Abstract
Edible insects present a potential solution to increasing global food insecurity. However, there is limited research on the microbial hazards they may pose. These include opportunistic pathogens like Cronobacter spp. (formerly Enterobacter sakazakii). In this study, nine types of ready-to-eat edible insect products purchased in the UK were examined for their microbial load (total aerobic count, total Enterobacteriaceae count), and screened for the presence of Cronobacter sakazakii(C. sakazakii) by selective enrichment and plating on chromogenic agar. While microbial load was generally low, presumptive Cronobacter spp. were detected in five of the edible insect products. Four of the isolates were identified as C. sakazakii, using the Remel RapID ONE biochemical test kit. Genotypic characterisation of the isolates by ITS-PCR, however, demonstrated that the isolates may be other species of Cronobacter instead. Further studies into understanding microbial hazards linked to edible insects for human consumption are required.
Key words: Cronobacter, entomophagy, food safety, PCR, RTE foods
Introduction
The consumption of insects (entomophagy) is considered as a potential solution to the increasing issues of food insecurity and malnutrition. Many insect species have been consumed worldwide, and have been shown to have protein and nutrient profiles comparable to meat1). However, consumer attitudes may present a barrier toward the acceptance of edible insects in Western countries2). There may also be microbial hazards linked to such edible insect products3,4).
Cronobacter spp. (formerly Enterobacter sakazakii) are Gram negative, oxidase negative, rod-shaped bacteria, which are members of the Enterobacteriaceae family. They are emerging opportunistic pathogens which can cause infections in adults and infants, including necrotising enterocolitis, bacteraemia, and meningitis5). The genus consists of seven species, most of which can cause human disease. Cronobacter sakazakii (C. sakazakii) is commonly associated with infant infections, while Cronobacter malonaticus is commonly linked to infections in adults, especially the elderly and immunocompromised6).
Cronobacter spp. are ubiquitous and have been isolated from a wide range of foods, such as vegetables, herbs, spices, meat products and ready-to-eat foods7). The presence of C. sakazakii in powdered infant formula is of major concern, due to its implications for infant health, and has been widely studied8).
Cronobacter spp. have also been isolated from the guts of insects such as fruit flies and stable flies, which may be considered as sources of environmental contamination in foods9). Studies involving culture-independent metagenomic analysis on edible insects suggest that Cronobacter may be associated with the natural microbiota of mealworms10,11).
Marketed edible insects may be whole insects, commonly processed by blanching, followed by drying12), or powdered3). These are considered to be ready-to-eat. To our knowledge, no studies have examined processed edible insect products for the presence of Cronobacter spp. Therefore, this study aimed to examine the microbial load of ready-to-eat edible insects purchased in the UK, and in particular, determine the presence of C. sakazakii, using culture-based methods. We also comparatively examined the use of phenotypic and genotypic methods in identifying and differentiating presumptive Cronobacter isolates.
Materials and Methods
Sample Collection
A total of nine edible insect products (Table 1) were purchased from an online UK-based retailer (two samples of each). All insects contained no additives or additional flavouring (except for the queen leafcutter ants which were lightly salted), and were stored at room temperature.
Table 1. Ready-to-eat insects used in this study, including country of origin, method of processing and bacterial counts (log CFU/g).
| Insect | Latin Name | Country of Origin | Processing Method | Total Aerobic Count | Enterobacteriaceae |
| Buffalo Worms | Alphitobius diaperinus | Netherlands | Freeze-dried | 4.59 | < 2.00 |
| Crickets | Acheta domesticus | Netherlands | Freeze-dried | 3.95 | < 2.00 |
| Cricket Flour | Acheta domesticus | Thailand | Dehydrated, finely milled | 4.00 | 2.57 |
| Giant Waterbugs | Lethocerus indicus | Thailand | Dehydrated | 2.24 | < 2.00 |
| Locust | Locusta migratoria | Netherlands | Freeze-dried | 3.72 | < 2.00 |
| Mealworms | Tenebrio molitor | Netherlands | Freeze-dried | 2.17 | < 2.00 |
| Queen Leafcutter Ants | Atta laevigata | Colombia | Brine boiled and air-dried | 3.35 | < 2.00 |
| Silkworm Pupae | Bombyx mori | Thailand | Pressure steam cooked then dehydrated | 2.00 | < 2.00 |
| Wild Black Ants | Lasius niger | Thailand | Pressure steam cooked then dehydrated | 2.24 | < 2.00 |
Reference Strains
C. sakazakii reference strains NCIMB 8272 and NCIMB 5920 were kindly provided by London Metropolitan University. Strains were confirmed by 16S rDNA sequencing13).
Microbiological Enumeration
A 10 g sample of each insect product was homogenized in 90 ml of buffered peptone water (BPW) (Oxoid CM1049) using a stomacher (Stomacher 400 Circulator, Seward) at 300 rpm for 1 minute. From the homogenized suspension, further serial dilutions were prepared up to 10−4 in BPW. Subsequently, 0.1 ml of each dilution was plated in duplicate onto nutrient agar (Oxoid CM0003) to determine total aerobic count, and violet red bile glucose (VRBG) agar (Oxoid CM1082) to determine total Enterobacteriaceae count. The inoculated plates were incubated at 37°C for 24 h. All enumeration experiments were carried out in two replicates.
Detection and Isolation of Presumptive Cronobacter spp.
To detect Cronobacter spp., the initial homogenized suspension of each insect sample in BPW, as made previously for enumeration, was incubated at 37°C for 24 h (pre-enrichment). Afterwards, 0.1 ml of the pre-enrichment was suspended in 10 ml of Cronobacter Screening Broth (CSB) (Sigma-Aldrich 38948) supplemented with vancomycin (Sigma-Aldrich 75423) according to manufacturer’s instructions, and incubated for 24-48 h at 37°C. If the CSB changed color from purple to yellow, then the sample was considered positive for the presence of Cronobacter spp. due to fermentation of sucrose14). A loopful of each positive broth was then inoculated onto chromogenic medium HiCrome Cronobacter spp. Agar (Sigma-Aldrich 92324) and incubated for 24 h at 37°C. Any presumptive Cronobacter spp. would grow as dark blue colonies on this agar, due to cleaving of the chromogenic substrate by α-glucosidase, which is produced by Cronobacter spp.15).
Phenotypic and Biochemical Characterisation
Dark blue colonies from the HiCrome Cronobacter spp. agar were purified by streaking onto tryptone soy agar (TSA) (Oxoid CM0131) and incubating at 22°C for 24-48 h13). Cultures were subjected to Gram staining, oxidase test (Sigma-Aldrich 40560) and catalase test. Biochemical identification was carried out using the Remel RapID ONE System (Thermo Fisher R8311006), according to manufacturer’s instructions.
Genotypic Characterisation
DNA was extracted from bacterial isolates using InstaGene Matrix (BioRad 7326030) according to manufacturer’s instructions. Extracted DNA samples were used in polymerase chain reactions (PCR). Genotypic characterisation of isolates was by 16S-23S rDNA internal transcribed spacer region PCR (ITS-PCR). ITS-PCR was carried out as described by Polit et al.16).
ITS-PCR products were visualized by electrophoresis on 1.2% (w/v) agarose gels (Bioline BIO-41025). Gels contained 2 µl GelRed stain (41003, Biotium, Fremont, CA, USA). 10 µl of PCR product was mixed with 2 µl of loading dye (R0611, Thermo Fisher Scientific, Loughborough, UK) and loaded into the wells. A DNA molecular size marker (SM1113, Thermo Fisher Scientific) was used to estimate the sizes of the PCR products. The gels were run in a 1x Tris-Borate-EDTA (TBE) buffer at 60 V for approximately 1 hour. Images of the gels were taken using a UV transilluminator (Gel Doc EZ Imager, BioRad).
Results and Discussion
It appeared that insect samples that originated from the Netherlands were processed by freeze-drying, whereas those originating from Thailand and Colombia were possibly processed by dehydration methods other than freeze-drying (Table 1). This may reflect the fact that freeze-drying is more expensive17), and as such, probably more affordable in more developed countries such as the Netherlands.
Total aerobic counts from the ready-to-eat insects ranged from 2.00 log CFU/g (silkworm pupae) to 4.59 log CFU/g (buffalo worms) (Table 1). No Enterobacteriaceae were recovered from any of the whole insect samples (Table 1). This may be the result of blanching in boiling water for a short time, which usually occurs prior to drying of edible insects3,18). Only the cricket flour showed any growth of Enterobacteriaceae on the VRBG agar. This could be because, unlike the other insect samples, the cricket flour is finely milled, which means that the crickets’ intestinal microbiota are distributed throughout the product3). Nevertheless, all the samples could be considered as having low microbial load and meeting recommended hygiene criteria4,19).
Out of the nine insect samples, five yielded positive results for both the CSB and HiCrome Cronobacter spp. agar. All five isolates were phenotypically characterized as gram negative, oxidase negative, catalase positive rods, forming yellow colonies on TSA at 22°C. Of these five samples that gave positive results in CSB after pre-enrichment, four gave no growth on VRBG agar, suggesting the presumptive Cronobacter spp. may have been stressed6). Stressed or injured cells may not grow on selective media and require recovery via an enrichment step20). This demonstrates that culture-based methods of hygiene determination may give misleading results. Therefore molecular methods are recommended, although they do not give an indication of the viability of the organisms detected, and should be used in combination with culture-based methods18).
The Remel RapID ONE kit identified both reference strains, and four isolates (buffalo worm, cricket flour, giant waterbug, queen leafcutter ant) as C. sakazakii. The mealworm isolate was identified as Pantoea agglomerans (Table 2). Non-Cronobacter spp. would generally be differentiated on selective chromogenic agar20,21). This therefore suggests misidentification of the mealworm isolate.
Table 2. Biochemical identification of reference strains and insect isolates.
| Sample | Remel RapID ONE identification (% probability) |
| NCIMB 8272 | Cronobacter sakazakii (99.9%) |
| NCIMB 5920 | Cronobacter sakazakii (99.9%) |
| Buffalo Worms | Cronobacter sakazakii (96.25%) |
| Cricket Flour | Cronobacter sakazakii (99.9%) |
| Giant Waterbug | Cronobacter sakazakii (99.9%) |
| Mealworms | Pantoea agglomerans (99.9%) |
| Queen Leafcutter Ants | Cronobacter sakazakii (99.9%) |
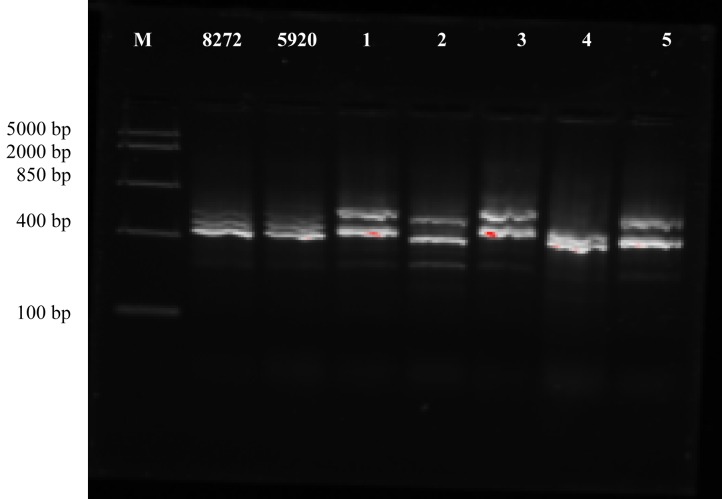
DNA sequence-based methods, such as 16S rDNA sequencing, are considered to be the most reliable for identifying and confirming Cronobacter spp.22). However, they are not always feasible, due to factors such as cost and time22,23). Other PCR-based methods are therefore an alternative. ITS-PCR allows discrimination at species level, due to variations in the ITS region within a genus24). Results of the ITS-PCR (Fig. 1) showed that both C. sakazakii reference strains had similar band patterns. None of the isolates exhibited similar band patterns as the C. sakazakii reference strains, suggesting they might not be C. sakazakii, but instead other species of Cronobacter. In addition, the mealworm, queen leafcutter ant and giant waterbug isolates appeared to have a similar genetic identity.
Fig. 1.
ITS-PCR band profiles of reference strains NCIMB 8272, NCIMB 5920 and insect isolates. M, marker; 1, mealworm; 2, buffalo worm; 3, queen leafcutter ant; 4, cricket flour; 5, giant waterbug.
The five isolates may therefore be other species of Cronobacter, but this did not correspond with the Remel RapID ONE identification, further suggesting misidentification of the isolates. Misidentification by the Remel RapID ONE system is a possibility because the subjective nature of the results of such biochemical kits makes them limited in their reliability25,26). Furthermore, the Remel RapID ONE system only has C. sakazakii on its database, meaning that other Cronobacter species cannot be reliably identified.
Presumptive Cronobacter spp. were detected in samples of ready-to-eat insects. Nevertheless, the risk of infection could be considered low for healthy individuals. Immunocompromised persons and the elderly may need to exercise caution when consuming ready-to-eat edible insects. ITS-PCR was useful for genotypic characterisation of the isolates. However, reference strains of other Cronobacter species should be included for effective identification of isolates. More studies that include a wider range of products from different companies are recommended.
Acknowledgments
The authors acknowledge the support from a Student Placement Scholarship grant awarded by the Society for Applied Microbiology, which funded JPG’s summer placement to carry out the research.
Abbreviations: C. sakazakii: Cronobacter sakazakii, PCR: polymerase chain reaction
Footnotes
Conflict of Interest: The authors have no conflict of interest.
References
- 1.Dobermann D,Swift JA,Field LM. Opportunities and hurdles of edible insects for food and feed. Nutr Bull. 2017; 42(4): 293–308. 10.1111/nbu.12291 [DOI] [Google Scholar]
- 2.Belluco S,Losasso C,Maggioletti M,Alonzi C,Ricci A,Paoletti MG. Edible insects: a food security solution or a food safety concern? Animal Front. 2015; 5(2): 25–30. 10.2527/af.2015-0016 [DOI] [Google Scholar]
- 3.Klunder HC,Wolkers-Rooijackers J,Korpela JM,Nout MJR. Microbiological aspects of processing and storage of edible insects. Food Control. 2012; 26(2): 628–631. 10.1016/j.foodcont.2012.02.013 [DOI] [Google Scholar]
- 4.Grabowski NT,Klein G. Microbiological analysis of raw edible insects. Journal of Insects as Food and Feed. 2017; 3(1): 7–14. 10.3920/JIFF2016.0004 [DOI] [Google Scholar]
- 5.Miranda N,Banerjee P,Simpson S,Kerdahi K,Sulaiman I. Molecular surveillance of Cronobacter spp. isolated from a wide variety of foods from 44 different countries by sequence typing of 16S rRNA, rpoB and O-antigen genes. Foods. 2017; 6(5): 36 10.3390/foods6050036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandão MLL,Umeda NS,Jackson E,Forsythe SJ,de Filippis I. Isolation, molecular and phenotypic characterization, and antibiotic susceptibility of Cronobacter spp. from Brazilian retail foods. Food Microbiol. 2017; 63: 129–138. 10.1016/j.fm.2016.11.011 [DOI] [PubMed] [Google Scholar]
- 7.Hochel I,Růžičková H,Krásný L,Demnerová K. Occurrence of Cronobacter spp. in retail foods. J Appl Microbiol. 2012; 112(6): 1257–1265. 10.1111/j.1365-2672.2012.05292.x [DOI] [PubMed] [Google Scholar]
- 8.Kalyantanda G,Shumyak L,Archibald LK. Cronobacter species contamination of powdered infant formula and the implications for neonatal health. Front Pediatr. 2015; 3: 56 10.3389/fped.2015.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton JV,Lehane MJ,Braig HR. Isolation of Enterobacter sakazakii from midgut of Stomoxys calcitrans. Emerg Infect Dis. 2003; 9(10): 1355–1356. 10.3201/eid0910.030218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osimani A,Milanović V,Cardinali F,et al. . The bacterial biota of laboratory-reared edible mealworms (Tenebrio molitor L.): From feed to frass. Int J Food Microbiol. 2018; 272: 49–60. 10.1016/j.ijfoodmicro.2018.03.001 [DOI] [PubMed] [Google Scholar]
- 11.Vandeweyer D,Crauwels S,Lievens B,Van Campenhout L. Metagenetic analysis of the bacterial communities of edible insects from diverse production cycles at industrial rearing companies. Int J Food Microbiol. 2017; 261: 11–18. 10.1016/j.ijfoodmicro.2017.08.018 [DOI] [PubMed] [Google Scholar]
- 12.Grabowski NT,Klein G. Microbiology of cooked and dried edible Mediterranean field crickets (Gryllus bimaculatus) and superworms (Zophobas atratus) submitted to four different heating treatments. Food Sci Technol Int. 2017; 23(1): 17–23. 10.1177/1082013216652994 [DOI] [PubMed] [Google Scholar]
- 13.Xu Y. Aspects of modeling variability of single cell lag time for Cronobacter spp. after exposure to sublethal heat treatment in normal and stressful environments. [PhD thesis]. London: London Metropolitan University; 2014.
- 14.Iversen C,Druggan P,Schumacher S,et al. . Development of a novel screening method for the isolation of “Cronobacter” spp. (Enterobacter sakazakii). Appl Environ Microbiol. 2008; 74(8): 2550–2553. 10.1128/AEM.02801-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iversen C,Druggan P Culture media for isolating Cronobacter species (Enterobacter sakazakii). In: Corry JEL, Curtis GDW, Baird RM, eds. Handbook of Culture Media for Food and Water Microbiology, 3rd ed. Cambridge, UK: Royal Society of Chemistry; 2012: 357-376.
- 16.Polit A,Yang H,Amund D. Investigating the transmissibility of tet(W) in bifidobacteria exposed to acid and bile stress. Biosci Microbiota Food Health. 2018; 37(2): 39–43. 10.12938/bmfh.17-017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenaerts S,Van Der Borght M,Callens A,Van Campenhout L. Suitability of microwave drying for mealworms (Tenebrio molitor) as alternative to freeze drying: Impact on nutritional quality and colour. Food Chem. 2018; 254: 129–136. 10.1016/j.foodchem.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 18.Garofalo C,Osimani A,Milanović V,et al. . The microbiota of marketed processed edible insects as revealed by high-throughput sequencing. Food Microbiol. 2017; 62: 15–22. 10.1016/j.fm.2016.09.012 [DOI] [PubMed] [Google Scholar]
- 19.Caparros Megido R,Desmedt S,Blecker C,et al. . Microbiological load of edible insects found in Belgium. Insects. 2017; 8(1): 12 10.3390/insects8010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Druggan P,Iversen C. Culture media for the isolation of Cronobacter spp. Int J Food Microbiol. 2009; 136(2): 169–178. 10.1016/j.ijfoodmicro.2009.09.008 [DOI] [PubMed] [Google Scholar]
- 21.Teramura H,Fukuda N,Okada Y,Ogihara H. Comparison of chromogenic selective media for the detection of Cronobacter spp. (Enterobacter sakazakii). Biocontrol Sci. 2018; 23(1): 27–33. 10.4265/bio.23.27 [DOI] [PubMed] [Google Scholar]
- 22.Jaradat ZW,Ababneh QO,Saadoun IM,Samara NA,Rashdan AM. Isolation of Cronobacter spp. (formerly Enterobacter sakazakii) from infant food, herbs and environmental samples and the subsequent identification and confirmation of the isolates using biochemical, chromogenic assays, PCR and 16S rRNA sequencing. BMC Microbiol. 2009; 9(1): 225 10.1186/1471-2180-9-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson EE,Forsythe SJ. Comparative study of Cronobacter identification according to phenotyping methods. BMC Microbiol. 2016; 16(1): 146 10.1186/s12866-016-0768-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu YZ,Anyogu A,Ouoba LII,Sutherland JP. Genotypic characterization of Brochothrix spp. isolated from meat, poultry and fish. Lett Appl Microbiol. 2010; 51(3): 245–251. 10.1111/j.1472-765X.2010.02886.x [DOI] [PubMed] [Google Scholar]
- 25.Becker B,Weiss C,Holzapfel WH. An evaluation of the use of three phenotypic test-systems for biochemical identification of Enterobacteriaceae and Pseudomonadaceae. Food Control. 2009; 20(9): 815–821. 10.1016/j.foodcont.2008.10.014 [DOI] [Google Scholar]
- 26.Cetinkaya E,Joseph S,Ayhan K,Forsythe SJ. Comparison of methods for the microbiological identification and profiling of Cronobacter species from ingredients used in the preparation of infant formula. Mol Cell Probes. 2013; 27(1): 60–64. 10.1016/j.mcp.2012.10.003 [DOI] [PubMed] [Google Scholar]