Abstract
Background
The aim of this study was to generate a prognostic model to predict survival outcome in pediatric Wilms tumor (WT).
Methods
The data including mRNA expression and clinical information of pediatric WT patients were downloaded from the Therapeutically Available Research to Generate Effective Treatments (TARGET) database. The differentially expressed genes were identified and a prognostic signature of pediatric WT was generated according to the results of univariate and multivariate Cox analysis. Receiver operating characteristic (ROC) curve was used to evaluate the five‐mRNA signature in pediatric Wilms tumor patients. Bootstrap test with 500 times was used to perform the internal validation.
Results
We identified 6,964 differentially expressed mRNAs associated with pediatric WT, including 3,190 downregulated mRNAs and 3,774 up‐regulated mRNAs. Univariate and multivariate Cox analysis identified five mRNAs (SPRY1, SPIN4, MAP7D3, C10orf71, and SPAG11A) to establish a predictive model. The risk score formula is as follows: Risk score = 0.3036*SPIN4 + 0.8576*MAP7D3 −0.1548*C10orf71 −0.7335*SPRY1 −0.2654*SPAG11A. The pediatric WT patients were divided into low‐risk group and high‐risk group based on the median risk score (value = 1.1503). The receiver operating characteristic (ROC) curve analysis revealed good performance of the 5‐mRNA prognostic model (the area under the curve [AUC] was 0.821). Bootstrap test (Bootstrap resampling times = 500) was used to perform the internal validation and revealed that the AUC was 0.822. REACTOME, KEGG, and BIOCARTA pathway analyses demonstrated that these survival‐related genes were mainly enriched in ErbB2 and ErbB3 signaling pathways, and calcium signaling pathway.
Conclusion
The five‐mRNA signature can predict the prognosis of patients with pediatric WT. It has significant implication in the understanding of therapeutic targets for pediatric WT patients. However, further study is needed to validate this five‐mRNA signature and uncover more novel diagnostic or prognostic mRNAs candidates in pediatric WT patients.
Keywords: bioinformatics, biomarkers, mRNAs, prognosis, Wilms tumor
The five‐mRNA signature can predict the prognosis of patients with pediatric WT. It has significant implication in the understanding of therapeutic targets for pediatric WT patients. However, further study is needed to validate this five‐mRNA signature and uncover more novel diagnostic or prognostic mRNAs candidates in pediatric WT patients.

1. INTRODUCTION
The incidence of Wilms tumor (WT) among children younger than 15 years is 7.1 cases per 1 million. WT, which accounts for more than 90% of renal tumors among pediatric patients, is the most common solid renal malignancy (Cone et al., 2016; Pastore et al., 2006; Stokes et al., 2018). The incidence is lower in Asians when compared with that in the United States. The male to female ratio is quite different between unilateral cases and bilateral cases of WT (0.92–1.00 in unilateral cases and 0.6–1.00 in bilateral cases). Also, the mean age at diagnosis between unilateral cases and bilateral cases are different (44 months in unilateral cases and 31 months in bilateral cases) (Breslow, Olshan, Beckwith, & Green, 1993; Phelps et al., 2019; Wang, Lou, & Ma, 2019). Scott RH et al. (Scott, Stiller, Walker, & Rahman, 2006) reported that about 10% of children with WT was suffering from congenital malformation syndrome. WT arise after a limited number of genetic aberrations, as reported by the Gadd S et al. (Gadd et al., 2017), including Wilms tumor 1 transcription factor (WT1; OMIM: 194070), catenin beta 1 (CTNNB1; OMIM: 116806), or APC membrane recruitment protein 1 (AMER1; OMIM: 300647) etc. Recently, some studies(Moch, Cubilla, Humphrey, Reuter, & Ulbright, 2016; Wegert et al., 2015) have revealed that approximately 15% of WT have microRNA‐processing gene mutations, including drosha ribonuclease III (DROSHA; OMIM: 608828), DGCR8 microprocessor complex subunit (DGCR8; OMIM: 609030), dicer 1 ribonuclease III (DICER1; OMIM: 606241), exportin 5 (XPO5; OMIM: 607845) and TARBP2 subunit of RISC loading complex (TARBP2; OMIM: 605053).
Besides stage and histology, a variety of clinical and biological factors was used to define treatment, including age, tumor size and volume, the loss of heterozygosity at chromosomes 1p and 16q, and response to chemotherapy (Dome et al., 2013; Dome, Perlman, & Graf, 2014). The treatment studies of children with WT have been evaluated by two different clinical groups, including COG Renal Tumor Committee (COG RTC) (D'Angio et al., 1989) and SIOP (Graf, Tournade, & de Kraker, 2000). The standard approach to WT treatment in the COG RTC group was immediate surgery, while the first step in treatment in SIOP was preoperative chemotherapy. Postoperative chemotherapy was used in both groups. The long‐term survival outcomes of pediatric WT patients has improved gradually in the recent years, however, the subsequent chronic health conditions, including renal failure, cardiac toxicity, and subsequent malignancies should not be ignored (Aldrink et al., 2018; Gratias et al., 2016; Wong et al., 2016).
High risk groups compose 25% of patients with WT, including those with unfavorable histological, bilateral disease, and recurrence disease (Dome et al., 2015). As we all know, the heterogeneity among individuals often makes conventional prognostic systems. For instance, the risk stratification of TNM staging system is not sufficient. Besides, it is also insufficient to provide an accurate estimation of survival outcome. Thus, it is urgent to generate an accurate prognostic model to predict the survival outcomes in pediatric WT patients. Prognostic model plays a crucial role in the management of tumors, such as prostate specific antigen, alpha fetoproteinca, and carcinoembryonic antigen. Although a meta‐analysis (Cone et al., 2016) reported that a large number of tumor biomarkers have been used to predict the prognostic outcomes in pediatric WT, there has been no prior study which has focused on an mRNA signature to predict the prognosis of WT patients.
The present study aimed to conduct an integrated study to develop a five‐mRNA signature for the prognostic predication of WT patients by analyzing pediatric WT patients from Therapeutically Available Research to Generate Effective Treatments (TARGET) database.
2. MATERIALS AND METHODS
2.1. Acquisition of TARGET pediatric WT data
The RNA‐seq data (level 3) and corresponding clinical information of pediatric WT in TARGET database were downloaded from Genomic Data Commons Data Portal (portal.gdc.cancer.gov/). We identified 136 cases investigated in this study, including 6 normal samples and 130 WT samples. No further normalization was needed for the expression data downloaded from TARGET database which have already been normalized. The data with no expression were deleted previously. The level 3 RNA‐seq data between normal tissues and WT tissues were analyzed by edgeR package based on R language for differential expression analysis. Genes with absolute log 2 fold change > 1 and p < .05 were regarded as differentially expressed mRNAs. Since the data come from the TARGET database, no further approval was required from the Ethics Committee.
2.2. Survival analysis
Clinical data were combined with those pediatric patients with WT in TARGET database to identify the prognostic differential expressed mRNAs signature. The survival curves of those samples with differential expressed mRNAs were plotted by using the “survival” package in R. The primary endpoint was overall survival. Univariate Cox analysis and multivariate Cox analysis were performed in this study. All identified differential expressed mRNAs were performed by univariate Cox analysis. The hazard ratio and P value of all differential expressed mRNAs were calculated. Receiver operating characteristic (ROC) curve has been used to prove the sensitivity and specificity of the calculated riskscore in predicting the overall survival of pediatric WT patients. The area under the curve (AUC) was generated and bootstrap was used to estimate 95%CI with the AUC.
2.3. Pathway analysis
The DAVID online tool (https://david.ncifcrf.gov/) was used to annotate the survival‐related mRNAs as previously described (Ke et al., 2019; Xu et al., 2019; Xu, Wu, Yin, Xue, & Gou, 2018). REACTOME (http://www.reactome.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/pathway.html), and BIOCARTA (https://cgap.nci.nih.gov/Pathways/BioCarta_Pathways) pathway databases were used to perform pathway analyses among survival‐related mRNAs screened by univariate Cox analysis.
2.4. Statistical analysis
Kaplan–Meier survival analyses were used to determine the overall survival of pediatric Wilms patients who were classified as high expression and low expression group based on the median expression level of each differentially expressed mRNA. Log‐rank test with the R package “survival” was used to determine the difference in the survival of pediatric patients. p < .05 was considered as statistically significant.
3. RESULTS
3.1. Survival analysis by Kaplan–Meier method among differentially expressed mRNAs in pediatric WT patients
We identified 6,964 differentially expressed mRNAs, including 3,190 downregulated mRNAs and 3,774 upregulated mRNAs. Survival analyses among each deferentially expressed mRNAs were performed by Kaplan–Meier method subsequently. The high expression and low expressed of those genes including chromosome 10 open reading frame 71 (C10orf71), EF‐hand calcium binding domain 5 (EFCAB5), hes‐related family bHLH transcription factor with YRPW motif 1(HEY1; OMIM: 602953), interleukin 20 receptor subunit alpha (IL20RA; OMIM: 605620), LINE1 type transposase domain containing 1(L1TD1), MAP7 domain containing 3 (MAP7D3; OMIM: 300930), polycomb group ring finger 3 (PCGF3; OMIM: 617543), pregnancy specific beta‐1‐glycoprotein 5 (PSG5; OMIM: 176394), RNA binding motif protein 15 (RBM15; OMIM: 606077), sarcoglycan delta (SGCD; OMIM: 601411), sperm associated antigen 11A (SPAG11A), spindlin family member 4 (SPIN4), sprouty RTK signaling antagonist 1 (SPRY1; OMIM: 602465), threonyl‐tRNA synthetase (TARS). The high expression of mRNAs, including C10orf71, EFCAB5, HEY1, SGCD, SPAG11A and SPRY1 were associated with favor overall survival in pediatric patients with WT (p < .05) (Figure 1a–f). Plus, the low expression of mRNAs, including IL20RA, L1TD1, MAP7D3, PCGF3, PSG5, RBM15, SPIN4, and TARS were associated with worse overall survival in pediatric patients with WT (p < .05) (Figure 1g–n).
Figure 1.
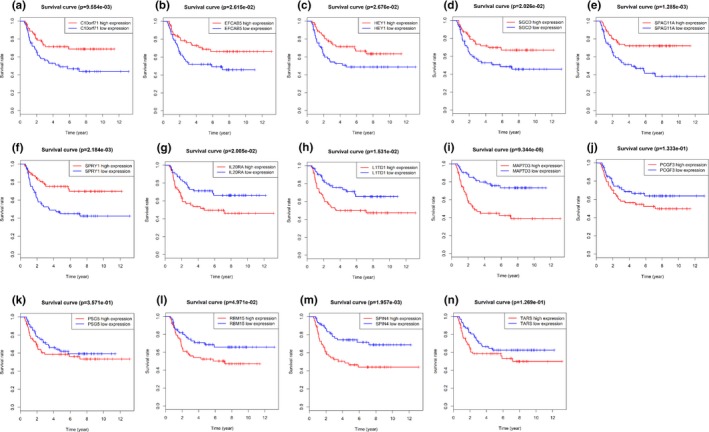
Kaplan–Meier survival curve analysis for overall survival of mRNAs in pediatric Wilms tumor patients. (a–f) mRNAs were associated with favor overall survival in pediatric patients with WT (p < .05). (g–n) mRNAs were associated with worse overall survival in pediatric patients with WT (p < .05)
3.2. Survival analysis by univariate cox analysis and multivariate cox analysis among differentially expressed mRNAs in pediatric WT patients
Univariate Cox analysis for all differentially expressed mRNAs was assessed to determine the survival‐related mRNAs (Table S1). The primary endpoint for survival analysis was overall survival. The significant level cutoff threshold was set as 0.001 (p < .001) to identify the candidate mRNAs (Table 1). Multivariate Cox analysis was then performed by using these candidate mRNAs identified by univariate Cox analysis. Finally, five mRNAs (SPRY1, SPIN4, MAP7D3, C10orf71, and SPAG11A) were identified (Table 1). The results of multivariate Cox analysis also revealed the independent prognostic value of these 5 hub mRNAs. Two were associated with high risk of death in pediatric WT (SPIN4 and MAP7D3). SPIN4 and MAP7D3 were associated with a poor overall survival of pediatric WT patients. Specifically, the risk of death in patient with high expression of SPIN4 was 1.355 times higher than patient with low expression of SPIN4. Plus, the risk of death in patient with higher expression of MAP7D3 was 2.358 times higher than patient with low expression of MAP7D3. Also, three were associated with low risk of death in pediatric WT (SPRY1, C10orf71, and SPAG11A). the risk of death in patients with low expression of SPRY1 was 2.083 times higher than patient with high expression of SPRY1. Plus, the risk of death in patients with low expression of C10orf71 was 1.167 times higher than patients with high expression of C10orf71. Also, the risk of death in patients with low expression of SPAG11A was 1.304 times higher than patients with high expression of SPAG11A.
Table 1.
Univariate and multivariate Cox analysis of overall survival
| Univariate Cox analysis | Multivariate Cox analysis | ||||
|---|---|---|---|---|---|
| mRNA | HR | p | mRNA | coef | HR |
| SPRY1 | 0.354022 | 6.96E‐07 | SPRY1 | −0.733483894 | 0.480232991 |
| SPIN4 | 2.035141 | 2.51E‐05 | SPIN4 | 0.303637441 | 1.35477778 |
| IL20RA | 1.295478 | 0.000108 | |||
| EFCAB5 | 0.442097 | 0.000172 | |||
| PSG5 | 1.525531 | 0.000218 | |||
| TARS | 4.606025 | 0.000239 | |||
| MAP7D3 | 4.359476 | 0.000297 | MAP7D3 | 0.857624458 | 2.357553568 |
| C10orf71 | 0.797138 | 0.000337 | C10orf71 | −0.154814358 | 0.856574179 |
| SPAG11A | 0.694749 | 0.000457 | SPAG11A | −0.265447689 | 0.766862557 |
| HEY1 | 0.608125 | 0.000547 | |||
| SGCD | 0.716995 | 0.000745 | |||
| L1TD1 | 1.258341 | 0.000811 | |||
| PCGF3 | 2.460722 | 0.000848 | |||
3.3. The development of the 5‐mRNA prognostic model
For each patient, a risk score analysis was conducted among the five mRNAs to determine the risk score (Table 1). The risk score formula is as follows: Risk score = 0.3036*SPIN4 + 0.8576*MAP7D3 −0.1548*C10orf71 −0.7335*SPRY1 −0.2654*SPAG11A. The distribution of survival risk score of these five mRNAs and mRNA‐related survival time were demonstrated in Figures 2a,b. The expression Heatmap of five‐mRNA signature was demonstrated in Figure 2c. The pediatric WT patients were divided into low‐risk group and high‐risk group based on the median risk score (value = 1.1503). Survival analysis between high‐risk group and low‐risk group was performed by using the log‐rank test (Figure 2d). The result revealed that low‐risk group was related to a better prognosis (p < .001).
Figure 2.
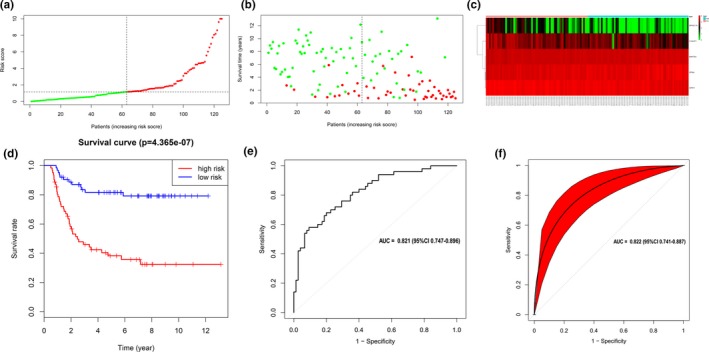
Prognostic evaluation of the five‐mRNA signature in pediatric Wilms tumor patients. (a) The distribution of mRNA‐related survival risk score. (b) The distribution of mRNA‐related survival time. (c) Gene expression heatmap of five identified genes between high‐risk and low‐risk groups. (d) Kaplan–Meier survival curve analysis for overall survival of pediatric Wilms tumor patients between low‐ and high‐risk groups. (e) Receiver operating characteristic (ROC) curve indicated that the area under receiver operating characteristic of 5‐mRNA model was 0.821. (f) Bootstrap test with 500 times was used to perform the internal validation indicated that the area under receiver operating characteristic of 5‐mRNA model was 0.822
The results of ROC demonstrated that AUC was 0.821 (95%CI [0.747, 0.896]) (Figure 2e). We have used bootstrap test (Bootstrap resampling times = 500) to perform the internal validation. The results demonstrated that the validated AUC was 0.822 (95%CI [0.741, 0.887]) (Figure 2f), which was consistent with primary results of AUC (0.821). The results demonstrated that the 5‐mRNA prognostic model had a promising sensitivity and specificity in predicting the survival outcomes of pediatric WT patients.
3.4. REACTOME, KEGG, and BIOCARTA pathway analyses among survival‐related mRNAs
We then included 466 survival‐related mRNAs screened by univariate Cox analysis (p < .05) into pathway analyses. A total of 47 pathway ways were enriched in this study, including 16 pathways enriched by KEGG database, 29 pathways enriched by REACTOME database, and 2 pathways enriched by BIOCARTA database. The top five enriched pathways ranked as the P value were demonstrated in Table 2. The results demonstrated that these survival‐related genes were mainly enriched in ErbB2 and ErbB3 signaling pathways, and calcium signaling pathway.
Table 2.
The results of pathway analyses including REACTOME, KEGG, and BIOCARTA pathway databases
| Category | Term | Count | p Value | Genes | FDR |
|---|---|---|---|---|---|
| REACTOME | R‐HSA‐1250196: SHC1 events in ERBB2 signaling | 5 | 9.62E‐04 | NRAS, ERBB3, ERBB2, EGF, NRG1 | 1.350958906 |
| REACTOME | R‐HSA‐419408: Lysosphingolipid and LPA receptors | 4 | 0.004144594 | S1PR1, PLPPR4, PLPPR5, LPAR1 | 5.702267239 |
| REACTOME | R‐HSA‐1306955: GRB7 events in ERBB2 signaling | 3 | 0.005525286 | ERBB3, ERBB2, NRG1 | 7.533745676 |
| REACTOME | R‐HSA‐1963640: GRB2 events in ERBB2 signaling | 4 | 0.006153572 | NRAS, ERBB2, EGF, NRG1 | 8.356172174 |
| REACTOME | R‐HSA‐1963642: PI3K events in ERBB2 signaling | 4 | 0.006153572 | ERBB3, ERBB2, EGF, NRG1 | 8.356172174 |
| KEGG | hsa04020: Calcium signaling pathway | 12 | 0.002926977 |
EDNRA, ADCY7, CHRM2, ERBB3, ERBB2, PHKA1, CACNA1G, PPP3CA, NTSR1, GRM1, CACNA1A, F2R |
3.649269099 |
| KEGG | hsa05202: Transcriptional misregulation in cancer | 11 | 0.005402032 |
PLAT, PRCC, RXRG, MDM2, IGF1, BCL6, WHSC1, NGFR, ZBTB16, HIST2H3D, MYCN |
6.638940309 |
| KEGG | hsa05200: Pathways in cancer | 18 | 0.009581263 |
BMP4, ADCY7, ERBB2, RXRG, TGFB3, IGF1, LPAR1, ZBTB16, MECOM, FZD7, EDNRA, CCNE1, NRAS, CCDC6, CBLB, MDM2, EGF, F2R |
11.49385516 |
| KEGG | hsa04068: FoxO signaling pathway | 9 | 0.012608779 |
NRAS, S1PR1, PRKAB2, TGFB3, MDM2, IGF1, BCL6, EGF, GRM1 |
14.86439542 |
| KEGG | hsa05215: Prostate cancer | 7 | 0.016293117 | CCNE1, NRAS, ERBB2, MDM2, IGF1, PDGFC, EGF | 18.80657177 |
| BIOCARTA | ErbB3 pathway | 3 | 0.008074119 | ERBB3, EGF, NRG1 | 8.793866109 |
| BIOCARTA | EGFR/SMRTE pathway | 3 | 0.039655032 | THRA, ZBTB16, EGF | 36.83536607 |
4. DISCUSSION
Evidence has proved that mRNAs play crucial roles in the tumorigenesis and progression of pediatric WT (Apelt et al., 2016; Martins, Pinto, Domingues, & Cavaco, 2018; Zhu et al., 2018). Although several previous studies have identified several mRNAs with prognostic value in pediatric WT, they were not focused on the correlations between mRNA signature model and prognosis of pediatric WT (Gadd et al., 2017; Ludwig et al., 2016; Wari et al., 2017). Moreover, with the development of detection technology, the single mRNA expression pattern was no longer sufficient for accurate predication of prognosis of pediatric WT.
To the best of our knowledge, it is the first time to screen out the DEGs between pediatric WT and paired tissues from TARGET database. A novel five‐mRNA signature (SPRY1, SPIN4, MAP7D3, C10orf71, and SPAG11A) was then generated. ROC curve proved that this five‐mRNA signature revealed a high sensitivity and specificity in predicating the survival outcomes of pediatric WT patients. The predictive value of the five‐mRNA signature was validated in TARGET dataset of 136 pediatric WT patients. Based on these five prognostic mRNAs, we established a five‐mRNA prognostic model which can classify pediatric WT patients into low‐risk and high‐risk groups with different survival outcomes.
Wilms tumor are most common types of childhood kidney cancers. It has been reported that for children younger than 15 years with Wilms tumor, the 5‐year survival rate has increased over the same time from 74% to 88% (Smith, Altekruse, Adamson, Reaman, & Seibel, 2014). The 5‐year survival rate for Wilms tumor with favorable histology has been consistently above 90% since the 1980s (Smith, Altekruse, Adamson, Reaman, & Seibel, 2014). The results of this manuscript demonstrated that among these five mRNAs, SPRY1 and SPIN4 were associated with high risk of development of pediatric WT, and MAP7D3, C10orf71, and SPAG11A were associated with low risk of development of pediatric WT.
In mammals, SPRY1 was reported to be consisted of four members and was inhibitor of receptor tyrosine kinase signaling (Rozen et al., 2009). In mice, SPRY1 plays an important role during kidney morphogenesis by antagonizing GDNF signaling (Basson et al., 2005). SPRY1 also plays an important role in the early steps of glomerulus formation and represents a physiologically associated target gene of WT1 during the development of kidney (Gross et al., 2003). SPRY1 was reported to be associated with many kinds of tumors, such as breast cancer (He et al., 2016), colorectal cancer (Zhang et al., 2016), and human epithelial ovarian cancer (Masoumi‐Moghaddam, Amini, Wei, Robertson, & Morris, 2015). The protein encoded by MAP7D3 belongs to the MAP7 family. There is little known about the role of MAP7 with respect to cancer progression (Blum et al., 2008). Many important cellular processes attributed to microtubules involvement, including cell division, motility, and changes in cell shape (Bhat & Setaluri, 2007). Yan et al. (Yan et al., 2013) showed that miR‐16 targeting MAP7 played an important role in regulating proliferation in cancer cells. Also, Blum et al. (Blum et al., 2008) demonstrated that the expression ratio of MAP7/B2M can be regarded as a prognostic factor for survival in patients with colon cancer. Peng et al. (Lin et al., 2019) demonstrated that SPAG11A was involved in the biological process of papillary thyroid cancer. However, the SPIN4 and C10orf71 have not been reported associated with the development and progression of cancer. To the best of our knowledge, given the potential molecular mechanism of the five mRNAs signature, no reports of the function and mechanism of these five mRNAs, SPRY1, SPIN4, MAP7D3, C10orf71, and SPAG11A, have been published concerning WT.
The development of pediatric WT is a multi‐step process. A large number of genetic alterations were involved in this multi‐step biological process (Morrison, Viney, Saleem, & Ladomery, 2008). For the sake of elucidating the effects and functions of these survival‐related mRNAs screened by univariate Cox analyses, we used REACTOME, KEGG, and BIOCARTA pathway databases to perform pathway analyses. The results demonstrated that these survival‐related genes were mainly enriched in ErbB2 and ErbB3 signaling pathways and calcium signaling pathway.
Both BIOCARTA and REACTOME pathway databases revealed that these survival‐related genes were mainly enriched in ErbB2 and ErbB3 signaling pathways. ErbB2 and ErbB3 belong to the family of human epidermal growth factor receptors consisting of EGFR (ErbB1), ErbB2, ErbB3, and ErbB4 (Vermeulen, Segers, & De Keulenaer, 2016). ErbB2 amplification plays a critical role in tumor growth. Amplified ErbB2 can bind to ErbB3 to form an oncogenic ErbB2/ErbB3 complex (Holbro et al., 2003). ErbB3 interacts with the regulatory p85 subunit of PIK3 in this complex to activate the PI3K/Akt pathway and intense cell growth and proliferation. (Schoeberl et al., 2009) Therefore, ErbB3 plays an important role in oncogenic ErbB2 signaling pathway. Rotter et al. (Rotter, Block, Busch, Thanner, & Hofler, 1992) reported that the expression of ErbB2 was downregulated in the renal cell carcinoma when compared with normal kidney tissue. To the best of our knowledge, the molecular mechanisms behind the alteration of ErbB2 in renal cell carcinoma compared with normal kidney was still unknown. Plus, the expression of ErbB3 has not been thoroughly studied in renal cell carcinoma. KEGG pathway database revealed that these survival‐related mRNAs were mainly enriched in calcium signaling pathway. Previous studies (Cole & Kohn, 1994; Soboloff, Zhang, Minden, & Berger, 2002; Sukumaran, Sun, Vyas, & Singh, 2015) have been reported that inhibition of calcium influx can cause either growth arrest or cell death in numbers of cancer cells. However, the role of calcium signaling pathway in the development and progression of WT has not been elucidated yet. Xu et al. (Xu, Chen, Ye, Zhong, & Chen, 2015) reported that calcium signaling pathway has been involved in inducing the apoptosis in non‐small cell lung cancer cells, for the overload of calcium has been reported to play a crucial role in the initiation and regulation of apoptosis.
There are some limitations in this study. The predictive value of the five‐mRNA signature was not validated in another independent dataset because it is very difficult for us to obtain tumor specimens, especially pediatric tumor samples.
5. CONCLUSION
In conclusion, the five‐mRNA signature can predict the prognosis of patients with pediatric WT. It has significant implication in the understanding of therapeutic targets for pediatric WT patients. However, further study is needed to validate this five‐mRNA signature and uncover more novel diagnostic or prognostic mRNA candidates in pediatric WT patients.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTION
Study conception and design: Xiao‐Dan Lin; Yong Wei; Qing‐Shui Zheng; Xue‐Yi Xue; Ning Xu. Data acquisition: Zhi‐Bin Ke; Dong‐Ning Chen; Yun‐Zhi Lin. Analysis and data interpretation: Xiao‐Dan Lin; Yu‐Peng Wu; Zhi‐Bin Ke; Xiong‐Lin Sun; Xiao‐Dong Li. Drafting of the manuscript: Xiao‐Dan Lin; Ning Xu; Yu‐Peng Wu; Shao‐Hao Chen; Xiong‐Lin Sun. Critical revision: Ning Xu; Yu‐Peng Wu; Shao‐Hao Chen; Qing‐Shui Zheng; Xue‐Yi Xue.
ETHICAL STATEMENT
Not applicable.
Supporting information
ACKNOWLEDGMENTS
None.
Lin X‐D, Wu Y‐P, Chen S‐H, et al. Identification of a five‐mRNA signature as a novel potential prognostic biomarker in pediatric Wilms tumor. Mol Genet Genomic Med. 2020;8:e1032 10.1002/mgg3.1032
Xiao‐Dan Lin, Yu‐Peng Wu, Shao‐Hao Chen and Xiong‐Lin Sun contributed equally to this work.
Funding Information
This study was supported by the Natural Science Foundation of Fujian Province (Grant number: 2018J01172), Startup Fund for scientific research, Fujian Medical University (Grant number: 2017XQ1063), Fujian educational research programs for young and middle‐aged teachers (Grant number: JT180187) and Guiding Project of Science and Technology Department of Fujian Province (Grant number: 2019Y0018).
Contributor Information
Ning Xu, Email: drxun@fjmu.edu.cn.
Xue‐Yi Xue, Email: xuexueyi@fjmu.edu.cn.
REFERENCES
- Aldrink, J. H. , Heaton, T. E. , Dasgupta, R. , Lautz, T. B. , Malek, M. M. , Abdessalam, S. F. , … Ehrlich, P. F. (2018). Summary article: Update on Wilms tumor. Journal of Pediatric Surgery, 54(3), 390–397. 10.1016/j.jpedsurg.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelt, N. , Hubertus, J. , Mayr, D. , Graf, N. , Furtwangler, R. , Von Schweinitz, D. , & Kappler, R. (2016). Association of FOXM1 expression with tumor histology and prognosis in Wilms tumor: Potential for a new prognostic marker. Oncology Letters, 12(4), 2854–2859. 10.3892/ol.2016.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson, M. A. , Akbulut, S. , Watson‐Johnson, J. , Simon, R. , Carroll, T. J. , Shakya, R. , … Licht, J. D. (2005). Sprouty1 is a critical regulator of GDNF/RET‐mediated kidney induction. Developmental Cell, 8(2), 229–239. 10.1016/j.devcel.2004.12.004 [DOI] [PubMed] [Google Scholar]
- Bhat, K. M. , & Setaluri, V. (2007). Microtubule‐associated proteins as targets in cancer chemotherapy. Clinical Cancer Research, 13(10), 2849–2854. 10.1158/1078-0432.Ccr-06-3040 [DOI] [PubMed] [Google Scholar]
- Blum, C. , Graham, A. , Yousefzadeh, M. , Shrout, J. , Benjamin, K. , Krishna, M. , … Mitas, M. (2008). The expression ratio of Map7/B2M is prognostic for survival in patients with stage II colon cancer. International Journal of Oncology, 33(3), 579–584. 10.3892/ijo_00000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow, N. , Olshan, A. , Beckwith, J. B. , & Green, D. M. (1993). Epidemiology of Wilms tumor. Medical and Pediatric Oncology, 21(3), 172–181. 10.1002/mpo.2950210305 [DOI] [PubMed] [Google Scholar]
- Cole, K. , & Kohn, E. (1994). Calcium‐mediated signal transduction: Biology, biochemistry, and therapy. Cancer and Metastasis Reviews, 13(1), 31–44. 10.1007/BF00690417 [DOI] [PubMed] [Google Scholar]
- Cone, E. B. , Dalton, S. S. , Van Noord, M. , Tracy, E. T. , Rice, H. E. , & Routh, J. C. (2016). Biomarkers for Wilms tumor: A systematic review. Journal of Urology, 196(5), 1530–1535. 10.1016/j.juro.2016.05.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angio, G. J. , Breslow, N. , Beckwith, J. B. , Evans, A. , Baum, H. , deLorimier, A. , … Othersen, H. B. (1989). Treatment of Wilms' tumor. Results of the third national Wilms' tumor study. Cancer, 64(2), 349–360. [DOI] [PubMed] [Google Scholar]
- Dome, J. S. , Fernandez, C. V. , Mullen, E. A. , Kalapurakal, J. A. , Geller, J. I. , Huff, V. , … Perlman, E. J. (2013). Children's Oncology Group's 2013 blueprint for research: Renal tumors. Pediatric Blood & Cancer, 60(6), 994–1000. 10.1002/pbc.24419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dome, J. S. , Graf, N. , Geller, J. I. , Fernandez, C. V. , Mullen, E. A. , Spreafico, F. , … Pritchard‐Jones, K. (2015). Advances in Wilms tumor treatment and biology: Progress through international collaboration. Journal of Clinical Oncology, 33(27), 2999–3007. 10.1200/jco.2015.62.1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dome, J. S. , Perlman, E. J. , & Graf, N. (2014). Risk stratification for Wilms tumor: Current approach and future directions. American Society of Clinical Oncology Educational Book, 34, 215–223, 10.14694/EdBook_AM.2014.34.215 [DOI] [PubMed] [Google Scholar]
- Gadd, S. , Huff, V. , Walz, A. L. , Ooms, A. H. A. G. , Armstrong, A. E. , Gerhard, D. S. , … Perlman, E. J. (2017). A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nature Genetics, 49(10), 1487–1494. 10.1038/ng.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, N. , Tournade, M. F. , & de Kraker, J. (2000). The role of preoperative chemotherapy in the management of Wilms' tumor. The SIOP studies. International Society of Pediatric Oncology. Urologic Clinics of North America, 27(3), 443–454. 10.1016/S0094-0143(05)70092-6 [DOI] [PubMed] [Google Scholar]
- Gratias, E. J. , Dome, J. S. , Jennings, L. J. , Chi, Y.‐Y. , Tian, J. , Anderson, J. , … Perlman, E. J. (2016). Association of chromosome 1q gain with inferior survival in favorable‐histology Wilms tumor: A report from the Children's Oncology Group. Journal of Clinical Oncology, 34(26), 3189–3194. 10.1200/jco.2015.66.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, I. , Morrison, D. J. , Hyink, D. P. , Georgas, K. , English, M. A. , Mericskay, M. , … Licht, J. D. (2003). The receptor tyrosine kinase regulator Sprouty1 is a target of the tumor suppressor WT1 and important for kidney development. Journal of Biological Chemistry, 278(42), 41420–41430. 10.1074/jbc.M306425200 [DOI] [PubMed] [Google Scholar]
- He, Q. , Jing, H. , Liaw, L. , Gower, L. , Vary, C. , Hua, S. , & Yang, X. (2016). Suppression of Spry1 inhibits triple‐negative breast cancer malignancy by decreasing EGF/EGFR mediated mesenchymal phenotype. Scientific Reports, 6, 23216 10.1038/srep23216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbro, T. , Beerli, R. R. , Maurer, F. , Koziczak, M. , Barbas, C. F. 3rd , & Hynes, N. E. (2003). The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proceedings of the National Academy of Sciences, 100(15), 8933–8938. 10.1073/pnas.1537685100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, Z.‐B. , Cai, H. , Wu, Y.‐P. , Lin, Y.‐Z. , Li, X.‐D. , Huang, J.‐B. , … Xu, N. (2019). Identification of key genes and pathways in benign prostatic hyperplasia. Journal of Cellular Physiology, 234(11), 19942–19950. 10.1002/jcp.28592 [DOI] [PubMed] [Google Scholar]
- Lin, P. , Guo, Y.‐N. , Shi, L. , Li, X.‐J. , Yang, H. , He, Y. , … Chen, G. (2019). Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY), 11(2), 480–500. 10.18632/aging.101754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, N. , Werner, T. , Backes, C. , Trampert, P. , Gessler, M. , Keller, A. , … Meese, E. (2016). Combining miRNA and mRNA expression profiles in Wilms tumor subtypes. International Journal of Molecular Sciences, 17(4), 475 10.3390/ijms17040475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, A. G. , Pinto, A. T. , Domingues, R. , & Cavaco, B. M. (2018). Identification of a novel CTR9 germline mutation in a family with Wilms tumor. European Journal of Medical Genetics, 61(5), 294–299. 10.1016/j.ejmg.2017.12.010 [DOI] [PubMed] [Google Scholar]
- Masoumi‐Moghaddam, S. , Amini, A. , Wei, A. Q. , Robertson, G. , & Morris, D. L. (2015). Sprouty 1 predicts prognosis in human epithelial ovarian cancer. American Journal of Cancer Research, 5(4), 1531–1541. [PMC free article] [PubMed] [Google Scholar]
- Moch, H. , Cubilla, A. L. , Humphrey, P. A. , Reuter, V. E. , & Ulbright, T. M. (2016). The 2016 WHO classification of tumours of the urinary system and male genital organs‐part A: Renal, penile, and testicular tumours. European Urology, 70(1), 93–105. 10.1016/j.eururo.2016.02.029 [DOI] [PubMed] [Google Scholar]
- Morrison, A. A. , Viney, R. L. , Saleem, M. A. , & Ladomery, M. R. (2008). New insights into the function of the Wilms tumor suppressor gene WT1 in podocytes. American Journal of Physiology‐Renal Physiology, 295(1), F12–17. 10.1152/ajprenal.00597.2007 [DOI] [PubMed] [Google Scholar]
- Pastore, G. , Znaor, A. , Spreafico, F. , Graf, N. , Pritchard‐Jones, K. , & Steliarova‐Foucher, E. (2006). Malignant renal tumours incidence and survival in European children (1978–1997): Report from the Automated Childhood Cancer Information System project. European Journal of Cancer, 42(13), 2103–2114. 10.1016/j.ejca.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Phelps, H. M. , Pierce, J. M. , Murphy, A. J. , Correa, H. , Qian, J. , Massion, P. P. , & Lovvorn, H. N. 3rd (2019). FXR1 expression domain in Wilms tumor. Journal of Pediatric Surgery, 54(6), 1198–1205. 10.1016/j.jpedsurg.2019.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter, M. , Block, T. , Busch, R. , Thanner, S. , & Hofler, H. (1992). Expression of HER‐2/neu in renal‐cell carcinoma. Correlation with histologic subtypes and differentiation. International Journal of Cancer, 52(2), 213–217. 10.1002/ijc.2910520210 [DOI] [PubMed] [Google Scholar]
- Rozen, E. J. , Schmidt, H. , Dolcet, X. , Basson, M. A. , Jain, S. , & Encinas, M. (2009). Loss of Sprouty1 rescues renal agenesis caused by Ret mutation. Journal of the American Society of Nephrology, 20(2), 255–259. 10.1681/asn.2008030267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeberl, B. , Pace, E. A. , Fitzgerald, J. B. , Harms, B. D. , Xu, L. , Nie, L. , … Nielsen, U. B. (2009). Therapeutically targeting ErbB3: A key node in ligand‐induced activation of the ErbB receptor‐PI3K axis. Science Signalling, 2(77), ra31 10.1126/scisignal.2000352 [DOI] [PubMed] [Google Scholar]
- Scott, R. H. , Stiller, C. A. , Walker, L. , & Rahman, N. (2006). Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. Journal of Medical Genetics, 43(9), 705–715. 10.1136/jmg.2006.041723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. A. , Altekruse, S. F. , Adamson, P. C. , Reaman, G. H. , & Seibel, N. L. (2014). Declining childhood and adolescent cancer mortality. Cancer, 120(16), 2497–2506. 10.1002/cncr.28748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff, J. , Zhang, Y. , Minden, M. , & Berger, S. A. (2002). Sensitivity of myeloid leukemia cells to calcium influx blockade: Application to bone marrow purging. Experimental Hematology, 30(10), 1219–1226. 10.1016/S0301-472X(02)00893-7 [DOI] [PubMed] [Google Scholar]
- Stokes, C. L. , Stokes, W. A. , Kalapurakal, J. A. , Paulino, A. C. , Cost, N. G. , Cost, C. R. , … Liu, A. K. (2018). Timing of radiation therapy in pediatric wilms tumor: A report from the national cancer database. International Journal of Radiation Oncology, Biology, Physics, 101(2), 453–461. 10.1016/j.ijrobp.2018.01.110 [DOI] [PubMed] [Google Scholar]
- Sukumaran, P. , Sun, Y. , Vyas, M. , & Singh, B. B. (2015). TRPC1‐mediated Ca(2)(+) entry is essential for the regulation of hypoxia and nutrient depletion‐dependent autophagy. Cell Death & Disease, 6, e1674 10.1038/cddis.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen, Z. , Segers, V. F. , & De Keulenaer, G. W. (2016). ErbB2 signaling at the crossing between heart failure and cancer. Basic Research in Cardiology, 111(6), 60 10.1007/s00395-016-0576-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Lou, C. , & Ma, N. (2019). miR‐140‐5p alleviates the aggressive progression of Wilms' tumor through directly targeting TGFBR1 gene. Cancer Management and Research, 11, 1641–1651. 10.2147/cmar.S177508 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wari, M. N. , Vallonthaiel, A. G. , Ahmed, A. , Saxena, D. , Iyer, V. K. , Mathur, S. R. , … Dinda, A. (2017). Glypican‐3 mRNA expression level in Wilms tumor: Correlation with histological type, stage, and outcome. Pediatric Surgery International, 33(6), 695–703. 10.1007/s00383-017-4087-2 [DOI] [PubMed] [Google Scholar]
- Wegert, J. , Ishaque, N. , Vardapour, R. , Geörg, C. , Gu, Z. , Bieg, M. , … Gessler, M. (2015). Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high‐risk blastemal type Wilms tumors. Cancer Cell, 27(2), 298–311. 10.1016/j.ccell.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Wong, K. F. , Reulen, R. C. , Winter, D. L. , Guha, J. , Fidler, M. M. , Kelly, J. , … Hawkins, M. M. (2016). Risk of adverse health and social outcomes up to 50 years after Wilms tumor: The British Childhood Cancer Survivor Study. Journal of Clinical Oncology, 34(15), 1772–1779. 10.1200/jco.2015.64.4344 [DOI] [PubMed] [Google Scholar]
- Xu, N. , Wu, Y.‐P. , Ke, Z.‐B. , Liang, Y.‐C. , Cai, H. , Su, W.‐T. , … Xue, X.‐Y. (2019). Identification of key DNA methylation‐driven genes in prostate adenocarcinoma: An integrative analysis of TCGA methylation data. Journal of Translational Medicine, 17(1), 311 10.1186/s12967-019-2065-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, N. , Wu, Y. P. , Yin, H. B. , Xue, X. Y. , & Gou, X. (2018). Molecular network‐based identification of competing endogenous RNAs and mRNA signatures that predict survival in prostate cancer. Journal of Translational Medicine, 16(1), 274 10.1186/s12967-018-1637-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Chen, D. , Ye, B. , Zhong, F. , & Chen, G. (2015). Curcumin induces the apoptosis of non‐small cell lung cancer cells through a calcium signaling pathway. International Journal of Molecular Medicine, 35(6), 1610–1616. 10.3892/ijmm.2015.2167 [DOI] [PubMed] [Google Scholar]
- Yan, X. , Liang, H. , Deng, T. , Zhu, K. , Zhang, S. , Wang, N. , … Chen, X. I. (2013). The identification of novel targets of miR‐16 and characterization of their biological functions in cancer cells. Molecular Cancer, 12, 92 10.1186/1476-4598-12-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Wei, T. , Shim, K. , Wright, K. , Xu, K. , Palka‐Hamblin, H. L. , … Khare, S. (2016). Atypical role of sprouty in colorectal cancer: Sprouty repression inhibits epithelial‐mesenchymal transition. Oncogene, 35(24), 3151–3162. 10.1038/onc.2015.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , Zhang, L. , Zhao, Z. , Fu, W. , Fu, K. , Liu, G. , & Jia, W. (2018). MicroRNA92a3p inhibits the cell proliferation, migration and invasion of Wilms tumor by targeting NOTCH1. Oncology Reports, 40(2), 571–578. 10.3892/or.2018.6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


