Abstract
Individual virions typically fail to infect cells. Such decoupling between virions and infectious units is most evident in multicomponent and other segmented viruses, but is also frequent in non-segmented viruses. Despite being a well-known observation, the causes and implications of low single-virion infectivity often remain unclear. In principle, this can originate from intrinsic genetic and/or structural virion defects, but also from host infection barriers that limit early viral proliferation. Hence, viruses may have evolved strategies to increase the per-virion likelihood of establishing successful infections. This can be achieved by adopting spread modes that elevate the multiplicity of infection at the cellular level, including direct cell-to-cell viral transfer, encapsulation of multiple virions in microvesicles or other intercellular vehicles, virion aggregation, and virion binding to microbiota. In turn, increasing the multiplicity of infection could favor the evolution of defective viruses, hence modifying the fitness value of these spread modes.
One is not enough: poor infectivity of individual virions
The virion or viral particle has been traditionally viewed as the minimal viral infectious unit. However, typically the majority of virions are non-infectious. Viral titers obtained by standard methods such as the plaque assay or the median infectious dose can be tens or even hundreds of times lower than the actual number of viral particles in a given suspension. This deviation can be measured using the particle to plaque forming unit (PFU) ratio [1]. High particle-to-PFU ratios are often attributed to lack of some genetic material inside the virion, structural defects in the capsid and/or envelope, or lethal mutations.
Segmented viruses are particularly prone to non-infectiousness. In principle, one may expect genome packaging processes to ensure the incorporation of all segments in each virion. Tight control of segment encapsidation has been indeed reported in some viruses such as bacteriophage ϕ6 [2], yet other viruses surprisingly appear to package segments quasi-randomly. For instance, in Rift Valley fever virus, FISH analysis of virions and infected cells revealed that up to 90% of viral particles lack at least one of the three segments, despite each segment being essential [3]. Another example of such apparent lack of regulation is provided by birnaviruses [4]. This issue is aggravated in multipartite viruses, because each segment is packaged in a different particle and, presumably, a high multiplicity of infection (MOI) is necessary for productive infection [5,6].
In recent years, the causes of influenza virus high particle-to-PFU ratios have been investigated in some detail. For instance, single-cell analysis and stochastic modelling suggested that up to 90% of cells infected by a single viral particle produce little or no progeny [7]. Similarly, it was found that approximately 90% of the particles fail to express at least one segment [8]. These appear to differ from classical defective interfering particles (DIPs) [9] in that they initiate cellular infection but fail to complete it, and have been termed semi-infectious particles (SIPs) [10]. Influenza virus infectivity increases strongly when the MOI is high enough to ensure coinfection of cells with multiple SIPs. However, surprisingly, the presence of SIPs does not seem to require a high MOI. This suggests that SIPs frequently appear de novo, or that they propagate across cells in association with other particles. The relevance of DIPs and/or SIPs is supported by in vivo work showing that influenza virus particles that lack segments can undergo multiple infection cycles in the upper respiratory tract of guinea pigs [11]. Sequencing of nasopharyngeal specimens from infected humans indicated frequent DIP production and suggested that DIPs can undergo inter-host transmission [12].
Per-virion cell invasion efficiencies have also been suggested to be unexpectedly low in non-segmented viruses, such as tomato mosaic virus (ToMV) undergoing cell-to-cell spread through plasmodesmata. ToMV was labeled with sequence tags or fluorescent markers to quantify the population bottleneck experienced during transfer between cells [13]. Whereas plasmodesmata should allow for the passage of hundreds or thousands of viral genomes, the authors suggested that the vast majority of these genomes fail to give progeny, and that each cellular infection is effectively initiated by only 2-7 viral genomes, on average. This sieving is a poorly understood phenomenon and could obey to lack of infectivity, but also to competition or even to altruistic interactions among viral genomes at the intracellular level [14]. Another study with vaccinia virus used microfluidics to place a specific number of viral particles in individual cells [15]. Most cells receiving a single particle were uninfected, whereas infection probability increased disproportionately (logistically) with the number of particles placed per cell, suggesting a cooperative initiation of the infection cycle.
Spatial clutering and MOIs
A monodisperse viral population (i.e. showing no spatial structure) will fail to reach sufficiently high MOIs during the early stages of population growth. This is because typically only a minuscule fraction of particles present in a given individual host colonize new hosts [16,17], and bottlenecks also operate at the intra-host level, as revealed by sequence analysis of well-studied pathogens such as HIV-1, influenza A virus, and hepatitis C virus [18–20]. Importantly, though, high MOIs are reached much earlier if the population exhibits spatial clustering. As a result of the diffusion process of free viral particles, most virus grow in the form of infection foci. Even stronger clustering can be achieved if the virus uses cell-to-cell spread, which allows for direct transfer of multiple viral genomes between cells and has been described in many viruses including most plant viruses, HIV-1, human T-cell leukemia virus, measles virus, vaccinia virus, and herpes virus [21,22]. However, in most cases, this spread mode is local, and systemic dissemination probably relies on free virions. Therefore, the high-MOI regime would be interrupted during systemic dissemination and inter-host transmission, purging out semi/non-infectious particles. In some cases, though, the cell-to-cell mode may also operate during systemic dissemination, notably in the case of blood-borne viruses such as HIV-1 [21,23]. Inter-host transmission in a cell-associated manner is an understudied process, and may be more common than often assumed. Again, a well-studied case is HIV-1, for which the cell-associated route is known to contribute significantly to inter-host transmission [24].
Despite a likely role of limited diffusion and cell-to-cell spread in the maintenance of semi/non-infectious particles, it was inferred in cell cultures and in humanized mice that cells co-infected with GFP- and mCherry-encoding HIV-1 had only a 6-14% chance of transferring both variants by the cell-to-cell route using virological synapses [25]. Although this was far more efficient than free virion-dependent coinfection, these data suggest that cell-to-cell spread may not allow for the sustained co-transmission of different virus variants throughout multiple cycles. However, further work is required in this area.
Collective infectious units as coinfection vehicles
If high cellular MOIs help overcome the low infectivity of individual particles, viruses might benefit from maintaining relatively high MOIs even in the presence of the strong population bottlenecks associated with dispersal. As outlined above, the case of multi-partite viruses is particularly extreme. Since very high viral population densities would be required for ensuring that a full set of independently diffusing segments is delivered to at least a fraction of cells, there should be mechanisms leading to the linked spread of segments, at least for viruses with more than three segments [5,6]. One possible such mechanism is inter-segment RNA-RNA interactions [26]. Interestingly, packaging does not appear to be necessary for systemic dissemination of brome mosaic virus, since uncoated viral RNAs can move long distances, probably in the form of ribonucleoprotein complexes involving cellular factors and the viral movement protein [27]. Systemic dissemination in the form of ribonucleoproteins has also been shown for potato mop top virus [28]. In some plant species, not all viral RNA segments are required for spread at the intra-host level, and it is therefore likely that the RNA-RNA-protein interactions mediating physical segment linkage involve only a subset of segments, mainly those encoding the replication machinery and other essential factors. However, this leaves unanswered the problem of how multipartite viruses undergo inter-host transmission, as this stage necessitates virions and the concurrence of all segments.
Polyploidy might be yet another strategy for increasing the chances of successful cellular infection. In segmented viruses, aneuploidy can be seen as a trivial consequence of non-selective segment encapsidation. However, and more interestingly, polyploidy might serve as a strategy to increase infectivity. This was first studied using infectious bursal disease virus [4]. The size of the icosahedral capsid of this virus is larger than required for packaging just one copy of each of the two segments, and can easily accommodate two copies of each. This might compensate for non-selective packaging. With room for only two RNA molecules and random packaging, 50% of the capsids would miss one of the two essential segments and hence would not be infectious. In contrast, with room for four RNA molecules, this chance drops to 12.5%. Similar findings were later reported in infectious pancreatic necrosis virus, another birnavirus [29]. Interestingly, polyploidy has also been shown in non-segmented viruses, where the problem of ensuring a full set of segments obviously does not exist. This includes filamentous viruses such as bacteriophage f1 [30] and Ebola virus [31], in which capsids are capable of accommodating extra genetic material, but also measles virus [32], which forms particles containing a flexible helicoidal nucleocapsid surrounded by an external spherical envelope.
The notion that viral intra-host spread and inter-host transmission rely on independently diffusing virions has been further challenged by the discovery of structures that mediate the collective transfer of groups of virions to the same target cells and, thus, increase the MOI at the cellular level [33,34]. Virion aggregation was reported long ago in electronic microscopy studies using a variety of viruses including tobacco mosaic virus, poxviruses, influenza virus, rhabdoviruses, and poliovirus, but was often interpreted as a laboratory artifact [33]. Recently, it has been shown that virion aggregates can be infectious, mediate the co-delivery of multiple viral genome copies to the same cell, and allow for functional interactions between different genetic variants in vesicular stomatitis virus [35] and poliovirus [36]. Other studies have detected aggregates, yet have not investigated their role in infection. For instance, examination of Junin virus by fluorescence-assisted flow virometry revealed three types of particles with different size: small (60 nm) and large (150 nm) individual particles, and aggregates. Large particles were more infectious than small particles because they were more likely to contain two essential components (the surface protein G and RNA), yet the infectivity of aggregates is unclear [37].
Lipid microvesicles constitute another instance of collective infectious units. In addition to releasing free virions by lysis, cells infected with enteroviruses such as poliovirus, coxsackie virus and rhinovirus secrete autophagosome-like vesicles of 200-400 nm containing multiple viral particles [38]. These vesicles, released before lysis, are internalized by recipient cells and are highly infectious. However, the actual number of genomes delivered by these vesicles to host cells, as well as the fraction of these genomes that effectively initiate infection, remain unclear. Whether vesicles are important for inter-host transmission is another open question. Expanding the role of lipid vesicles in viral spread, it has been found that the large DNA marseilleviruses can enter their amoebal hosts by two alternative routes: endocytosis of free virions and phagocytosis of multi-virion vesicles [39].
Collective modes of viral spread can take additional forms. A well-known case is that of baculoviruses, which undergo inter-host transmission inside polyhedrin crystals that wrap up tens of virions (occlusion bodies). These crystals are dissolved under alkaline pH in the insect mid gut following their ingestion and release occlusion-derived viruses. In turn, in so-called multiple baculoviruses, occlusion-derived viruses are made of small aggregates of a few virions that share an external envelope, but the significance of this additional level of aggregation remains unclear [40].
Finally, it has been recently shown that enteroviruses bind bacterial cells of the gut microbiota, which leads to clustering of viral particles in space and hence constitutes yet another way of co-delivering multiple particles to the same target cells [41]. In the future, similar processes might be unveiled in other medically relevant fecal-orally transmitted viruses, such as noroviruses and rotaviruses.
Pros and cons of collective spread
The selective pressures (if any) that have promoted high-MOI spread modes remain largely unexplored. Two types of benefit can be envisaged: “safety in numbers” (or “mass effect”), whereby the probability of successful infection increases in cells receiving multiple genome copies even if these are identical, and “heterotypic cooperation” whereby the advantage of collective infection resides precisely in bringing together different genetic variants (for instance, by genetic complementation). Most of the above discussion revolves around the more popular view that heterotypic cooperation is responsible for increasing per-virion infectivity at high MOIs. However, the mass effect seems also plausible and should probably be considered as a null hypothesis because it relies only on quantity, whereas heterotypic cooperation relies on both quantity and quality. One possible explanation for this mass effect could be that innate immunity is overwhelmed in cells receiving multiple particles. Another possible scenario is that, during the very early stages of infection, such as translation or transcription of the first incoming molecules, viruses are subject to stochastic failures due to lack of some required cellular components, dilution, and/or degradation. The risks of such stochastic loss would be diminished by initiating infection with multiple copies of each element.
The higher infectivity of collective infectious units is supported by some empirical evidence. For instance, in infectious bursal disease virus, it was found that polyploid capsids are more infectious than haploid capsids [4], which could arguably be explained in terms of complementation. In enteroviruses, it has been found that cells inoculated with virion-containing vesicles produce more progeny than those inoculated with free virions in the short term [38]. However, whether this was due to a mass effect, heterotypic cooperation, or other processes (such as different entry cellular entry route, for instance) remains unknown. Other experiments support the heterotypic cooperation hypothesis. For instance, it was found that coencapsidation of measles virus genomes allowed complementation of deleterious mutations, and even favored the emergence of new phenotypes such as extended cell tropism [42]. Baculovirus occlusion bodies contain genetically heterogeneous genomes, which have been suggested to act cooperatively [43,44]. However, in most cases how heterotypic cooperation occurs remains poorly understood mechanistically. Several possibilities exist, though. As shown for coxsackievirus [45], some beneficial mutations cannot be combined in the same genome because they exhibit strongly negative epistasis, but this antagonism may disappear if mutations are present in different genomes. Cooperation might also involve division of labor. In influenza A virus, it was found that the hemagglutinin variant of one strain provided efficient attachment to host cells, whereas the neuraminidase variant from another strain provided efficient release of virions from cells. Sharing of viral proteins in cells infected by both strains led to faster viral spread than in singly infected cells [46].
Collective spread also entails costs, because pooling multiple virions (or genomes in the case of polyploids) in the same structure reduces dispersal. K virions encapsulated in a single unit will reach one cell at best, whereas free virions could reach up to K cells. Hence, for collective infectious units to be selectively advantageous, they should increase progeny production on a per-particle basis. This means that, on average, K free virions should produce less progeny than a collective unit made of K virions. However, whether this condition is fulfilled remains unassessed. An additional possible downside of collective spread is that genetically heterogeneous groups could lead to the evolution of social cheaters, i.e. variants that benefit from others without reciprocating. Indeed, semi/non-infectious particles might not necessarily be the causal factor promoting the evolution of mechanisms for increasing MOIs locally but, instead, many of these particles could be cheaters thriving as a result of such mechanisms. For instance, baculovirus populations typically contain multiple genetic variants, including defective genomes with large deletions [44,47,48], but whether these are cooperative variants or social cheaters is unclear. A fitness cost has been demonstrated for factors involved in host-to-host transmission, such as per oral infection factors [43].This suggests that mutants defective for these genes are selectively advantageous at the intra-host level, yet have to use functional viruses for transmission, fitting the definition of social cheaters.
Conclusions
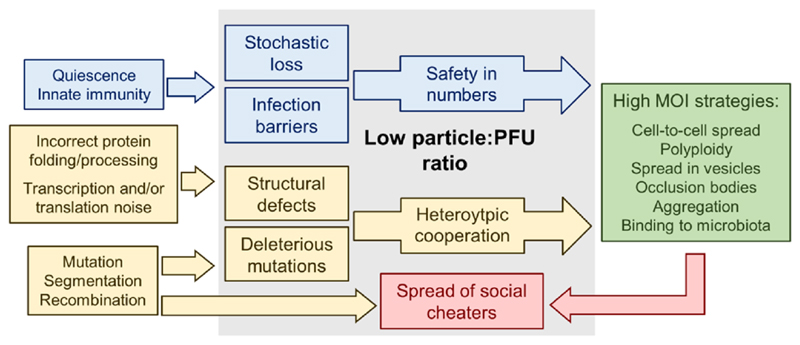
The concepts discussed are schematically outlined in Figure 1. High particle:PFU ratios are often viewed as the result of an intrinsically poor infectivity with a structural or genetic origin. However, infectivity is not absolute because it depends at least on two extrinsic factors: the host and the presence of other viruses. First, it is well known that a given viral stock can show widely different specific infectivity values, depending on the host cells assayed. Second, defects in one virion can be compensated by other virions entering the same cell. Yet, the per-particle chance of successful infection may increase in groups even if these contain identical virions. Whether collective infection increases viral fitness by promoting heterotypic cooperation, mass effects, or other processes remains an opened question. In either case, these interactions depend on a high cellular MOI. This can be achieved simply via spatial clustering of free virions or by adopting a spread mode in which virions do not diffuse independently. It is also well known that high MOIs relax competition and promote the maintenance of low-fitness variants, or even DIPs, reducing average viral fitness. Therefore, there might be mechanisms preventing access of such non-cooperative variants to collective infectious units. These mechanisms are currently unknown, and could vary depending on how viral particles cluster. For instance, if grouping occurs before cell exit, such as in vesicles, cooperator assortment might be achieved by compartmentalization of viruses within discrete viral factories.
Figure 1. Summary of possible cause-effect relationships between poor virion infectivity and viral spread modes that elevate the MOI.
See text for details.
Acknowledgments
This work was financially supported by ERC Consolidator Grant 724519 (Vis-a-Vis).
References
- 1.Klasse PJ. Molecular determinants of the ratio of inert to infectious virus particles. Prog Mol Biol Transl Sci. 2015;129:285–326. doi: 10.1016/bs.pmbts.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol Mol Biol Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wichgers Schreur PJ, Kortekaas J. Single-molecule FISH reveals non-selective packaging of Rift Valley Fever virus genome segments. PLoS Pathog. 2016;12:e1005800. doi: 10.1371/journal.ppat.1005800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luque D, Rivas G, Alfonso C, Carrascosa JL, Rodríguez JF, Castón JR. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci USA. 2009;106:2148–2152. doi: 10.1073/pnas.0808498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sicard A, Michalakis Y, Gutierrez S, Blanc S. The strange lifestyle of multipartite viruses. PLoS Pathog. 2016;12:e1005819. doi: 10.1371/journal.ppat.1005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iranzo J, Manrubia SC. Evolutionary dynamics of genome segmentation in multipartite viruses. Proc Biol Sci. 2012;279:3812–3819. doi: 10.1098/rspb.2012.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heldt FS, Kupke SY, Dorl S, Reichl U, Frensing T. Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection. Nat Commun. 2015;6 doi: 10.1038/ncomms9938. 8938. [••Used single-cell analysis to demonstrate that most cells infected by individual particles of influenza A virus fail to produce viruses, and suggested that this was in part due to early stochastic loss of components essential to infection] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, Bennink JR, Yewdell JW. Most influenza a virions fail to express at least one essential viral protein. J Virol. 2013;87:3155–3162. doi: 10.1128/JVI.02284-12. [•Proposed the concept of semi-infectious particles] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marriott AC, Dimmock NJ. Defective interfering viruses and their potential as antiviral agents. Rev Med Virol. 2010;20:51–62. doi: 10.1002/rmv.641. [DOI] [PubMed] [Google Scholar]
- 10.Brooke CB. Population diversity and collective interactions during influenza virus infection. J Virol. 2017;91:e01164–17. doi: 10.1128/JVI.01164-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooke CB, Ince WL, Wei J, Bennink JR, Yewdell JW. Influenza A virus nucleoprotein selectively decreases neuraminidase gene-segment packaging while enhancing viral fitness and transmissibility. Proc Natl Acad Sci USA. 2014;111:16854–16859. doi: 10.1073/pnas.1415396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saira K, Lin X, DePasse JV, Halpin R, Twaddle A, Stockwell T, Angus B, Cozzi-Lepri A, Delfino M, Dugan V, et al. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J Virol. 2013;87:8064–8074. doi: 10.1128/JVI.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyashita S, Ishibashi K, Kishino H, Ishikawa M. Viruses roll the dice: the stochastic behavior of viral genome molecules accelerates viral adaptation at the cell and tissue levels. PLoS Biol. 2015;13:e1002094. doi: 10.1371/journal.pbio.1002094. [•Cell-to-cell transfer of tomato mosaic virus through plasmodesmata surprisingly imposes a strong genetic bottleneck] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Díaz-Munoz SL, Sanjuán R, West S. Sociovirology: conflict, cooperation, and communication among viruses. Cell Host Microbe. 2017;22:437–441. doi: 10.1016/j.chom.2017.09.012. [•Discussion of how social evolution concepts apply to viruses] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stiefel P, Schmidt FI, Dorig P, Behr P, Zambelli T, Vorholt JA, Mercer J. Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett. 2012;12:4219–4227. doi: 10.1021/nl3018109. [••The probability of successful infection increases disproportionately with the number of vaccinia virions entering a cell] [DOI] [PubMed] [Google Scholar]
- 16.Zwart MP, Elena SF. Matters of size: genetic bottlenecks in virus infection and their potential impact on evolution. Annu Rev Virol. 2015;2:161–179. doi: 10.1146/annurev-virology-100114-055135. [DOI] [PubMed] [Google Scholar]
- 17.Gutiérrez S, Michalakis Y, Blanc S. Virus population bottlenecks during within-host progression and host-to-host transmission. Curr Opin Virol. 2012;2:546–555. doi: 10.1016/j.coviro.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Richard M, Herfst S, Tao H, Jacobs NT, Lowen AC. Influenza A virus reassortment is limited by anatomical compartmentalization following co-infection via distinct routes. J Virol. 2017 doi: 10.1128/JVI.02063-17. (ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandenberghe Y, Morel F, Foriers A, Ketterer B, Vercruysse A, Guillouzo A, Rogiers V. Effect of phenobarbital on the expression of glutathione S-transferase isoenzymes in cultured rat hepatocytes. FEBS Lett. 1989;251:59–64. doi: 10.1016/0014-5793(89)81428-0. [DOI] [PubMed] [Google Scholar]
- 20.Salemi M, Rife B. Phylogenetics and phyloanatomy of HIV/SIV intra-host compartments and reservoirs: the key role of the central nervous system. Curr HIV Res. 2016;14:110–120. doi: 10.2174/1570162x13666151029102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mothes W, Sherer NM, Jin J, Zhong P. Virus cell-to-cell transmission. J Virol. 2010;84:8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pais-Correia AM, Sachse M, Guadagnini S, Robbiati V, Lasserre R, Gessain A, Gout O, Alcover A, Thoulouze MI. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat Med. 2010;16:83–89. doi: 10.1038/nm.2065. [•Extracellular matrix components are used to concentrate virions at the cellular surface and facilitate transfer to neighbor cells] [DOI] [PubMed] [Google Scholar]
- 23.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, Tager AM, Luster AD, Mempel TR. HIV-infected T cells are migratory vehicles for viral dissemination. Nature. 2012;490:283–287. doi: 10.1038/nature11398. [•Demonstrates the importance of cell-to-cell transfer for systemic spread of a blood-borne virus] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson DJ, Le Grand R. Cell-associated HIV mucosal transmission: the neglected pathway. J Infect Dis. 2014;210(Suppl 3):S606–608. doi: 10.1093/infdis/jiu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law KM, Komarova NL, Yewdall AW, Lee RK, Herrera OL, Wodarz D, Chen BK. In vivo HIV-1 cell-to-cell transmission promotes multicopy micro-compartmentalized infection. Cell Rep. 2016;15:2771–2783. doi: 10.1016/j.celrep.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 26.Dall'Ara M, Ratti C, Bouzoubaa SE, Gilmer D. Ins and outs of multipartite positive-strand RNA plant viruses: packaging versus systemic Spread. Viruses. 2016;8:E228. doi: 10.3390/v8080228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopinath K, Kao CC. Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell. 2007;19:1179–1191. doi: 10.1105/tpc.107.050088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savenkov EI, Germundsson A, Zamyatnin AA, Sandgren M, Jr, Valkonen JP. Potato mop-top virus: the coat protein-encoding RNA and the gene for cysteine-rich protein are dispensable for systemic virus movement in Nicotiana benthamiana. J Gen Virol. 2003;84:1001–1005. doi: 10.1099/vir.0.18813-0. [DOI] [PubMed] [Google Scholar]
- 29.Lago M, Rodríguez JF, Bandin I, Dopazo CP. Aquabirnavirus polyploidy: A new strategy to modulate virulence? J Gen Virol. 2016;10 doi: 10.1099/jgv.0.000434. [DOI] [PubMed] [Google Scholar]
- 30.López J, Webster RE. Morphogenesis of filamentous bacteriophage f1: orientation of extrusion and production of polyphage. Virology. 1983;127:177–193. doi: 10.1016/0042-6822(83)90382-3. [DOI] [PubMed] [Google Scholar]
- 31.Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS ONE. 2012;7:e29608. doi: 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rager M, Vongpunsawad S, Duprex WP, Cattaneo R. Polyploid measles virus with hexameric genome length. EMBO J. 2002;21:2364–2372. doi: 10.1093/emboj/21.10.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanjuán R. Collective infectious units in viruses. Trends Microbiol. 2017;22:402–412. doi: 10.1016/j.tim.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altan-Bonnet N. Extracellular vesicles are the Trojan horses of viral infection. Curr Opin Microbiol. 2016;32:77–81. doi: 10.1016/j.mib.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuevas JM, Durán-Moreno M, Sanjuán R. Emergence of supra-virion infectious units from free viral particles in an enveloped virus. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.78. 17078. [•Shows that certain fluids such as saliva promote virion aggregation, permitting the joint transfer of multiple viral genomes to cells] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguilera ER, Erickson AK, Jesudhasan PR, Robinson CM, Pfeiffer JK. Plaques formed by mutagenized viral populations have elevated coinfection frequencies. MBio. 2017;8:e02020–02016. doi: 10.1128/mBio.02020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudin RB, NS Sorting of small infectious virus particles by flow virometry reveals distinct infectivity profiles. Nat Commun. 2015;6 doi: 10.1038/ncomms7022. 6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [•Shows that extracellular vesicles can be used for the joint transfer of multiple virions between cells] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arantes TS, Rodrigues RA, Dos Santos Silva LK, Oliveira GP, de Souza HL, Khalil JY, de Oliveira DB, Torres AA, da Silva LL, Colson P, et al. The large Marseillevirus explores different entry pathways by forming giant infectious vesicles. J Virol. 2016;90:5246–5255. doi: 10.1128/JVI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohrmann GF. Baculovirus nucleocapsid aggregation (MNPV vs SNPV): an evolutionary strategy, or a product of replication conditions? Virus Genes. 2014;49:351–357. doi: 10.1007/s11262-014-1113-5. [DOI] [PubMed] [Google Scholar]
- 41.Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe. 2018;23:77–88 e75. doi: 10.1016/j.chom.2017.11.007. [•Shows that enteric bacteria can function as attractors of viral particles, promoting their joint transfer to cells] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirogane Y, Watanabe S, Yanagi Y. Cooperation between different RNA virus genomes produces a new phenotype. Nat Commun. 2012;3 doi: 10.1038/ncomms2252. 1235. [DOI] [PubMed] [Google Scholar]
- 43.Simon O, Williams T, Cerutti M, Caballero P, Lopez-Ferber M. Expression of a peroral infection factor determines pathogenicity and population structure in an insect virus. PLoS ONE. 2013;8:e78834. doi: 10.1371/journal.pone.0078834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clavijo G, Williams T, Muñoz D, Caballero P, López-Ferber M. Mixed genotype transmission bodies and virions contribute to the maintenance of diversity in an insect virus. Proc Biol Sci. 2010;277:943–951. doi: 10.1098/rspb.2009.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bordería AV, Isakov O, Moratorio G, Henningsson R, Aguera-González S, Organtini L, Gnadig NF, Blanc H, Alcover A, Hafenstein S, et al. Group selection and contribution of minority variants during virus adaptation determines virus fitness and phenotype. PLoS Pathog. 2015;11:e1004838. doi: 10.1371/journal.ppat.1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue KSH KA, Ollodart AR, Dingens AS, Bloom JD. Cooperation between distinct viral variants promotes growth of H3N2 influenza in cell culture. Elife. 2016;5:e13974. doi: 10.7554/eLife.13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon O, Williams T, Caballero P, López-Ferber M. Dynamics of deletion genotypes in an experimental insect virus population. Proc Biol Sci. 2006;273:783–790. doi: 10.1098/rspb.2005.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chateigner A, Bezier A, Labrousse C, Jiolle D, Barbe V, Herniou EA. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses. 2015;7:3625–3646. doi: 10.3390/v7072788. [DOI] [PMC free article] [PubMed] [Google Scholar]