Abstract
Background
Null mutations within the filaggrin gene (FLG) are associated with moderate-severe atopic eczema; their role in the mild-moderate eczema in the general population is unknown.
Objective
To investigate the significance of five common FLG null mutations in childhood atopic eczema in an unselected population cohort.
Methods
811 English children aged 7-9 years were screened for FLG mutations. Eczema cases were defined by UK diagnostic criteria and skin examination. Asthma and seasonal rhinitis cases were defined by parental questionnaire. Association between phenotype and genotype was investigated using Fisher’s exact test and logistic regression analysis.
Results
The 12-month period prevalence of atopic eczema was 24.2% (95% confidence interval 21.2-27.2%) with 96% (115/120) of cases having mild-moderate disease. The combined null genotype (carriage of one or more FLG mutations) was significantly associated with atopic eczema (p=1.2x10-4). The odds ratio for individuals carrying two null mutations was 26.9 (95% CI 3.3-217.1), but heterozygote carriers showed no significant increase in risk (odds ratio 1.2, 95% confidence interval 0.7-1.9). 8/190 (4.2%) of eczema cases carried two FLG null mutations and thus may be attributed to filaggrin deficiency. Asthma in the context of eczema showed significant association with the FLG null mutations (p=7.1x10-4, OR for carriers of two mutant alleles 11.9, 95% CI 3.1-45.6). There was no association of FLG with asthma independent of eczema (p=0.15) and no association with seasonal rhinitis (p=0.66).
Conclusion
FLG null mutations are significantly associated with mild-moderate atopic eczema in childhood with a recessive pattern of inheritance.
Keywords: Asthma, seasonal rhinitis, atopic eczema, complex trait, filaggrin, ichthyosis vulgaris, skin barrier function
Introduction
Atopic eczema1 is an itchy inflammatory skin condition which follows a chronic relapsing course. It can cause significant morbidity as a result of pruritus, sleep deprivation and emotional distress.2
Eczema is a complex trait ie multiple genetic and environmental factors contribute to the phenotype. There is an increasing recognition of the role of epithelial barrier dysfunction in the pathogenesis of atopic eczema3, 4 and it has been suggested that allergen penetration may predate the development of asthma and allergic rhinitis3 in the so-called ‘atopic march’.5
Filaggrin (filament-aggregating-protein) plays a key role in epidermal differentiation and skin barrier function. Filaggrin aggregates the keratin cytoskeleton to facilitate the collapse and flattening of keratinocytes in the outermost skin layer.6 The protein-lipid cornified envelope which replaces the plasma membrane of terminally-differentiated keratinocytes is extensively cross-linked and forms an important barrier to prevent water loss and minimise the entry of allergens and micro-organisms.7 Filaggrin is subsequently degraded to produce a mixture of hygroscopic amino acids which may also contribute to barrier function.8 Genotypes resulting in relative or absolute filaggrin deficiency may therefore contribute to epidermal barrier dysfunction by more than one mechanism.
At least fifteen different loss-of-function (null) mutations in the filaggrin gene (FLG) have been reported to date, of which five are prevalent in the European population.9 FLG null mutations cause the dry, scaly skin condition ichthyosis vulgaris;10 the five most common European mutations are also significantly associated with atopic eczema when analysed individually and as a combined null genotype.9 The association with atopic eczema has been replicated in at least twelve separate studies, most of which contain further within-study replication, with no negative or equivocal results,11 making this an unusually well-replicated finding in the field of complex genetics.
The clinical phenotype of ‘eczema’ is heterogeneous and likely to encompass considerable aetiological heterogeneity.12 The studies published to date looking at FLG null mutations have focussed on moderate-severe atopic eczema cases recruited via specialist clinics9, 13–20 and on children with eczema as part of atopy-related birth cohort studies.13, 15 We therefore aimed to investigate whether the five most prevalent FLG null mutations make a significant contribution to the mild-moderate atopic eczema that is common in the English population. We also aimed to assess the association of FLG null mutations with other signs of dry skin -- xerosis, ichthyosis, keratosis pilaris and hyperlinear palms -- in order to further define the filaggrin-deficiency phenotype.
Methods
Study design and case definition
We used a population cohort (n=7737) from the northwest of England for whom DNA samples had been collected at birth,21 representing over 85% of all deliveries in one hospital (West Cumberland Hospital, Whitehaven) between 1996 and 2003. This geographical area is particularly useful for epidemiological research because of relatively low rates of population movement. Children within the birth cohort catchment region who had not previously been included in the cohort were offered the opportunity to give a saliva sample for DNA extraction so that they could be included in this study.
Children between 7 and 9 years of age (n ≈ 4000) were chosen for this study as an optimal group in which to assess atopic disease (eczema, asthma and seasonal rhinitis). Eczema is most prevalent in infancy and early childhood; 85% of children are affected before the age of 5 years22 and 50-70% of children ‘grow out’ of their eczema by the age of 10 years23, 24 although they have an increased risk of recurrence in adulthood,25 as well as an increased incidence of asthma and rhinitis.5 Asthma shows a peak age of onset of 5 years so the majority of cases should be captured in our study age-group.
The study protocol was approved by the Local Research Ethics Committee and a parent or guardian of each child gave written informed consent.
All 57 Primary and Junior Schools within the birth cohort catchment area were requested to participate in the study, to allow distribution of questionnaires to all children aged 7 to 9 years. Questionnaire design utilised the well-validated UK Diagnostic Criteria26 to assess the 12 month period prevalence of eczema. [The full questionnaire is available in the Online Repository.] Additional, more detailed phenotype data was gathered by a single dermatologist (SJB) who had experience of working in a paediatric dermatology clinic. The skin on each child’s face, limbs and abdomen was examined. Eczema cases were defined according to the UK diagnostic criteria (representing a 12 month period prevalence) and/or flexural eczema on examination by the dermatologist (a point prevalence). Children not included in either of these categories were classified as controls.
Eczema was graded using the ‘Three Item Severity’ score,27 a simple, validated scale which allows the rapid assessment and recording of clinically-significant features.28 1-2 represents mild eczema, 3-5 moderate and 6-9 severe disease.29
The association between FLG mutations and the other atopic disorders, asthma and seasonal rhinitis, was investigated as a secondary analysis. Asthma and seasonal rhinitis prevalence were defined by parental replies to the questions “Has your child ever suffered from asthma? By asthma we mean bouts of wheezing and coughing” and “Has your child ever suffered from hay fever? By hay fever we mean bouts of sneezing with a runny nose or itchy eyes in the summer”.26
Genotype analysis
DNA was extracted from umbilical cord blood or umbilical cord tissue from the birth cohort individuals using standard procedures. Saliva samples were collected from the remainder using Oragene™ DNA self-collection kits.
DNA samples were screened for the five FLG null mutations most prevalent in the European population,9 as previously described: TaqMan allelic discrimination assay (Applied Biosystems, Foster City, CA) for R501X, 2282del4, R2447X and S3247X; size analysis of fluorescently-labelled PCR products using an Applied Biosystems 3100 DNA sequencer for 3702delG;30 homozygote and heterozygote results were confirmed by restriction enzyme digest10, 30 or sequencing.9
Statistical analysis
Calculations based on an estimated 11% prevalence of eczema25 and combined null allele frequency of 0.07813 predicted that recruitment of 1000 children would give a 91% power at p=0.001 to detect an allelic odds ratio of 3 (at the lower end of published data13, 15). A similar calculation predicted that there would be insufficient power to detect an association between each individual mutation and eczema.
The rationale for counting the five screened FLG null mutations as a single null allele is based on biochemical and immunohistochemical studies demonstrating that each of these null mutations produce truncated forms of profilaggrin which result in a marked reduction or absence of processed filaggrin when present in the homozygote or compound heterozygote state.9, 10
Allele and genotype frequencies in case and control groups and different phenotype groups were compared by Fisher’s exact test, under the null hypothesis that there is no association with genotype. Logistic regression analysis was used to estimate the odds ratio and penetrance of the FLG null allele using both allele- and genotype-based models of disease, to investigate the pattern of inheritance. Analysis was performed using the statistical analysis package Stata (version 9, Stata for Linux, StataCorp LP, College Station, Texas, USA).
Results
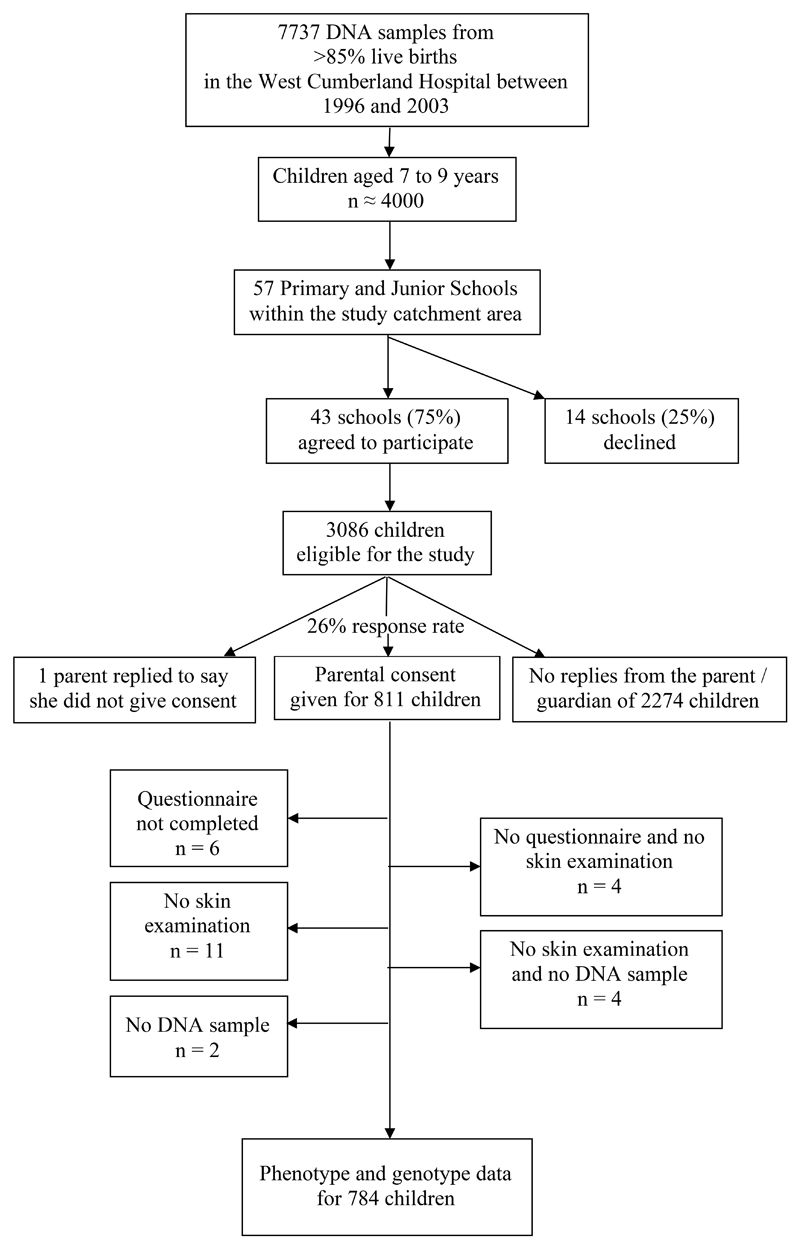
75% (43/57) of eligible schools agreed to take part in the research by distributing study literature and facilitating skin examinations by the visiting dermatologist. An estimated 3086 children in these schools were within the target age-range, and consent to participate was given by the parents/guardians of 811 children (26%). 801 questionnaires were completed, 792 children underwent skin examination and 805 DNA samples were located from the birth cohort stores (n=577) or extracted from saliva (n=228), giving complete phenotype and genotype information on 784 individuals (Figure 1). Individuals with incomplete phenotype and/or genotype data were included in the statistical analyses unless the missing data precluded this (eg insufficient criteria to define UK diagnosis or insufficient genotyping results to identify the combined genotype).
Figure 1. Flow diagram showing recruitment of children to the study.
The demographic data for this cohort plus clinical features relating to eczema and atopy are summarised in Table I.
Table I. Demographic data and clinical features of 811 children aged 7 to 9 years.
| Demographic data | Number | Percentage |
|---|---|---|
| Gender | 417 male | 51 |
| 394 female | 49 | |
| Prevalence of atopic eczema | 195/806 | 24 |
| Eczema severity | 70/120 mild | 58 |
| 45/120 moderate | 38 | |
| 5/120 severe | 4 | |
| Prevalence of asthma | 205/794 | 26 |
| Children with eczema & asthma | 83/793 | 11 % of total cohort |
| 43 % of eczema cases | ||
| 41 % of asthma cases | ||
| Prevalence of seasonal rhinitis | 152/793 | 19 |
The prevalence of atopic eczema was defined using a questionnaire based on the UK diagnostic criteria26 (12 month period prevalence) plus skin examination by a dermatologist (point prevalence). Eczema severity was assessed using the Three Item Severity score.27 The lifetime prevalence of asthma and seasonal rhinitis were defined by parental questionnaire. The denominator varies in order to maximise the use of available data.
Screening for all 5 mutations achieved results for 789/805 DNA samples (Table II). 14.2% of the study cohort, representing approximately 1 in 7 children, are carriers of one or more of the FLG mutations. 18.4% of eczema cases carried one/more FLG null mutations, compared with 12.9% of controls. This combined null genotype is significantly associated with atopic eczema (p=1.2x10-4).
Table II. FLG genotypes in a cohort of English school children, compared using the Fisher’s exact test.
| Atopic eczema |
|||
|---|---|---|---|
| Number (%) cases | Number (%) controls | ||
| FLG null genotype | AA | 155 (81.6) | 522 (87.1) |
| Aa | 27 (14.2) | 76 (12.7) | |
| aa | 8 (4.2) | 1 (0.2) | |
| Total | 190 | 599 | |
| Fisher’s exact test | p=1.2x10-4 | ||
Complete genotype data (results on screening for all 5 mutations) was obtained on a total of 789 children. ‘A’ refers to the wild-type form of FLG and ‘a’ refers to a null mutant form, either R501X, 2282del4, R2447X, S3247X or 3702delG. ‘aa’ individuals include 1 homozygote (R501X/R501X) and 8 compound heterozygotes (6 are R501X/2282del4 including the aa in the control group, 1 is 2282del4/R2447X and 1 is R501X/R2447X). Each of the five variants are in Hardy-Weinberg equilibrium within the cohort.
[The genotyping results for each individual mutation are available as a supplementary table, Table E1, in the Online Repository.]
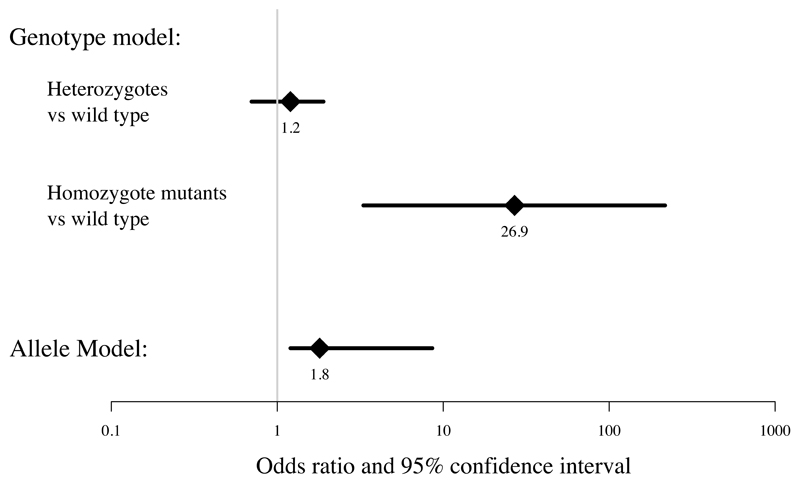
Genotyping results can be analysed using either a genotype model (comparing frequencies of wild-type with heterozygote and homozygote mutants) or using an allele-based model (which presumes that carriage of two mutations results in twice the effect of one mutation on a log odds scale). A comparison of these two models for our data is shown in Figure 2. The genotype model fits the data significantly better (likelihood ratio test p=0.0018) and hence this model is preferable because it makes fewer assumptions about the mode of inheritance. This indicates that FLG null mutations have a recessive mode of inheritance, since the heterozygotes have no significant increased risk of eczema (odds ratio 1.2, 95% confidence interval 0.7-1.9) but individuals carrying two copies of the FLG null mutations (homozygotes and compound heterozygotes) have an approximately 27 times greater risk of having childhood eczema (odds ratio 26.9, CI 3.3-217.1).
Figure 2. Logistic regression analysis to estimate odds of disease (atopic eczema) with FLG null mutations comparing genotype and allele-based models.
The 95% confidence intervals are as follows: Genotype model heterozygotes vs wild type 0.7-1.9; homozygote mutants vs wild type 3.3-217.1. Allele model 1.2-8.6.The likelihood ratio test, to test whether the allele model can be nested in full within the genotype model of disease, shows that the genotype model fits the data significantly better (p=1.8x10-3).
There was no association between parental report of asthma (p=0.15) or seasonal rhinitis (p=0.66) and FLG null mutations, but there was evidence of an association between the phenotype of asthma with eczema, which was significant even after allowing for multiple testing (p=7.1x10-4, odds ratio for carriers of two null mutations 11.9, 95% confidence interval 3.1-45.6). 23.2% of children with eczema and asthma carry one/more of the FLG null mutations, compared with 11.8% of controls. The results of logistic regression analysis and Fisher’s exact test to test association of these and other phenotypes with the FLG genotype are summarised in Table III.
Table III. Prevalence, logistic regression odds ratios and p values for Fisher’s exact test for different phenotypes and the combined FLG null genotype.
| Phenotype or clinical feature | Number (denominator) | Odds ratio (95% confidence interval) | Fisher’s exact test |
|---|---|---|---|
| Atopic eczema (UKcriteria+flexural) | 195 (806) | Hets: 1.2 (0.7 – 1.9) Homs: 26.9 (3.3 – 217.1) |
p=1.2x10-4 |
| Non-flexural eczema* | 35 (792) | Hets: 0.4 (0.1 – 1.8) Homs: § |
p=0.52 |
| Asthma | 205 (794) | Hets: 1.0 (0.6 – 1.6) Homs: 3.6 (1.0 – 13.7) |
p=0.15 |
| Atopic eczema with asthma | 83 (793) | Hets: 1.5 (0.8 – 2.8) Homs: 11.9 (3.1 – 45.6) |
p=7.1x10-4 |
| Seasonal rhinitis | 152 (793) | Hets: 1.2 (0.7 – 2.0) Homs: 0.5 (0.1 – 4.4) |
p=0.66 |
| Ichthyosis vulgaris | 10 (792) |
Hets: >100
# Homs: >>100 # |
p=4.1x10-16 |
| Milder ichthyosis | 54 (792) |
Hets: 5.5 (3.0 – 10.0) Homs: 5.8 (1.2 – 29.1) |
p=1.7x10-7 |
| Xerosis | 193 (792) | Hets: 1.5 (0.9 – 2.4) Homs: § |
p=0.04 |
| Hyperlinear palms (marked) | 52 (792) |
Hets: 17.5 (9.0 – 34.2) Homs: 152.1 (29.1–794.3) |
p=1.2x10-23 |
| Hyperlinear palms (any) | 167 (792) |
Hets: 19.3 (11.7 – 31.7) Homs: § |
p=6.8x10-42 |
| Keratosis pilaris (marked) | 77 (792) |
Hets: 8.9 (5.2 – 15.2) Homs: 35.1 (8.4 – 146.1) |
p=2.8x10-18 |
| Keratosis pilaris (any) | 273 (792) |
Hets: 3.6 (2.3 – 5.5) Homs: § |
p=2.2x10-12 |
Atopic eczema is defined using the UK diagnostic criteria26 and skin examination by a dermatologist; asthma and seasonal rhinitis are defined by parental questionnaire. The diagnosis of ichthyosis vulgaris (scaly skin on extensor surfaces plus hyperlinear palms and keratosis pilaris), milder ichthyosis (scaly skin) and xerosis (dry skin) are based on clinical examination by a dermatologist on a single occasion for each child during the winter months. ‘Hets’ refers to individuals who are heterozygote for any of the five FLG null mutations (R501X, 2282del4, R2447X, S3247X, 3702delG); ‘Homs’ refers to individuals with any two FLG mutations, including homozygotes and compound heterozygotes. These analyses use the genotype model of disease which is less powerful than the allele-based model but makes no assumption about the mode of inheritance, which is currently unknown. Statistically significant results (p<0.01) are highlighted in bold
cases with non-flexural eczema only (ie no flexural involvement)
odds ratio cannot be estimated because there are no homozygotes in the control group
insufficient data for reliable estimation of odds ratio.
In our cohort, 10 individuals out of 792 children had classical ichthyosis vulgaris (fine grey-white scaling on extensor skin surfaces plus hyperlinear palms and keratosis pilaris) on clinical examination, a prevalence of approximately 1 in 80. One was homozygous for the R501X mutation, six were compound heterozygotes and three were heterozygotes (each carrying the R501X mutation). Three out of the ten ichthyosis vulgaris cases had no eczema by either of our definitions and no parental report of eczema at any time in the child’s life and interestingly these were the three R501X heterozygotes. There were a further 54 children with milder ichthyosis (scaling of the skin) as well as 193 with xerosis (dryness of the skin).
Looking at the genotype-phenotype correlation, all 9 of the homozygote/compound heterozygote individuals in our cohort had ichthyosis vulgaris or milder ichthyosis and 8 out of these 9 had eczema. Conversely, in the 103 heterozygote individuals, 75 (73%) did not have ichthyosis vulgaris or milder ichthyosis but 20 (27%) of this subgroup (heterozygotes without any ichthyosis) did have eczema. However, using logistic regression to model the effect of genotype on eczema status having controlled for the effect of ichthyosis vulgaris showed that any additional effect of FLG genotype is not statistically significant (p=0.43).
The physical signs of hyperlinear palms and keratosis pilaris in childhood are each strongly and significantly associated with the combined FLG null genotype (Table III). Marked hyperlinear palms also show significant association with the four most prevalent FLG null mutations (R501X, 2282del4, R2447X and S3247X) when they are analysed individually (p<0.01 for each variant). Similarly, marked keratosis pilaris shows significant and independent association with the two most prevalent null mutations (R501X and 2282del4, p<0.01 for each). These figures translate into a positive predictive value of 71% for marked hyperlinear palms, ie 71% of children with marked hyperlinear palms carry one/more of the five common FLG null mutations; the negative predictive value is 90%, meaning 90% of children without hyperlinear palms are not carriers of these mutations. The predictive values for marked keratosis pilaris in this age-group are 53% (positive) and 90% (negative predictive value).
Discussion
There have, to date, been no published studies investigating the role of FLG mutations in the mild-moderate eczema that predominates in our society. We used a unique resource in the northwest of England -- an unselected birth cohort with DNA samples -- to investigate this question. We were careful to define eczema using a well-validated method (the UK modification of Hanifin and Rajka’s diagnostic criteria)26 in addition to skin examination by an experienced dermatologist. This allowed us to gather more detailed information on phenotypic features such as xerosis, ichthyosis and keratosis pilaris, which have not been documented in previous studies. Diagnostic accuracy and careful phenotype documentation are particularly important in genetic studies if individual genetic factors are to be identified on the background of complex environmental effects.31
The low response rate of only 26% reflects our method of recruitment, requiring informed consent prior to questionnaire-completion.32 This raises the possibility that bias has been introduced by self-selection in to our study cohort; we addressed this question in two ways. Firstly, comparison of the prevalence of eczema and asthma in our cohort with a recently published study of atopy prevalence in the northeast of England, using a case definition based on the ISAAC questionnaire (90% response rate, n=3000, 6-7 year olds)33 showed very similar findings: ‘Rash with typical distribution’ 21.1% for boys and 23.8% for girls, ‘current rash’ 23.3% for boys and 25.0% for girls, compared with a prevalence of 24.2% in our study (boys and girls combined). Similarly, asthma lifetime prevalence from this comparable study is reported as 29.8% for boys and 24.1% of girls, compared with 26.2% in our study.
Secondly, we compared the allele frequencies for each of the five FLG mutations in our study cohort with an unselected 1000 samples from age-matched children in the same birth cohort (screened as part of another study34) and found no significant difference (p>0.05, Fisher’s exact test). [This data is available as a supplementary table, Table E2, in the Online Repository.] We therefore conclude that our cohort is representative of the local population and hence we can use it to estimate the true risk (or penetrance) associated with FLG mutations, not just their relative risk, which would be estimated from a selected case-control study. From our data, we calculate that the probability of disease (ie having mild-moderate eczema) is 22.9% in wild-type individuals (95% CI 19.7-26.1%), 26.2% in heterozygotes (17.7-34.7%) and 88.9% (68.4-109.4%) for homozygotes.
In our cohort the prevalence of ichthyosis vulgaris is approximately 1 in 80. This is significantly higher than that reported in the only previous population cohort study, when 1 in 250 English school children were found to have the disorder.35 This higher prevalence may reflect more careful disease ascertainment, using a dermatology specialist. Furthermore, the prevalence of FLG null mutations in the population (9/789 individuals carrying two mutations) would support these examination findings, with a predicted prevalence of ichthyosis vulgaris of approximately 1 in 88 (presuming 100% penetrance and not including the effect of semi-dominant inheritance). Ichthyosis vulgaris and milder ichthyosis were each significantly associated with FLG status in our cohort. This is consistent with data from previous studies that FLG shows semi-dominant inheritance (ie heterozygote carriers tend to have a milder phenotype than homozygotes).13
This study adds to our understanding of the genetic basis of eczema in several ways. The phenotype studied (mild-moderate atopic eczema in 7-9 year olds) is significantly associated with FLG loss-of-function mutations in the population of the northwest of England. This contrasts with the cases of non-flexural eczema within the cohort, a group likely to include discoid eczema and contact dermatitis, which do not show an association with FLG null mutations, although the numbers are too small to draw firm conclusions.
In these cases of predominantly mild-moderate atopic eczema, FLG shows a recessive pattern of inheritance, rather than the semi-dominant pattern reported in the original group of eczema cases in families with ichthyosis vulgaris.13 The fact that we only have 9 individuals in our cohort that are homozygous for FLG null mutations is reflected in the wide confidence intervals around the odds ratios in Figure 2.
However, we can still be confident that the data demonstrate a recessive pattern of inheritance since 8 out of the 190 cases (4.0%) carry two FLG null mutations compared to only 1 out of 599 controls (0.2%) and hence these 9 individuals provide good evidence for a strong homozygote effect. The confidence intervals do not overlap and there is a very small chance (p=1.8x10-3) of observing these data under an allele-based model (semi-dominant inheritance) rather than a genotype model (recessive inheritance).
The association of mild-moderate eczema with FLG is closely related to the presence of ichthyosis vulgaris, since logistic regression modelling showed no additional effect of genotype, having allowed for the presence/absence of ichthyosis vulgaris. This is perhaps unsurprising, given that FLG mutations are known to cause ichthyosis vulgaris and that 50-70% of ichthyosis vulgaris patients also have eczema. This chain of reasoning emphasises the role of epidermal barrier dysfunction, characteristic of filaggrin deficiency, in the pathogenesis of eczema. An alternative explanation is that FLG is associated with eczema only via linkage disequilibrium with another gene. However this seems unlikely given the independent association of five different FLG mutations with atopic eczema in the European population9, 30 and Asian-specific FLG null mutations which have arisen independently from the European variants,36 as well as a reduction in filaggrin expression in atopic skin37 and biochemical studies demonstrating a similar reduction/absence of processed filaggrin as a result of FLG null mutations.10
The odds ratio of having moderate-severe eczema given the presence of one/more FLG mutations has varied between 2.03 and 13.4 in previous reports.11, 13, 20 The equivalent odds ratio figure for our predominantly mild-moderate cases would be 1.53 (95% confidence interval 0.99-2.37) if we had used a dominant model of analysis (combining the heterozygotes with the homozygote mutants, to compare carriers of one/more FLG null mutations with wild-type individuals). This lower odds ratio indicates a lesser effect of the FLG mutations in mild-moderate eczema compared with moderate-severe eczema. Possible explanations for this observation include an intuitive ‘dilution’ of effect in the less severe phenotype, or a greater effect of other factors -- genetic and environmental -- which are relatively more important than filaggrin insufficiency in milder eczema. The dominant model gives a non-significant result for our data (95% confidence interval overlapping 1.0) because of the large number of heterozygotes and small number of homozygotes in the cohort. Recognition of the relative importance of different aetiological factors in the mild-moderate compared with the moderate-severe eczema phenotype may be helpful in designing more rational therapies aimed at targeting specific pathology.
Interestingly, the frequency of carriage of FLG mutations in our cases (18.4%) is similar to that reported in children from a Danish birth cohort whose mothers had asthma (n=372) in which 17.5% of eczema cases were carriers of one or more FLG null alleles.13 In these children there was a strikingly high penetrance: 63% of carriers had developed atopic eczema by the age of 3 years, suggesting that other influential factors (genetic and/or environmental) are associated with the maternal history of asthma. Our cohort shows an association of FLG mutations with asthma only in those children who also have eczema, replicating earlier reports13, 15 and emphasising the existence of a subgroup of asthma in association with eczema.
This study demonstrates the usefulness of careful physical examination of the skin, since hyperlinear palms and keratosis pilaris are features that can help to predict the presence of a FLG null mutation. The mechanisms by which filaggrin insufficiency produces these clinical signs remain to be elucidated.
Finally, in spite of the significant effect demonstrated in this cohort, FLG null mutations can only explain a small proportion of the total burden of childhood eczema on a population scale. Eight out of 190 (4.2%) of atopic eczema cases in our cohort carry two null mutations and thus can be attributed to FLG deficiency. The residual 95.8% of cases remain to be explained by other genetic and environmental factors. However, FLG is the single most significant genetic factor in atopic eczema that has been identified to date, and our study supports a significant role for filaggrin insufficiency in the pathogenesis of a small proportion of common, mild-moderate atopic eczema cases.
Clinical implications.
This finding emphasises the role of ichthyosis vulgaris and filaggrin deficiency in the pathogenesis of approximately 4% of eczema cases in the English population.
Capsule summary.
This population-based case-control study shows that filaggrin null mutations are significantly associated with common, mild-moderate atopic eczema in childhood, with a recessive pattern of inheritance.
Acknowledgements
We are very grateful to the schools in West Cumbria, the children and their parents for participating in this research.
Sources of funding
The study was funded by the Newcastle Healthcare Charity, the British Skin Foundation and Action Medical Research. SJB is supported by the Lily Ross Fellowship and a British Society for Paediatric Dermatology training fellowship. Support for HJC and partial support for SJB is provided by a Senior Fellowship in Basic Biomedical Science (grant ref 074524) from the Wellcome Trust. Filaggrin research in the McLean laboratory is supported by grants from The British Skin Foundation; The National Eczema Society; The Medical Research Council (Reference number G0700314) and donations from anonymous families affected by eczema in the Tayside Region of Scotland.
Abbreviations used
- FLG
filaggrin gene
- OR
odds ratio
- CI
confidence interval
- aa
homozygote or compound heterozygote genotype for filaggrin mutations R501X, 2282del4, R2447X, S3247X and 3702delG
- AA
homozygote for the wild-type filaggrin genotype
- Aa
heterozygote genotype for filaggrin mutations R501X, 2282del4, R2447X, S3247X or 3702delG and wild-type
Footnotes
Conflict of interest statement
IMcL has filed patents relating to genetic testing and therapy development aimed at the filaggrin gene.
The other authors have no conflict of interest.
References
- 1.Johansson SG, Bieber T, Dahl R, Friedmann PS, Lanier BQ, Lockey RF, et al. Revised nomenclature for allergy for global use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol. 2004;113:832–6. doi: 10.1016/j.jaci.2003.12.591. [DOI] [PubMed] [Google Scholar]
- 2.Chamlin SL, Cella D, Frieden IJ, Williams ML, Mancini AJ, Lai JS, et al. Development of the Childhood Atopic Dermatitis Impact Scale: initial validation of a quality-of-life measure for young children with atopic dermatitis and their families. J Invest Dermatol. 2005;125:1106–11. doi: 10.1111/j.0022-202X.2005.23911.x. [DOI] [PubMed] [Google Scholar]
- 3.Hudson TJ. Skin barrier function and allergic risk. Nat Genet. 2006;38:399–400. doi: 10.1038/ng0406-399. [DOI] [PubMed] [Google Scholar]
- 4.Taieb A, Hanifin J, Cooper K, Bos JD, Imokawa G, David TJ, et al. J Allergy Clin Immunol; Proceedings of the 4th Georg Rajka International Symposium on Atopic Dermatitis; Arcachon, France. September 15-17, 2005; 2006. pp. 378–90. [DOI] [PubMed] [Google Scholar]
- 5.Hahn EL, Bacharier LB. The atopic march: the pattern of allergic disease development in childhood. Immunol Allergy Clin North Am. 2005;25:231–46, v. doi: 10.1016/j.iac.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Manabe M, Sanchez M, Sun TT, Dale BA. Interaction of filaggrin with keratin filaments during advanced stages of normal human epidermal differentiation and in ichthyosis vulgaris. Differentiation. 1991;48:43–50. doi: 10.1111/j.1432-0436.1991.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 7.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 8.Rawlings AV, Harding CR. Moisturization and skin barrier function. Dermatol Ther. 2004;17(Suppl 1):43–8. doi: 10.1111/j.1396-0296.2004.04s1005.x. [DOI] [PubMed] [Google Scholar]
- 9.Sandilands A, Terron-Kwiatkowski A, Hull PR, O'Regan GM, Clayton TH, Watson RM, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007;39:650–4. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- 10.Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–42. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 11.Irvine AD. Fleshing out filaggrin phenotypes. J Invest Dermatol. 2007;127:504–7. doi: 10.1038/sj.jid.5700695. [DOI] [PubMed] [Google Scholar]
- 12.Brown S, Reynolds NJ. Atopic and non-atopic eczema. Bmj. 2006;332:584–8. doi: 10.1136/bmj.332.7541.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 14.Weidinger S, Illig T, Baurecht H, Irvine AD, Rodriguez E, Diaz-Lacava A, et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–9. doi: 10.1016/j.jaci.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Marenholz I, Nickel R, Ruschendorf F, Schulz F, Esparza-Gordillo J, Kerscher T, et al. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J Allergy Clin Immunol. 2006;118:866–71. doi: 10.1016/j.jaci.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Ruether A, Stoll M, Schwarz T, Schreiber S, Folster-Holst R. Filaggrin loss-of-function variant contributes to atopic dermatitis risk in the population of Northern Germany. Br J Dermatol. 2006;155:1093–4. doi: 10.1111/j.1365-2133.2006.07500.x. [DOI] [PubMed] [Google Scholar]
- 17.Barker JN, Palmer CN, Zhao Y, Liao H, Hull PR, Lee SP, et al. Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J Invest Dermatol. 2007;127:564–7. doi: 10.1038/sj.jid.5700587. [DOI] [PubMed] [Google Scholar]
- 18.Stemmler S, Parwez Q, Petrasch-Parwez E, Epplen JT, Hoffjan S. Two common loss-of-function mutations within the filaggrin gene predispose for early onset of atopic dermatitis. J Invest Dermatol. 2007;127:722–4. doi: 10.1038/sj.jid.5700579. [DOI] [PubMed] [Google Scholar]
- 19.Weidinger S, Rodriguez E, Stahl C, Wagenpfeil S, Klopp N, Illig T, et al. Filaggrin mutations strongly predispose to early-onset and extrinsic atopic dermatitis. J Invest Dermatol. 2007;127:724–6. doi: 10.1038/sj.jid.5700630. [DOI] [PubMed] [Google Scholar]
- 20.Morar N, Cookson WO, Harper JI, Moffatt MF. Filaggrin mutations in children with severe atopic dermatitis. J Invest Dermatol. 2007;127:1667–72. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 21.Chase DS, Tawn EJ, Parker L, Jonas P, Parker CO, Burn J. The North Cumbria Community Genetics Project. J Med Genet. 1998;35:413–6. doi: 10.1136/jmg.35.5.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kay J, Gawkrodger DJ, Mortimer MJ, Jaron AG. The prevalence of childhood atopic eczema in a general population. J Am Acad Dermatol. 1994;30:35–9. doi: 10.1016/s0190-9622(94)70004-4. [DOI] [PubMed] [Google Scholar]
- 23.Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P, et al. Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Report. J Allergy Clin Immunol. 2006;118:152–69. doi: 10.1016/j.jaci.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 24.Illi S, von Mutius E, Lau S, Nickel R, Gruber C, Niggemann B, et al. The natural course of atopic dermatitis from birth to age 7 years and the association with asthma. J Allergy Clin Immunol. 2004;113:925–31. doi: 10.1016/j.jaci.2004.01.778. [DOI] [PubMed] [Google Scholar]
- 25.Williams HC, Wuthrich B. The natural history of atopic dermatitis. In: Williams HC, editor. Atopic Dermatitis The epidemiology, causes and prevention of atopic eczema. Cambridge University Press; 2000. pp. 41–59. [Google Scholar]
- 26.Williams HC, Burney PG, Pembroke AC, Hay RJ. Validation of the U.K. diagnostic criteria for atopic dermatitis in a population setting. U.K. Diagnostic Criteria for Atopic Dermatitis Working Party. Br J Dermatol. 1996;135:12–7. [PubMed] [Google Scholar]
- 27.Wolkerstorfer A, de Waard van der Spek FB, Glazenburg EJ, Mulder PG, Oranje AP. Scoring the severity of atopic dermatitis: three item severity score as a rough system for daily practice and as a pre-screening tool for studies. Acta Derm Venereol. 1999;79:356–9. doi: 10.1080/000155599750010256. [DOI] [PubMed] [Google Scholar]
- 28.Charman CR, Venn AJ, Williams H. Measuring atopic eczema severity visually: which variables are most important to patients? Arch Dermatol. 2005;141:1146–51. doi: 10.1001/archderm.141.9.1146. discussion 51. [DOI] [PubMed] [Google Scholar]
- 29.Oranje AP, Glazenburg EJ, Wolkerstorfer A, de Waard-van der Spek FB. Practical issues on interpretation of scoring atopic dermatitis: the SCORAD index, objective SCORAD and the three-item severity score. Br J Dermatol. 2007;157:645–8. doi: 10.1111/j.1365-2133.2007.08112.x. [DOI] [PubMed] [Google Scholar]
- 30.Sandilands A, O'Regan GM, Liao H, Zhao Y, Terron-Kwiatkowski A, Watson RM, et al. Prevalent and rare mutations in the gene encoding filaggrin cause ichthyosis vulgaris and predispose individuals to atopic dermatitis. J Invest Dermatol. 2006;126:1770–5. doi: 10.1038/sj.jid.5700459. [DOI] [PubMed] [Google Scholar]
- 31.Morar N, Willis-Owen SA, Moffatt MF, Cookson WO. The genetics of atopic dermatitis. J Allergy Clin Immunol. 2006;118:24–34. doi: 10.1016/j.jaci.2006.03.037. quiz 5-6. [DOI] [PubMed] [Google Scholar]
- 32.Angus VC, Entwistle VA, Emslie MJ, Walker KA, Andrew JE. The requirement for prior consent to participate on survey response rates: a population-based survey in Grampian. BMC Health Serv Res. 2003;3:21. doi: 10.1186/1472-6963-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamssain M. Trends in the prevalence and severity of asthma, rhinitis and atopic eczema in 6- to 7- and 13- to 14-yr-old children from the north-east of England. Pediatr Allergy Immunol. 2007;18:149–53. doi: 10.1111/j.1399-3038.2006.00498.x. [DOI] [PubMed] [Google Scholar]
- 34.Brown SJ, Sandilands A, Zhao Y, Liao H, Relton CL, Meggitt SJ, et al. Prevalent and low-frequency null mutations in the filaggrin gene are associated with early-onset and persistent atopic eczema. Accepted for publication in J Invest Dermatol. 2007 doi: 10.1038/sj.jid.5701206. [DOI] [PubMed] [Google Scholar]
- 35.Wells RS, Kerr CB. Clinical features of autosomal dominant and sex-linked ichthyosis in an English population. Br Med J. 1966;1:947–50. doi: 10.1136/bmj.1.5493.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura T, Sandilands A, Akiyama M, Liao H, Evans AT, Sakai K, et al. Unique mutations in the filaggrin gene in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J Allergy Clin Immunol. 2007;119:434–40. doi: 10.1016/j.jaci.2006.12.646. [DOI] [PubMed] [Google Scholar]
- 37.Seguchi T, Cui CY, Kusuda S, Takahashi M, Aisu K, Tezuka T. Decreased expression of filaggrin in atopic skin. Arch Dermatol Res. 1996;288:442–6. doi: 10.1007/BF02505232. [DOI] [PubMed] [Google Scholar]