Abstract
The well-orchestrated multicellular life cycle of Dictyostelium discoideum has fascinated biologists for over a century. Self-organisation of its amoebas into aggregates, migrating slugs and fruiting structures by pulsatile cAMP signalling and their ability to follow separate differentiation pathways in well-regulated proportions continue to be topics under investigation. A striking aspect of D. discoideum development is the recurrent use of cAMP as chemoattractant, differentiation inducing signal and second messenger for other signals that control the developmental programme. D. discoideum is one of >150 species of Dictyostelia and aggregative life styles similar to those of Dictyostelia evolved many times in eukaryotes. Here we review experimental studies investigating how phenotypic complexity and cAMP signalling co-evolved in Dictyostelia. In addition, we summarize comparative genomic studies of multicellular Dictyostelia and unicellular Amoebozoa aimed to identify evolutionary conservation and change in all genes known to be essential for D. discoideum development.
Keywords: sorocarpic multicellularity, evolution of phenotype, evolution of cAMP signalling, encystation
Aggregative multicellularity evolved many times independently
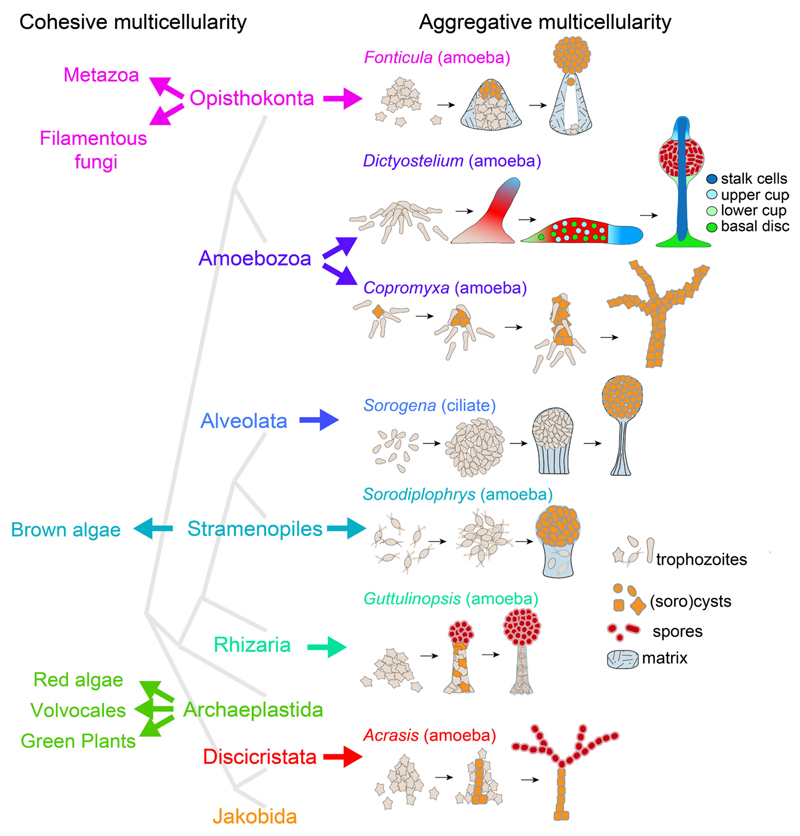
While the naked eye perceives only three types of multicellular organisms – plants, animals and fungi –, at the microscopic level or just at up-close inspection there are many more. In fact, multicellularity evolved at many times independently in prokaryotes (Lyons and Kolter 2015), and in seven of the eight major divisions of eukaryotes (Fig. 1). The most common route to multicellularity is the one taken by sorocarpic organisms, which alternate between a unicellular feeding stage and a multicellular stage where starving cells aggregate to build a fruiting structure (sorocarp) with spores. This type of multicellularity evolved independently in the Discicristata (Brown, et al., 2012b), Rhizaria (Brown, et al., 2012a), Stramenopiles (Tice, et al., 2016), Alveolata (Sugimoto and Endoh 2006) and Holozoa (Brown, et al., 2009) and twice within the Amoebozoa (Brown, et al., 2011). Sorocarpic multicellularity or aggregative multicellularity differs from all other forms of multicellularity, where cells stick together after division and feed in the multicellular stage. This type of cohesive multicellularity is common to green plants, red and brown algae and fungi, and encompasses all animals, which altogether have a more limited phylogenetic distribution (Fig. 1).
Fig. 1. Evolution of multicellularity in eukaryotes.
Multicellular organisms can be subdivided into two types. Type I – cohesive multicellularity -starts off from a spore or fertilized egg that undergoes repeated divisions and morphogenesis, with cells remaining together and feeding in the multicellular stage. In Type II – aggregative multicellularity – cells feed and proliferate as single cells and only come together when stressed, usually by starvation. The aggregates next transform into a fruiting body where the cells enter dormancy and encapsulate into either cysts or spores. Aggregative multicellularity evolved independently in most major eukaryotic divisions. Phylogeny (grey lines) after He, et al. (2014).
Multicellular development of most sorocarpic organisms has remained relatively simple. We think that this is due to the fact that their proliferation takes place in the unicellular stage, where they go through many generations, while competing for food. This stage is likely to be under much more stringent natural selection than a single generation of cells going through sorocarp formation. Acrasis amoebae aggregate and some encyst to form a stalk. Others move to the top of stalk, arrange themselves into chains or form a globular mass, and then differentiate into round spores (Brown, et al., 2012b). Guttulinopsis amoebae construct a stalk consisting of encysted and decaying cells surrounded by an elastic sheath. Other amoebae move to the top of this mass and differentiate into spores (Brown, et al., 2012a). The ciliate Sorogena aggregates under water and while the aggregate compacts by cell adhesion and cells start to secrete matrix to form a sheath, it rises to the surface. Continued matrix production and contraction of the sheath causes a stalk to form which pushes up the cells, which then encyst (Blanton and Olive 1983). Sorodiplophrys amoebae construct a stalk consisting of gelatinous matrix with embedded decaying amoebas with most cells moving to the top to differentiate into sorocysts (Tice, et al., 2016). Copromyxa amoebae move towards an encysted founder cell, then crawl on top and also encapsulate to form sorocysts. This process continues until a tall branched fruiting structure has formed (Brown, et al., 2011). Fonticula amoebae aggregate to form a mound surrounded by a slime sheath, and continue to secrete matrix between the cell mass and sheath. The amoebae then differentiate into spores that are similar to their unicellular cysts, which are expulsed through the top of the structure (Brown, et al., 2009).
Among aggregating eukaryotes, the Dictyostelia display the most sophisticated form of multicellularity. In addition to spores, they can differentiate in up to four somatic cell types and aggregates can transform into an intermediate motile “slug” stage that brings the cell mass to an optimal site for spore dispersal. As it also by far the most thoroughly studied sorocarpic organism, its road to multicellularity is the topic of this review.
Evolution of phenotype in Dictyostelia
The evolutionary history of any biological process can only be meaningfully investigated if the genetic relationships between the organisms under study are known. The original subdivision of Dictyostelia in three genera – Dictyostelids, Acytostelids and Polysphondylids – was based on differences in fruiting body morphology. The first use of the popular molecular marker gene small subunit ribosomal DNA (SSU-rDNA) for phylogenetic inference subdivided Dictyostelia into four major and some minor groups, with the former three genera being distributed over more than one group each (Schaap, et al., 2006). This indicated that similarities in fruiting body morphology are not a reliable marker for genetic similarity in Dictyostelia.
The availability of group-representative genome sequences made it possible to infer phylogenies from many concatenated protein sequences. Three different studies robustly placed the root of the phylogeny between two major branches that contained groups 1+2 and groups 3+4, respectively (Heidel, et al., 2011; Romeralo, et al., 2013; Sheikh, et al., 2015), instead of a weakly resolved position between groups 1 and 2 in the SSU-rDNA phylogeny. Sequencing of 5 genomes of early branching species resolved the position of the minor groups and highlighted a set of genes that either individually or concatenated as a pair fully reproduced a phylogeny based on 47 concatenated genes (Singh, et al., 2016). Six of these genes were amplified by PCR across 34 species, of which 22 resided in group 4, with the main goal to infer species relationships in group 4 (Schilde, et al., 2019), which were poorly resolved in the SSU-rDNA phylogeny. Compared to the SSU-rDNA tree, nodes in the 6-protein tree were better or equally well resolved and branch lengths between groups were more homogeneous (Fig. 2). This highlights a subdivision of group 4 into two divisions, which each contain two or three clades. Only some of these clades can be recognized in the SSU-rDNA tree. In contrast to the slow rate of evolution of SSU-rDNA in group 4 (as evident from the very short branches), SSU-rDNA in group 1 underwent very rapid evolution, which likely distorted the deep topology of the tree.
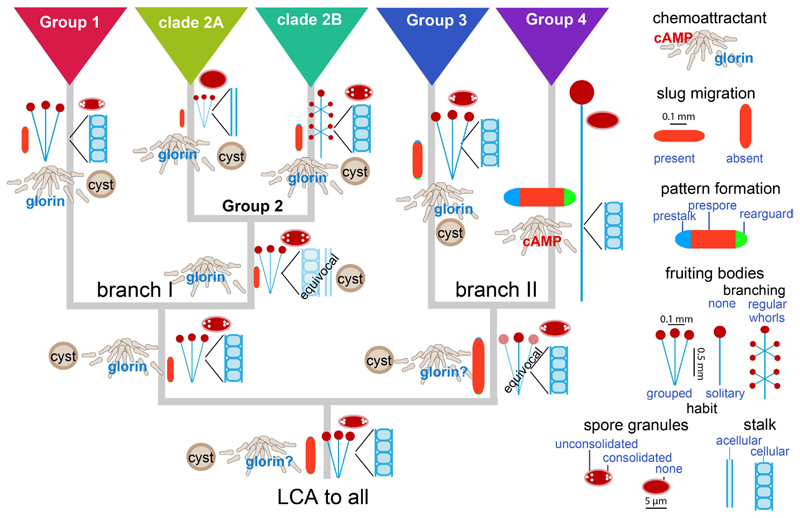
Fig. 2. Multi-protein based phylogenies improve poor node resolution and branch length anomalies of single gene trees.
Comparison of phylogenetic trees of based on six concatenated protein sequences (A) (Schilde, et al., 2019) or small subunit ribosomal DNA (B). Fast evolution of SSU rDNA in the group 1 Cavenderia distorts the position of the root, while slow SSU rDNA evolution in group 4 Dictyostelia prevents resolution of species positions. Taxon names in (B) follow the recent re-classification of Dictyostelids (Sheikh, et al., 2018).
To study the evolution of phenotype, 25 morphological and behavioural characters were scored over 99 species, such as the size and shape of sorogens, fruiting bodies and spores, the ability of sorogens to migrate and of amoebas to encyst individually, the likely identity of the chemoattractant and the proportion of prespore, prestalk and rear-guard cells in sorogens (Romeralo, et al., 2013; Schilde, et al., 2014). Phylogeny-based statistical methods were applied to infer trait evolution, i.e. the probability of the state of the character at the interior nodes of the tree. These studies show that the last common ancestor (LCA) of all Dictyostelia, formed small clustered fruiting bodies with cellular stalks and spores with polar granules (Fig. 3). Amoebas likely used glorin to aggregate, but could also encyst individually. Its sorogens did not migrate and consisted almost entirely of prespore cells. This phenotype was retained in the LCA to branches I and II and the LCAs to groups 1, 2 and 3. The LCA to clade 2A lost its cellular stalk, while the LCA to clade 2B gained whorls of side branches on its fruiting structures. The largest change occurred in the LCA to group 4, which gained use of cAMP as attractant, sorogen migration, differentiation of regulated proportions of prestalk, prespore and rear-guard/basal disc cells, and construction of large solitary fruiting bodies, but lost spore granules and encystation. In short, many of the phenotypic characters that make D. discoideum such a popular developmental model system only evolved in group 4.
Fig. 3. Evolution of phenotype in Dictyostelia.
Phylogenetic comparative methods were applied to reconstruct ancestral states for a set of 25 phenotypic characters measured over 99 Dictyostelium species (Romeralo, et al., 2013; Schilde, et al., 2014). Ancestral states for characters that showed a clear evolutionary progression are shown at the root and interior nodes of the Dictyostelium phylogeny. Quantitative traits such as spore-, slug-, stalk- and sorus size and contributions of prestalk, prespore and rear-guard regions to slug pattern are shown at correct relative values.
Prevalence of cAMP signalling in D. discoideum development
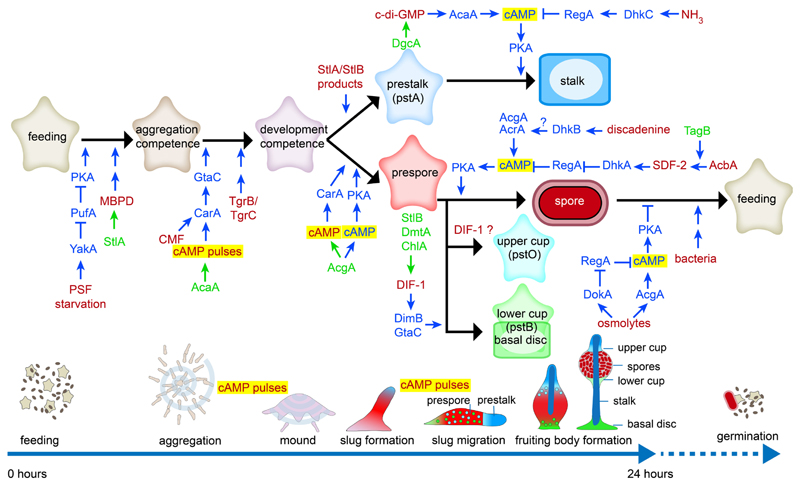
Among Dictyostelia, D. discoideum owes its original popularity to its well-regulated morphogenesis (Raper 1940), combined with the identification of its chemo-attractant as cAMP (Konijn, et al., 1967), then just becoming more widely known as a second messenger for hormone action. Development of genetic transformation (Nellen, et al., 1984) and tagged mutagenesis (Kuspa and Loomis 1992) consolidated its usage as the model of choice. Developmental studies identified several secreted signals that regulate cell differentiation morphogenesis as well as the enzymes that synthesize and degrade these signals and components of their signal transduction pathways (see (Du, et al., 2015; Loomis 2014; Williams 2006) for reviews and (Chen, et al., 2017; Narita, et al., 2014; Sato, et al., 2016) for recent research). A striking aspect of pathways downstream of most developmental signals is that they all converge on regulation of intracellular cAMP levels by controlling the activity of the cAMP phosphodiesterase RegA or the adenylate cyclases AcrA, AcgA and AcaA, and thereby regulate the activation of cAMP dependent protein kinase (PKA) (Fig. 4). PKA triggers the transition from growth to development and is required for the differentiation of prespore cells, the maturation of spores and stalk cells and the maintenance of spore dormancy in the spore head.
Fig. 4. Developmental signalling in Dictyostelium discoideum.
Extracellular signals and the enzymes that synthesize them are in red and green text, respectively. Proteins and small molecules involved in signal processing are in blue text.
In addition to their role in coordinating the aggregation process, secreted pulses of cAMP also organize cell movement in slugs and fruiting bodies (Singer, et al., 2019) and induce the expression of aggregation genes. Together with intracellular cAMP acting on PKA, secreted cAMP also induces the differentiation of prespore cells. The extracellular roles of cAMP are mediated by G-protein coupled cAMP receptors (Cars) and regulated by the cAMP phosphodiesterase PdsA and the adenylate cyclases AcaA, AcgA and AcrA. This dominant role for both intra- and extracellular cAMP signalling suggests that it is derived from deeper origins in Dictyostelia or Amoeboza.
Conservation of cAMP signalling genes and their function
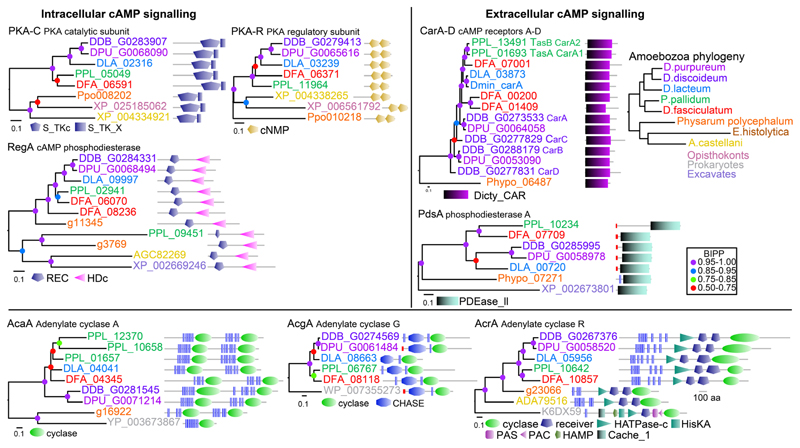
Sequencing of genomes of taxon-group representative Dictyostelia (Eichinger, et al., 2005; Gloeckner, et al., 2016; Heidel, et al., 2011; Urushihara, et al., 2015) and unicellular Amoebozoa (Clarke, et al., 2013; Loftus, et al., 2005; Schaap, et al., 2015) provides opportunities to investigate the evolutionary history of cAMP signalling. PKA is well conserved in most eukaryotes and also in Amoebozoa (Fig. 5). RegA, the other enzyme with a strictly intracellular role is present in Amoebozoa and in Excavates. Homologs of CarA and PdsA, diagnostic for extracellular roles of cAMP, are present throughout Dictyostelia and in Physarum, while PdsA-like enzymes are also present outside of Amoebozoa. The adenylate cyclase AcgA is in eukaryotes only found in Dictyostelia, while AcaA and AcrA are more broadly conserved in Amoebozoa. Because Dictyostelia secrete most of the cAMP that these adenylate cyclases synthesize (Alvarez-Curto, et al., 2007), they cannot be attributed with a strictly intra- or extracellular role.
Fig. 5. Conservation of genes involved in cAMP synthesis, detection and degration.
The closest homologs of D. discoideum cAMP signalling proteins in each of eight amoebozoan genomes were aligned with each other and with the closest homolog outside Amoebozoa. Phylogenetic trees were inferred and annotated with the functional domain architecture of the proteins. Bayesian posterior probabilities (BIPP) of the nodes are represented by coloured dots.
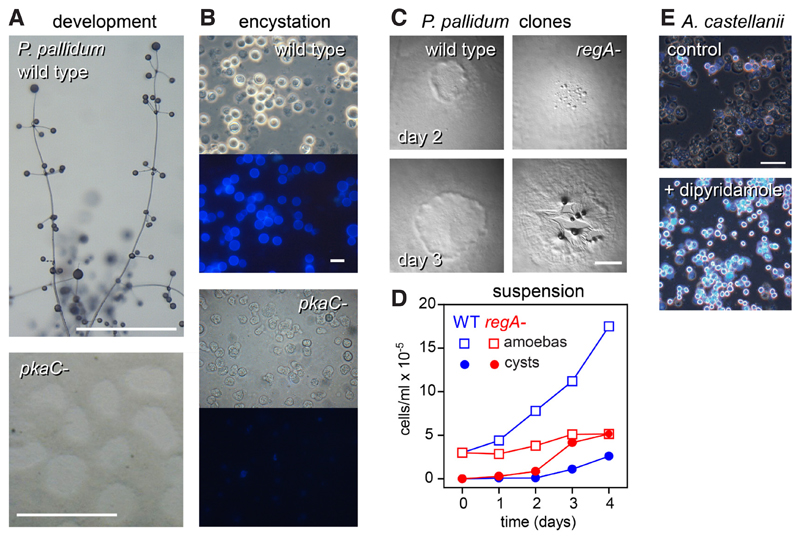
Since conserved genes may have acquired novel or different roles, more in depth analysis is required to unravel ancestral roles and identify more recent innovations. Gene knock-out and tagged mutagenesis were developed for Polysphondylium pallidum (Heterostelium album in the recent re-classification (Sheikh, et al., 2018)) in group 2 (Kawabe, et al., 1999), a species that uses glorin for aggregation (Asghar, et al., 2011) and can also encyst when conditions are unfavourable for aggregation. Inhibition of PKA function either by expression of a dominant-negative PKA inhibitor or deletion of the pkaC gene blocked P. pallidum development into fruiting bodies as is also the case in D. discoideum (Funamoto, et al., 2003; Kawabe, et al., 2015; Ritchie, et al., 2008), but also blocked their ability to encyst (Fig. 6). Deletion of both the acrA and acgA genes also prevented encystation, but did not affect fruiting body formation (Kawabe, et al., 2015), possibly because P. pallidum has three acaA genes (Fig. 5). As in D. discoideum (Shaulsky, et al., 1998), deletion of P. pallidum regA causes acceleration of development into fruiting bodies, but also strongly inhibits axenic growth, because the cells precociously encyst in growth medium (Fig. 6C,D) (Du, et al., 2014). Heterologously expressed RegA of the solitary Amoebozoan Acanthamoeba castellani is effectively inhibited by the PDE inhibitor dipyridamole. Addition of dipyridamole to its growth medium also causes precocious encystation of A. castellani amoebas (Fig. 6E). Combined, these data show that the roles of PKA and RegA in multicellular development of Dictyostelia are derived from an ancestral role in controlling encystation. While AcrA is also conserved in Acanthamoeba and Physarum, these Amoebozoa have many other adenylate cyclases (Clarke, et al., 2013; Schaap, et al., 2015), which likely also provide cAMP for PKA activation.
Fig. 6. Roles of PKA and RegA in encystation and development.
A/B. Deletion of pkaC in P. pallidum prevents amoebas from forming fruiting bodies on solid substratum (A) and from encysting when starved in suspension (B). (C,D) Deletion of regA in P. pallidum causes precocious aggregation when cells develop as clones on bacterial lawns (C), and precocious encystation, preventing proliferation, when growing in axenic suspension (D).(E) The RegA inhibitor dipyridamole induces precocious encystation in Acanthamoeba castellanii. Cellulose walls of cysts fluoresce when stained with Calcofluor in (E,B), lower panels. Simplified images from (Kawabe, et al., 2015) and (Du, et al., 2014). Bar, (A,C) 1 mm; (B,E) 10 μ.
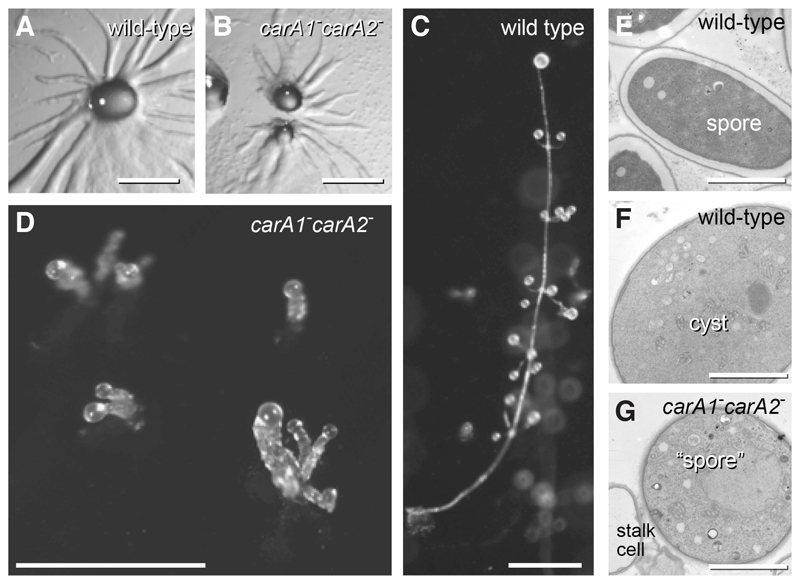
Deletion of car and pdsA genes in P. pallidum highlight that here extracellular cAMP signalling is only used in the multicellular stage. Based on synteny (Alvarez-Curto, et al., 2005) and phylogenetic evidence (Fig. 4), all car genes in non-group 4 species are orthologs and homologs of D. discoideum carA, which implies that the duplications that gave rise to carB, carC and carD only occurred in group 4. P. pallidum has duplicate carA genes, named tasA (carA1) and tasB (carA2). Loss of carA1 causes P. pallidum to form fruiting bodies with thick irregular stalks (Kawabe, et al., 2002), while loss of both carA1 and carA2 reduces the regularly whorled fruiting bodies of P.pallidum to irregularly club-shaped structures (Fig. 7). These structures contain disorganized stalk cells and “spores” that are morphologically identical to cysts (Fig. 7E-G). Unlike wild-type P. pallidum and D. discoideum, the carA1-carA2- mutant does not express prespore genes in response to cAMP stimulation (Kawabe, et al., 2009). Cell aggregation still proceeds normally, as expected from P. pallidum’s use of glorin as attractant (Asghar, et al., 2011). Deletion of pdsA yields similar disorganized fruiting bodies, with cells that were round like cysts, but otherwise ultrastructurally identical to the elliptical spores (Kawabe, et al., 2012).
Fig. 7. Roles of cAMP receptors in P. pallidum.
A knock-out of the duplicate carA-like receptors of P. pallidum shows normal aggregation (A,B), but defective fruiting body formation (C,D). Cells encapsulate in the carA1-carA2- spore heads, which closely resemble cysts and not spores (E-G). Simplified image from (Kawabe, et al., 2009). Bars A-D: 0.5 mm; E-G, 3 μ.
The loss of post-aggregative morphogenesis in carA or pdsA defective P. pallidum suggests that P. pallidum uses extracellular cAMP signalling to organize sorogen and fruiting body formation. Spore differentiation in D. discoideum requires both extracellular cAMP acting on CarA and intracellular cAMP acting on PKA, while P. pallidum encystation only requires PKA activity. P. pallidum therefore returns to the ancestral process of encystation in its fruiting bodies, when deprived of carA genes. In D. discoideum CarC-mediated activation of glycogen synthase kinase (GSK3) by cAMP favors spore differentiation by inhibiting basal disc differentiation (Harwood 2008). P. pallidum has no basal disc cells and here deletion of gsk3 has no effect on spore formation, once cells have aggregated. However, P. pallidum gsk3- cells encyst readily under conditions where wild-type cells aggregate and are hypersensitive to osmolytes, which promote encystation, and to cyst-inducing factors that are secreted during starvation (Kawabe, et al., 2018). GSK3 may therefore ancestrally have triggered aggregation in favour of encystation and gained a novel role in basal disc formation, a group 4 novelty, later in dictyostelid evolution.
Evolution of cAMP signalling in Dictyostelia
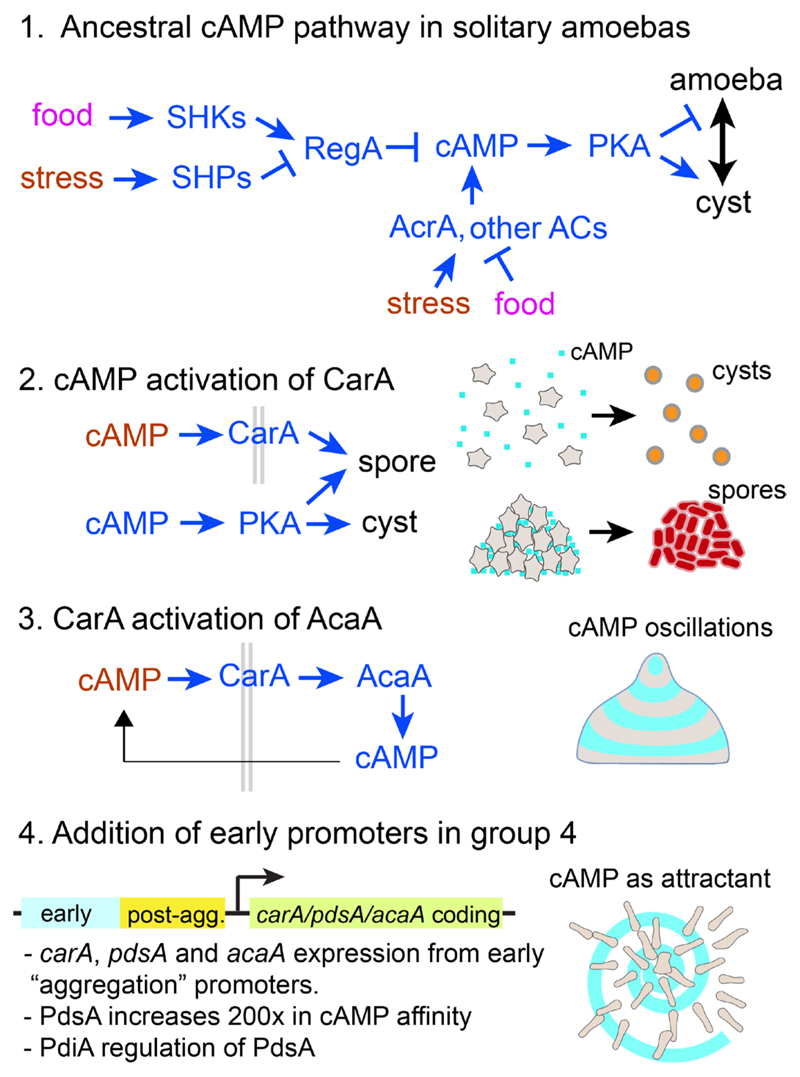
When combined the studies described above suggest the following scenario for the evolution of cAMP signalling (Fig. 8).
Fig. 8. Evolution of cAMP signalling in Dictyostelia.
Possile scenario for the evolution of intra- and extracellular cAMP signalling in D. discoideum from an amoebozoan stress response. See main text for explanation.
In unicellular Amoebozoa, environmental stress increases intracellular cAMP by increasing the activity of AcrA and/or other adenylate cyclases and inhibiting the activity of RegA. Like Dictyostelia, free-living Amoebozoa have many sensor histidine kinases/phosphatases (SHKPs) in their genomes (Table 2). In D. discoideum RegA activity is controlled by several SHKPs that act as sensors for developmental signals that regulate the differentiation and dormancy of spores (Fig. 4), such as SDF2 (Wang, et al., 1999), NH3 (Singleton, et al., 1998), discadenine (Zinda and Singleton 1998) and osmolytes (Ott, et al., 2000). In solitary Amoebozoa they likely detect environmental stress signals, such as drought (which increases osmolyte levels) or food availability, which control encystation and excystation, respectively.
The first role of extracellular cAMP probably emerged once the dictyostelid ancestor evolved aggregation and spore formation. Dictyostelids secrete most of the cAMP that they synthesize (Alvarez-Curto, et al., 2007). While this would not raise extracellular levels much when the cells are dispersed, once they are aggregated it might yield the micromolar levels required for prespore differentiation (Schaap and Van Driel 1985). In this scenario, secreted cAMP acting on CarA could be a signal for the aggregated state, instructing cells to form spores and not cysts.
While all extant Dictyostelids form a well-structured stalk starting from the organizing tip of the sorogen, this process was unlikely to have been in place when their sorocarpic habit first emerged. The proto-dictyostelid ancestor is more likely to have formed the simple structures with decaying cells and/or extensive matrix production that characterize most other sorocarpic organisms (Fig. 1). The current well-organized mode of stalk formation in D. discoideum is the result of interaction between the function of the tip as the organizer that secretes cAMP pulses to organize cell movement (Singer, et al., 2019), and the stalk-inducing signal c-di-GMP, which hyper-activates the adenylate cyclase, AcaA, that produces the cAMP pulses, thereby activating PKA, which turns the tip cells into stalk cells (Chen and Schaap 2012; Chen, et al., 2017). We propose that the interaction of CarA with AcaA was a third step in the evolution of cAMP signalling that set up the conditions for oscillatory cAMP signalling and well-organized morphogenesis (Fig. 8).
Finally, extension of the usage of pulsatile cAMP signalling to the aggregative stage of development in the last common ancestor to group 4, caused use of cAMP as chemoattractant in this group. The structure of the regulatory regions of the carA (Louis, et al., 1993), pdsA (Faure, et al., 1990) and acaA (Galardi-Castilla, et al., 2010) genes, where the promoters for post-aggregative expression are proximal to the coding sequence and the promoters for pre-aggregative expression more distal (Fig. 8), suggest that this novel usage was, if not caused, at least correlated with the addition of distal promoters to existing cAMP signalling genes. Aggregation and oscillatory signalling of a D. discoideum carA-carC- mutant can be restored by a group 3 carA (Alvarez-Curto, et al., 2005), indicating the CarA function did not change in group 4. However, PdsA, which in group 3 binds cAMP and cGMP equally well with low affinity, increased its affinity for cAMP 200-fold in group 4 (Kawabe, et al., 2012), likely to be able to hydrolyse the lower cAMP concentrations present in a dispersed field of amoeba, than the higher concentration in the group 3 aggregates. The secreted PdsA inhibitor, PdiA, which favours generation of spiral cAMP waves that organize large territories over concentric waves that control smaller territories (Palsson and Cox 1996), only emerged in group 4 (Kawabe, et al., 2012). This may explain the ability of group 4 species to form larger slugs and fruiting bodies than species in the other groups (Fig. 3).
Table 2. Validated cases of lateral gene transfer in Dictyostelia.
| Gene | Function | Biological role |
|---|---|---|
| chlA | DIF-1 chlorination | induces basal disc |
| dgcA | c-di-GMP synthesis | induces stalk formation |
| dokA | osmolyte sensing | osmotic stress resistance, spore viability |
| iptA | discadenine synthesis | induces spore maturation and dormancy |
Reproduced from (Gloeckner, et al., 2016).
While speculative, this evolutionary scenario explains how and why D. discoideum came to use cAMP in so many different roles. It also shows the power of the comparative approach to distinguish between core regulatory mechanisms and recent innovations that only occurred in a small group of species. It is a strategy that should be applied to infer the broader relevance of any new gene function or mechanism uncovered in a single organism approach. Additionally, changes in gene function or regulation may reveal how genotypic change caused phenotypic innovations, one of the most fundamental problems in biology.
Are there specific multicellularity genes?
While the above paragraphs focus on cAMP signalling, broader comparative analysis of the genomes of multicellular Dictyostelia and unicellular Amoebozoa allows discovery of more features that are unique to the multicellular forms. Since the regulation of multicellular development requires intensive intercellular communication, such a feature might be the number of genes involved in signal detection and processing. However, here a comparison of uni- and multcellular amoebozoan genomes showed that the multicellular Dictyostelia actually have overall less members in major families of signal transduction proteins than unicellular Amoebozoa (Table 1), indicating that simply more signal transducers are not a prerequisite for multicellularity.
Table 1. Signal transduction proteins in uni- and multicellular Amoebozoa.
| Category | Dictyostelium discoideum | Protostelium aurantium | Physarum polycephalum | Acanthamoeba castellanii |
|---|---|---|---|---|
| G-protein coupled receptors | 55 | 17 | 146 | 35 |
| Heterotrimeric G-proteins | ||||
| Alpha | 12 | 9 | 26 | 6 |
| Beta | 1 | 1 | 1 | n.d. |
| Gamma | 1 | 1 | 1 | n.d. |
| Histidine kinases/phosphatases | 16 | 71 | 51 | 48 |
| Cyclic nucleotide signaling | ||||
| Adenylate/guanylate cyclases | 5 | 52 | 64 | 67 |
| cNMP binding domains | 5 | 27 | 28 | 7 |
| cNMP phosphodiesterases | 7 | 16 | 11 | 10 |
| Protein kinases | ||||
| All (S/T, S/T/Y, Y) | 295 | 827 | 447 | 377 |
| Sensor tyrosine kinases (Y) | 0 | 167 | 4 | 21 |
| SH2 domain proteins | 15 | 85 | 18 | 48 |
Retrieved from (Hillmann, et al., 2018).
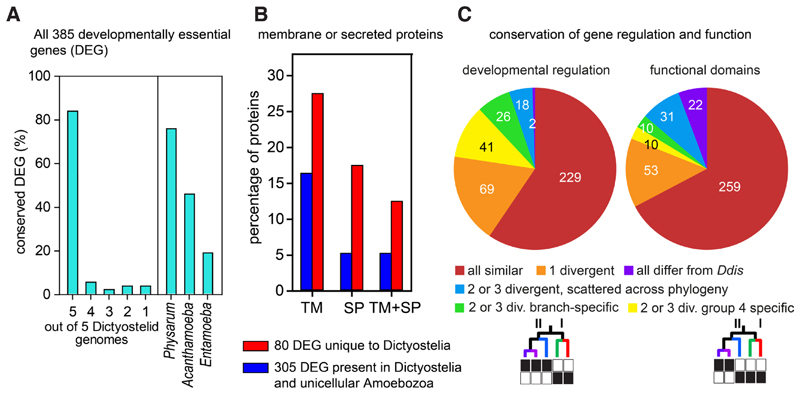
In D. discoideum knock-outs in ~385 genes cause a defect in multicellular development (Basu, et al., 2015). About 84% of these developmentally essential genes (DEG) are conserved across all Dictyostelium taxon groups, while 76% are present in at least one of the genomes of solitary Amoebozoa (Fig. 9A). Strikingly, the 80 genes that were unique to Dictyostelia were strongly enriched in proteins with transmembrane domains and/or signal peptides (Fig. 9B). Gene ontology enrichment analysis showed that DEG unique to Dictyostelia were enriched in G-protein coupled receptors, sensor histidine kinases and cell-cell recognition proteins, while the DEG that were also present in solitary Amoebozoa were enriched in protein kinases, transferase activity and nucleotide binding (Gloeckner, et al., 2016). This suggests that the proteins shared with solitary amoebas tend to be involved in intracellular signal processing, while those unique to Dictyostelia are involved in signal sensing, signal exposure or secretion and cell-cell recognition. In other words, the multicellular forms needed new signals and sensors, but the signal processing components were mostly already there. Four of the genes unique to Dictyostelia entered their genomes by horizontal gene transfer. One, dokA, encodes an osmosensor, while the other three chlA, dgcA and iptA are enzymes that synthesize three out of the five known small molecules that regulate cell differentiation (Table 2), highlighting the need of emerging multicellular organisms for novel signals and their sensors.
Fig. 9. Conservation of D. discoideum developmentally essential genes across Amoebozoa.
(A) Conservation of 385 genes that are essential for D. discoideum multicellular development (DEG) across 5 group-representative Dictyostelium genomes and 3 solitary amoebozoan genomes. (B) Percentages of proteins with transmembrane (TM) domains, signal peptides (SP) or both among 80 DEG that are unique to Dictyostelia (blue bars) or the 305 DEG that are also present in solitary Amoebozoa. (C) Phylogenetic distribution of DEG with conserved developmental regulation or conserved protein functional domains. Reproduced in simplified form from (Gloeckner, et al., 2016).
Across Dictyostelia, both the developmental regulation of DEG and the functional domain architecture of the encoded proteins is conserved for 59% and 67% of genes, respectively. When functional domains were not conserved for 2 or 3 genes, their occurrence was mostly scattered across the phylogeny. However, differences in developmental expression mostly occurred between group 4 and groups 1-3, followed by differences between the genetically more distant branches I and II of the phylogeny (Fig. 8C). The group 4 specific changes in developmental regulation correlate with the major phenotypic change that occurs in group 4 (Fig. 3), indicating that changes in gene expression are more likely to cause phenotypic change than dramatic changes in gene function, caused by loss or gain of functional domains. In the case of the novel role of cAMP as chemoattractant in group 4, as outlined in the previous paragraph, it appeared that changes in both gene regulation and gene function were important, as exemplified by the addition of early promoters to the carA, acaA and pdsA genes and the increased affinity of the PdsA protein for cAMP. The increase in PdsA affinity may however be due to changes in just a few amino-acids. The use of loss of protein functional domains, the only practicable metric available for large scale genome comparison may be too crude to assess such subtle change. This illustrates the importance of experimentation to consolidate the broad but shallow evidence base of bioinformatic approaches.
Conclusions
Aggregative or sorocarpic multicellularity, as displayed by Dictyostelium, is the most common form of multicellularity in eukaryotes
Within Dictyostelia the most extensive phenotypic innovation occurred in the last common ancestor to group 4.
The multiple roles of extracellular and intracellular cAMP signalling in the group 4 species D. discoideum likely evolved from an ancestral role of cAMP as second messenger for environmental stress in solitary amoebas.
Multicellularity requires new signals and receptors, but most signal transduction pathways are already present in the unicellular ancestors
Abbreviations used in this paper
- cAMP
3’,5’-cyclic adenosine monophosphate
- DEG
developmentally essential genes
- LCA
last common ancestor
- PKA
cAMP dependent protein kinase
- SHKP
sensor histidine kinases/phosphatases
- SSU-rDNA
small subunit ribosomal DNA
Footnotes
Pauline Schaap: https://orcid.org/0000-0003-4500-2555
References
- Alvarez-Curto E, Rozen DE, Ritchie AV, Fouquet C, Baldauf SL, Schaap P. Evolutionary origin of cAMP-based chemoattraction in the social amoebae. Proc Natl Acad Sci USA. 2005;102:6385–6390. doi: 10.1073/pnas.0502238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Curto E, Weening KE, Schaap P. Pharmacological profiling of the Dictyostelium adenylate cyclases ACA, ACB and ACG. Biochem J. 2007;401:309–316. doi: 10.1042/BJ20060880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar A, Groth M, Siol O, Gaube F, Enzensperger C, Glockner G, Winckler T. Developmental gene regulation by an ancient intercellular communication system in social amoebae. Protist. 2011:25–37. doi: 10.1016/j.protis.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Basu S, Fey P, Jimenez-Morales D, Dodson RJ, Chisholm RL. dictyBase 2015: Expanding data and annotations in a new software environment. Genesis. 2015;53:523–534. doi: 10.1002/dvg.22867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton RL, Olive LS. Stalk function during sorogenesis by the ciliated protozoan sorogena-stoianovitchae. Protoplasma. 1983;116:136–144. [Google Scholar]
- Brown Matthew W, Kolisko M, Silberman Jeffrey D, Roger Andrew J. Aggregative Multicellularity Evolved Independently in the Eukaryotic Supergroup Rhizaria. Curr Biol. 2012a;22:1123–1127. doi: 10.1016/j.cub.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Brown MW, Silberman JD, Spiegel FW. “Slime molds” among the Tubulinea (Amoebozoa): molecular systematics and taxonomy of Copromyxa. Protist. 2011;162:277–287. doi: 10.1016/j.protis.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Brown MW, Silberman JD, Spiegel FW. A contemporary evaluation of the acrasids (Acrasidae, Heterolobosea, Excavata) Eur J Protistol. 2012b;48:103–123. doi: 10.1016/j.ejop.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 2009;26:2699–2709. doi: 10.1093/molbev/msp185. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Singh R, Cole C, Lawal HM, Schilde C, Febrer M, Barton GJ, Schaap P. Adenylate cyclase A acting on PKA mediates induction of stalk formation by cyclic diguanylate at the Dictyostelium organizer. Proc Natl Acad Sci U S A. 2017;114:516–521. doi: 10.1073/pnas.1608393114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Burglin TR, Frech C, et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013;14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Kawabe Y, Schilde C, Chen ZH, Schaap P. The evolution of aggregative multicellularity and cell-cell communication in the Dictyostelia. J Molec Biol. 2015;427:3722–3733. doi: 10.1016/j.jmb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Schilde C, Birgersson E, Chen ZH, McElroy S, Schaap P. The cyclic AMP phosphodiesterase RegA critically regulates encystation in social and pathogenic amoebas. Cell Signal. 2014;26:453–459. doi: 10.1016/j.cellsig.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure M, Franke J, Hall AL, Podgorski GJ, Kessin RH. The cyclic nucleotide phosphodiesterase gene of Dictyostelium discoideum contains three promoters specific for growth, aggregation, and late development. Mol Cell Biol. 1990;10:1921–1930. doi: 10.1128/mcb.10.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funamoto S, Anjard C, Nellen W, Ochiai H. cAMP-dependent protein kinase regulates Polysphondylium pallidum development. Differentiation. 2003;71:51–61. doi: 10.1046/j.1432-0436.2003.700605.x. [DOI] [PubMed] [Google Scholar]
- Galardi-Castilla M, Garciandia A, Suarez T, Sastre L. The Dictyostelium discoideum acaA gene is transcribed from alternative promoters during aggregation and multicellular development. PLoS ONE. 2010;5:e13286. doi: 10.1371/journal.pone.0013286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner G, Lawal HM, Felder M, Singh R, Singer G, Weijer CJ, Schaap P. The multicellularity genes of dictyostelid social amoebas. Nat Commun. 2016;7 doi: 10.1038/ncomms12085. 12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood AJ. Dictyostelium development: a prototypic Wnt pathway? Meth Mol Biol. 2008;469:21–32. doi: 10.1007/978-1-60327-469-2_2. [DOI] [PubMed] [Google Scholar]
- He D, Fiz-Palacios O, Fu CJ, Fehling J, Tsai CC, Baldauf SL. An alternative root for the eukaryote tree of life. Curr Biol. 2014;24:465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- Heidel A, Lawal H, Felder M, Schilde C, Helps N, Tunggal B, Rivero F, John U, Schleicher M, Eichinger L, Platzer M, et al. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 2011:1882–1891. doi: 10.1101/gr.121137.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmann F, Forbes G, Novohradska S, Ferling I, Riege K, Groth M, Westermann M, Marz M, Spaller T, Winckler T, Schaap P, et al. Multiple roots of fruiting body formation in Amoebozoa. Genome Biol Evol. 2018;10(2):591–606. doi: 10.1093/gbe/evy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Enomoto T, Morio T, Urushihara H, Tanaka Y. LbrA, a protein predicted to have a role in vesicle trafficking, is necessary for normal morphogenesis in Polysphondylium pallidum. Gene. 1999;239:75–79. doi: 10.1016/s0378-1119(99)00379-0. [DOI] [PubMed] [Google Scholar]
- Kawabe Y, Kuwayama H, Morio T, Urushihara H, Tanaka Y. A putative serpentine receptor gene tasA required for normal morphogenesis of primary stalk and branch structure in Polysphondylium pallidum. Gene. 2002;285:291–299. doi: 10.1016/s0378-1119(01)00887-3. [DOI] [PubMed] [Google Scholar]
- Kawabe Y, Morio T, James JL, Prescott AR, Tanaka Y, Schaap P. Activated cAMP receptors switch encystation into sporulation. Proc Natl Acad Sci USA. 2009;106:7089–7094. doi: 10.1073/pnas.0901617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Morio T, Tanaka Y, Schaap P. Glycogen synthase kinase 3 promotes multicellular development over unicellular encystation in encysting Dictyostelia. EvoDevo. 2018;9:12. doi: 10.1186/s13227-018-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Schilde C, Du Q, Schaap P. A conserved signalling pathway for amoebozoan encystation that was co-opted for multicellular development. Sci Rep. 2015;5 doi: 10.1038/srep09644. 9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Weening KE, Marquay-Markiewicz J, Schaap P. Evolution of self-organisation in Dictyostelia by adaptation of a non-selective phosphodiesterase and a matrix component for regulated cAMP degradation. Development. 2012;139:1336–1345. doi: 10.1242/dev.077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konijn TM, Van de Meene JG, Bonner JT, Barkley DS. The acrasin activity of adenosine-3’,5’-cyclic phosphate. Proc Natl Acad Sci USA. 1967;58:1152–1154. doi: 10.1073/pnas.58.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- Loomis WF. Cell signaling during development of Dictyostelium. Dev Biol. 2014;391:1–16. doi: 10.1016/j.ydbio.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis JM, Saxe CL, III, Kimmel AR. Two transmembrane signaling mechanisms control expression of the cAMP receptor gene cAR1 during Dictyostelium development. Proc Natl Acad Sci USA. 1993;90:5969–5973. doi: 10.1073/pnas.90.13.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons NA, Kolter R. On the evolution of bacterial multicellularity. Current Opinion in Microbiology. 2015;24:21–28. doi: 10.1016/j.mib.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita TB, Chen ZH, Schaap P, Saito T. The Hybrid Type Polyketide Synthase SteelyA Is Required for cAMP Signalling in Early Dictyostelium Development. PLoS ONE. 2014;9:e106634. doi: 10.1371/journal.pone.0106634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W, Silan C, Firtel RA. DNA-mediated transformation in Dictyostelium discoideum: Regulated expression of an actin gene fusion. Mol Cell Biol. 1984;4(12):2890–8. doi: 10.1128/mcb.4.12.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott A, Oehme F, Keller H, Schuster SC. Osmotic stress response in Dictyostelium is mediated by cAMP. EMBO J. 2000;19:5782–5792. doi: 10.1093/emboj/19.21.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson E, Cox EC. Origin and evolution of circular waves and spirals in Dictyostelium discoideum territories. Proc Natl Acad Sci USA. 1996;93:1151–1155. doi: 10.1073/pnas.93.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper KB. Pseudoplasmodium formation and organization in Dictyostelium discoideum. J Elisha Mitchell Scient Soc. 1940;56:241–282. [Google Scholar]
- Ritchie AV, Van Es S, Fouquet C, Schaap P. From drought sensing to developmental control: evolution of cyclic AMP signaling in social amoebas. Mol Biol Evol. 2008;25:2109–2118. doi: 10.1093/molbev/msn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeralo M, Skiba A, Gonzalez-Voyer A, Schilde C, Lawal H, Kedziora S, Cavender JC, Glockner G, Urushihara H, Schaap P. Analysis of phenotypic evolution in Dictyostelia highlights developmental plasticity as a likely consequence of colonial multicellularity. Proc Biol Sci / Royal Soc. 2013;280 doi: 10.1098/rspb.2013.0976. 20130976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato YG, Suarez T, Saito T. Stalk cell differentiation without polyketides in the cellular slime mold. Biosci Biotechnol Biochem. 2016;80:1368–1374. doi: 10.1080/09168451.2016.1162087. [DOI] [PubMed] [Google Scholar]
- Schaap P, Barrantes I, Minx P, Sasaki N, Anderson RW, Benard M, Biggar KK, Buchler NE, Bundschuh R, Chen X, Fronick C, et al. The Physarum polycephalum Genome Reveals Extensive Use of Prokaryotic Two-Component and Metazoan-Type Tyrosine Kinase Signaling. Genome Biol Evol. 2015;8:109–125. doi: 10.1093/gbe/evv237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap P, Van Driel R. Induction of post-aggregative differentiation in Dictyostelium discoideum by cAMP. Evidence of involvement of the cell surface cAMP receptor. Exp Cell Res. 1985;159:388–398. doi: 10.1016/s0014-4827(85)80012-4. [DOI] [PubMed] [Google Scholar]
- Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, Cavender J, Milano-Curto A, Rozen DE, Dingermann T, Mutzel R, et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilde C, Lawal HM, Kin K, Shibano-Hayakawa I, Inouye K, Schaap P. A well supported multi gene phylogeny of 52 dictyostelia. Mol Phylogenet Evol. 2019;134:66–73. doi: 10.1016/j.ympev.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilde C, Skiba A, Schaap P. Evolutionary reconstruction of pattern formation in 98 Dictyostelium species reveals that cell-type specialization by lateral inhibition is a derived trait. EvoDevo. 2014;5:34. doi: 10.1186/2041-9139-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulsky G, Fuller D, Loomis WF. A cAMP-phosphodiesterase controls PKA-dependent differentiation. Development. 1998;125:691–699. doi: 10.1242/dev.125.4.691. [DOI] [PubMed] [Google Scholar]
- Sheikh S, Gloeckner G, Kuwayama H, Schaap P, Urushihara H, Baldauf SL. Root of Dictyostelia based on 213 universal proteins. Mol Phylogenet Evol. 2015;92:53–62. doi: 10.1016/j.ympev.2015.05.017. [DOI] [PubMed] [Google Scholar]
- Sheikh S, Thulin M, Cavender JC, Escalante R, Kawakami SI, Lado C, Landolt JC, Nanjundiah V, Queller DC, Strassmann JE, Spiegel FW, et al. A New Classification of the Dictyostelids. Protist. 2018;169:1–28. doi: 10.1016/j.protis.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Singer G, Araki T, Weijer CJ. Oscillatory cAMP cell-cell signalling persists during multicellular Dictyostelium development. Commun Biol. 2019;2:139. doi: 10.1038/s42003-019-0371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Schilde C, Schaap P. A core phylogeny of Dictyostelia inferred from genomes representative of the eight major and minor taxonomic divisions of the group. BMC Evol Biol. 2016;16:251. doi: 10.1186/s12862-016-0825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton CK, Zinda MJ, Mykytka B, Yang P. The histidine kinase dhkC regulates the choice between migrating slugs and terminal differentiation in Dictyostelium discoideum. Dev Biol. 1998;203:345–357. doi: 10.1006/dbio.1998.9049. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Endoh H. Analysis of fruiting body development in the aggregative ciliate Sorogena stoianovitchae (Ciliophora, Colpodea) J Eukaryot Microbiol. 2006;53:96–102. doi: 10.1111/j.1550-7408.2005.00077.x. [DOI] [PubMed] [Google Scholar]
- Tice AK, Silberman JD, Walthall AC, Le KN, Spiegel FW, Brown MW. Sorodiplophrys stercorea: Another Novel Lineage of Sorocarpic Multicellularity. J Eukaryot Microbiol. 2016;63(5):623–8. doi: 10.1111/jeu.12311. [DOI] [PubMed] [Google Scholar]
- Urushihara H, Kuwayama H, Fukuhara K, Itoh T, Kagoshima H, Shin IT, Toyoda A, Ohishi K, Taniguchi T, Noguchi H, Kuroki Y, et al. Comparative genome and transcriptome analyses of the social amoeba Acytostelium subglobosum that accomplishes multicellular development without germ-soma differentiation. BMC Genomics. 2015;16:80. doi: 10.1186/s12864-015-1278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Soderbom F, Anjard C, Shaulsky G, Loomis WF. SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol Cell Biol. 1999;19:4750–4756. doi: 10.1128/mcb.19.7.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JG. Transcriptional regulation of Dictyostelium pattern formation. EMBO Rep. 2006;7:694–698. doi: 10.1038/sj.embor.7400714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinda MJ, Singleton CK. The hybrid histidine kinase dhkB regulates spore germination in Dictyostelium discoideum. Dev Biol. 1998;196:171–183. doi: 10.1006/dbio.1998.8854. [DOI] [PubMed] [Google Scholar]