google Scholar]
- 23.Zhang HY, Li JH, Li G, Wang SR. Activation of ARK5/miR-1181/HOXA10 axis promotes epithelial-mesenchymal transition in ovarian cancer. Oncol Rep. 2015;34(3):1193–202. doi: 10.3892/or.2015.4113. [DOI] [PubMed] [Google Scholar]
- 24.Bonfim-Silva R, Ferreira Melo FU, Thome CH, et al. Functional analysis of HOXA10 and HOXB4 in human medulloblastoma cell lines. Int J Oncol. 2017;51(6):1929–40. doi: 10.3892/ijo.2017.4151. [DOI] [PubMed] [Google Scholar]
- 25.Han Y, Lu S, Wen YG, et al. Overexpression of HOXA10 promotes gastric cancer cells proliferation and HOXA10(+)/CD44(+) is potential prognostic biomarker for gastric cancer. Eur J Cell Biol. 2015;94(12):642–52. doi: 10.1016/j.ejcb.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Shao M, Yang Q, Zhu W, et al. LncHOXA10 drives liver TICs self-renewal and tumorigenesis via HOXA10 transcription activation. Mol Cancer. 2018;17(1):173. doi: 10.1186/s12943-018-0921-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao ZD, Jiao CY, Huang HT, et al. miR-218 modulate hepatocellular carcinoma cell proliferation through PTEN/AKT/PI3K pathway and HoxA10. Int J Clin Exp Pathol. 2014;7(7):4039–44. [PMC free article] [PubMed] [Google Scholar]
- 28.Deng J, Bai X, Feng X, et al. Inhibition of PI3K/Akt/mTOR signaling pathway alleviates ovarian cancer chemoresistance through reversing epithelial-mesenchymal transition and decreasing cancer stem cell marker expression. BMC Cancer. 2019;19(1):618. doi: 10.1186/s12885-019-5824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayanankutty A. PI3K/Akt/mTOR pathway as a therapeutic target for colorectal cancer: A review of preclinical and clinical evidence. Curr Drug Targets. 2019;20(12):1217–26. doi: 10.2174/1389450120666190618123846. [DOI] [PubMed] [Google Scholar]
- 30.Tao K, Yin Y, Shen Q, et al. Akt inhibitor MK-2206 enhances the effect of cisplatin in gastric cancer cells. Biomed Rep. 2016;4(3):365–68. doi: 10.3892/br.2016.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bang YJ, Kang YK, Ng M, et al. A phase II, randomised study of mFOLFOX6 with or without the Akt inhibitor ipatasertib in patients with locally advanced or metastatic gastric or gastroesophageal junction cancer. Eur J Cancer. 2019;108:17–24. doi: 10.1016/j.ejca.2018.11.017. [DOI] [PubMed] [Google Scholar]