Abstract
Background
Consanguineous marriages are common in the Middle East including the Gulf countries. The rate of consanguinity in Qatar is approximately 54%, which are mainly first cousins’ marriages. Previous studies showed that consanguinity increases the prevalence of birth defects and other genetic disorders. Thus, we studied the effects of consanguinity in a cohort of subjects with certain genetic disorders in Qatar.
Methods
This cross‐sectional study was conducted at two centers in Qatar (Hamad Medical Corporation “HMC” and Shafallah “SC”) including 599 Qatari families with certain types of genetic and nongenetic anomalies.
Results
Consanguineous marriages were seen in 397 of 599 (66.2%) Qatari families and first cousin group counts for 65% in Qatari population. In the total cohort and at HMC, all consanguineous marriages had a significantly higher risk of Autosomal Recessive disorders than nonconsanguineous marriages (total cohort: odds ratio (OR) = 1.72; 95% CI: 1.10, 2.71; p = .02; HMC: OR = 2.98; 95% CI: 1.37, 6.09; p = .005). On the other hand, at HMC, nonconsanguinity was significantly related to chromosomal abnormality (OR = 6.36; 95% CI: 1.13, 35.85; p = .036).
Conclusion
Our data suggest a significant role of parental consanguinity in increasing the prevalence of genetic disorders; mainly Autosomal Recessive disorders. Chromosomal abnormality disorders were significantly higher among nonconsanguineous marriages. These results help better inform policy makers on social, educational, and public health initiatives that might mitigate the impact of genetic disease in the Qatari society.
Keywords: Arab, Autosomal Recessive, consanguinity, genetic disorders, Qatar
The study highlights the significant role of parental consanguinity and specific relatedness (first cousin) in increasing the risk of developing genetic disorders, in particular Autosomal Recessive.
![]()
1. INTRODUCTION
Consanguinity and endogamy are high in the Middle East including the Gulf countries and range between 20% and 50% (Al‐Gazali et al., 1997; Al‐Gazali et al., 1999; Hamamy et al., 2011; Tadmouri et al., 2009; Teebi & El‐Shanti, 2006). In many Middle Eastern countries, consanguineous marriages are culturally favored with longstanding traditions (Hamamy, Masri, Al‐Hadidy, & Ajlouni, 2007). The significant improvement in the health care system in Qatar and other Gulf States has led to the eradication of infectious and malnutrition disorders; however, it has made genetic disorders the leading cause of mortality and morbidity in those countries (Hamamy et al., 2007; Majeed‐Saidan et al., 2015).
In Qatar, Bener et al reported that the rate of consanguinity is relatively high with a rate of 54%, and predominantly first cousin marriages comprising 26.7% of all marriages (Bener & Alali, 2006). This significantly raises the risk for genetic disorders in the Qatari population among consanguineous marriages. The study indicated that this is due to the strong relationship to the social and cultural contexts that reproduce those conditions (Bener & Alali, 2006).
The marriage is an influential element in the family, and it is the basis of family configuration in ancient and modern societies. Society regulates marriage in multiple ways through the development of specific rules regarding the selection of a partner. In some societies, individuals are prevented from marrying anyone who is not a member of the large family group; also known as internal marriage (Endogamy). On the other hand, other societies prevent a member of the group from marrying anyone within the group; also known as external marriage (Exogamy). In such case, an individual is only allowed to marry from outside the community kinship. These rules are not contradictory as long as they relate to two different groups. Thus, internal and external marriages may appear within the same society (Bener & Alali, 2006; Bittles & Black, 2010; Hamamy, 2012; Tadmouri et al., 2009).
This pattern is still widespread in many societies despite differing cultures, though it is a marked feature of traditional societies.
Furthermore, despite the emergence of multiple forms of marriage and even relationships outside marriage, there are studies showing that variables such as social and economic characteristics, class and social status, race, age, parents, and friends, still influence the choice of a spouse or partner (Ross, 1997).
A study by Bener et al showed also that consanguinity had a slightly higher risk for diseases such as diabetes mellitus, cancer, blood and mental disorders, heart diseases, asthma, gastro‐intestinal disorders, hypertension, hearing deficit, Glucose 6 Phosphate Dehydrogenase (G6PD), and common eye diseases (Bener & Mohammad, 2017).
Several studies showed that consanguinity leads and contributes to an increase in the rate of Autosomal Recessive genetic disorders (Abdulrazzaq et al., 1997; El‐Shafei, Rao, & Sandhu, 1986; El Mouzan, Al Salloum, Al Herbish, Qurachi, & Al Omar, 2008; Hamamy et al., 2007; Hoodfar & Teebi, 1996; Kerkeni, Monastiri, Saket, Guediche, & Ben Cheikh, 2007).
In contrast to international databases, the updated Catalogue for Transmission Genetics in Arabs (CTGA) showed that the vast proportion of the disorders in the CTGA Database follow a recessive mode of inheritance (61.7%) compared to the minor proportion of dominantly inherited traits (30.8%, Catalogue for Transmission Genetics in Arabs, 2017). However, as far as we know there is a paucity of studies worldwide and specifically in Qatar that looks at the relationship between consanguineous marriages and specific relatedness on one hand, and genetic disorders on the other hand.
Thus, in this study we will investigate the effects of consanguinity and specific relatedness in a cohort of subjects with certain genetic disorders in two centers in the state of Qatar.
2. MATERIALS AND METHODS
2.1. Participants
All participants that met the inclusion criteria and consented to participate into the study were recruited between August 2012 and August 2013. As part of this cross‐sectional study, etiological categories and consanguinity rates were studied among 599 Qatari families with certain anomalies seen at two centers: the Clinical and Metabolic Genetic Section (CMGS), Department of Pediatrics at Hamad Medical Corporation (HMC), which is the only referral center in Qatar, and Shafallah Center (SC), which is a rehabilitation school. We present here a comparative analysis for each center because they receive different cohort of patients, where HMC is the main tertiary care center for diagnosis and management for genetic disorders, whereas SC is mainly for rehabilitation that also includes cases with nongenetic disorders. Most patients seen at SC were diagnosed with a variety of disorders ranging from developmental delay/intellectual disability, autism, Down syndrome, cerebral palsy, and so on. In this study, the patients with age less than or equals to 14 years are classified as pediatric population (56%) and patients aged more than 14 years as adult population (44%).
The participant's ages ranged between 1 and 58 years (with a mean of 13.2 ± 7.4 years) with male to female ratio 1.4:1 (58.8% male, 41.2% female). Father's and mother's education more than secondary school was found to be 28.1% and 28.5%, respectively, and approximately 80% of fathers and 29% of mothers were employed (Table 1).
Table 1.
Patients characteristics at HMC and SC and the total sample (N = 599)
| Characteristics | HMC (n = 200) | SC (n = 399) | Total (n = 599) | p‐valuea |
|---|---|---|---|---|
| Age (years, n = 599) | ||||
| Mean ± SD | 8.04 ± 7.8 | 15.7 ± 5.6 | 13.2 ± 7.4 | <.0001 |
| [median (min‐max)] | [6 (1–58)] | [15 (1–32)] | [13(1–58)] | |
| Age (years, n = 599) | ||||
| ≤14 years | 172 (86.0%) | 163 (40.9%) | 335 (56.0%) | <.0001 |
| >14 years | 28 (14.0%) | 236 (59.1%) | 264 (44.0%) | |
| Gender (n = 599) | ||||
| Male | 110 (55.0%) | 242 (60.7%) | 352 (58.8%) | .185 |
| Female | 90 (45.0%) | 157 (39.3%) | 247 (41.2%) | |
| Parental consanguinity (n = 586) | ||||
| Yes | 145 (74.7%) | 252 (64.3%) | 397 (67.7%) | .011 |
| No | 49 (25.3%) | 140 (35.7%) | 189 (32.3%) | |
| Diagnosis (n = 599) | ||||
| Autosomal Recessive | 82 (41.0%) | 64 (16.0%) | 146 (24.4%) | <.001 |
| Autosomal Dominant | 9 (4.5%) | — | 9 (1.5%) | |
| X‐linked | 3 (1.5%) | 2 (0.5%) | 5 (0.8%) | |
| Chromosomal disorder | 6 (3.0%) | 188 (47.1%) | 194 (32.4%) | |
| Undiagnosed | 104 (52.0%) | 146 (36.6%) | 250 (41.7%) | |
| Degree of relation (n = 172) | ||||
| First cousin | 22 (16.1%) | 13 (37.1%) | 35 (20.3%) | <.001 |
| First cousin (paternal) | 38 (27.7%) | 4 (11.4%) | 42 (24.4%) | |
| First cousin (maternal) | 21 (15.3%) | 0 | 21 (12.2%) | |
| Double first cousin | 8 (5.8%) | 0 | 8 (4.7%) | |
| Second cousin | 22 (16.1%) | 0 | 22 (12.8%) | |
| Same family tribe | 6 (4.4%) | 18 (51.4%) | 24 (14.0%) | |
| Others | 20 (14.6%) | 0 | 20 (11.6%) | |
| Father’ education (n = 523) | ||||
| Postgraduate degree | 9 (5.0%) | 10 (2.9%) | 19 (3.6%) | .009 |
| University degree | 43 (23.6%) | 85 (24.9%) | 128 (24.5%) | |
| Completed secondary school | 71 (39.0%) | 86 (25.2%) | 157 (30.0%) | |
| Completed primary school | 30 (16.5%) | 78 (22.9%) | 108 (20.7%) | |
| Primary school | 17 (9.3%) | 52 (15.2%) | 69 (13.2%) | |
| Illiterate | 12 (6.6%) | 30 (8.8%) | 42 (8.0%) | |
| Mother's education (n = 546) | ||||
| Postgraduate degree | 1 (0.5%) | 3 (0.8%) | 4 (0.7%) | .003 |
| University degree | 38 (20.7%) | 113 (31.2%) | 151 (27.8%) | |
| Completed secondary school | 73 (39.7%) | 87 (24.0%) | 160 (29.5%) | |
| Completed primary school | 35 (19.0%) | 62 (17.1%) | 97 (17.9%) | |
| Primary school | 16 (8.7%) | 43 (11.9%) | 59 (10.9%) | |
| Illiterate | 21 (11.4%) | 54 (14.9%) | 75 (13.8%) | |
| Fathers’ Employment (n = 554) | .011 | |||
| Employed | 158 (87.3%) | 287 (76.9%) | 445 (80.3%) | |
| Unemployed | 4 (2.2%) | 24 (6.4%) | 28 (5.1%) | |
| Retired | 19 (10.5%) | 62 (16.6%) | 81 (14.6%) | |
| Mothers’ employment (n = 560) | ||||
| Employed | 56 (31.1%) | 107 (28.2%) | 163 (29.1%) | .121 |
| Unemployed | 121 (67.2%) | 253 (66.6%) | 374 (66.8%) | |
| Retired | 3 (1.7%) | 20 (5.3%) | 23 (4.1%) | |
Some values were observed to be either missing or unknown for some parameters and therefore sums are not equal to n = 599 for each variable. All percentages calculations were based on nonmissing values.
Abbreviation: HMC, Hamad Medical Corporation; SC, Shafallah Center.
p‐value is computed using unpaired t test, Pearson Chi‐square and Yates corrected Chi‐Squared tests for general comparisons between variables’ categories among the two institutions.
2.2. Data collection
We collected and analyzed a large set of clinical data, genetic disorders, and other related data at HMC and SC. We interviewed all the 200 patients/families from HMC and recorded and documented several factors and collected data from the patients’ medical files. We also collected relevant data from 399 patients/families from SC, but only from the patients’ medical record file. Consanguineous marriages as demonstrated through both a quantitative analysis and through a set of detailed case studies have significant impact on hereditary diseases.
Patients were classified mainly into four etiological groups: Group 1—Included patients with Autosomal Recessive (AR) disorders, all AR disorders have been confirmed by molecular studies and follow AR inheritance; Group 2—Included patients with Autosomal Dominant (AD) disorders; Group 3—Included X‐linked; and Group 4 included chromosomal disorders. However, Group 5 included a large “unclassified” patient. The phenotypes in the group of undiagnosed disorders were heterogeneous, and in most of them thorough investigations were done including exome sequencing (see Table 1).
Consanguineous marriages were divided into six categories according to the degree of consanguinity: first cousin maternal; first cousin paternal, first cousin unknown, double first cousin, second cousin, same family tribe.
The data collected from these patients are divided into two main sections. The first section discusses the general patient characteristics and the relationship between consanguinity and demographic factors. The second section discusses the relationship between consanguineous marriages and level of relatedness, and specific categories of genetic disorders for each center (HMC and SC) separately and for the total cases together.
We incorporated both quantitative and qualitative analyses into the research design and have explored the association between consanguinity and genetic disorders using data obtained from medical records and in‐depth interviews. Between August 2012 and August 2013 all Qatari families who attended the outpatient Genetic Clinics at HMC, and patients attending SC, who agreed to consent for the study were recruited. Some families refused to disclose some of the information about their demographic and marriages background (e.g., paternal or maternal cousins), thus, the classification of relatedness among consanguineous families included nondefined cousins. The study was approved by the ethics committee of HMC (Ref No. RC/64512/2012).
2.3. Statistical analysis
A well‐structured data capture form, in view of the research study design and the objectives, was designed and created to collect all required data. Quality of data (review of completeness, data verification, validation and accuracy, security, and confidentiality of data) were performed and maintained by the lead research investigators. Categorical and continuous data values were expressed as frequency (percentage) and mean ± SD or median and inter quartile range (IQR) as appropriate. Descriptive statistics were used to summarize demographic, genetic disorder, and disease and other clinical characteristics of the participants. The association of specific categories of genetic disorders with consanguineous and nonconsanguineous marriages were analyzed and compared using Chi‐squared (χ 2) test or Fisher exact test as appropriate. All the results were presented with the associated 95% confidence interval.
The associations of the various genetic disorders and participants’ characteristics with consanguinity were assessed applying univariate logistic regression analysis using dichotomous outcome variable: consanguineous marriages and nonconsanguineous marriages in one hand, and various genetic disorders (Autosomal Recessive, Autosomal Dominant, X‐linked, chromosomal disorder, undiagnosed), age and gender as independent variables or covariates on the other hand. Multinominal logistic regression analysis was also employed to test for relatedness (first cousin maternal; first cousin paternal, first cousin unknown, double first cousin, second cousin, same family tribe) as an independent variable, where the reference point was nonconsanguineous, using dichotomous outcome variable (being diagnosed with genetic disorder vs. not) as the dependent variable. Univariate and logistic regression results were presented in terms of odds ratio (OR) along with the corresponding 95% CI.
All p values presented were two‐tailed, and p < .05 was considered as statistically significant. All Statistical analyses were done using statistical package SPSS 26.0 (SPSS Inc).
To investigate the differences between consanguinity and nonconsanguinity we calculated the pooled odds ratio of both studies in both institutions, HMC and Shafallah, for each genetic disorder, age, and gender. Meta‐analysis was conducted using Comprehensive Meta‐Analysis (CMA) software (Borenstein, Hedges, Higgins, & Rothstein, 2005). Effect sizes are reported as Odds ratio with 95% confidence intervals for each study and the pooled Odds ratio with 95% confidence intervals for the total. The difference (Cohen's d) compares the individual study's odds ratio to the overall. A d of 0.20 is a small, 0.50 medium, and 0.80 or more a large effect (Cohen, 1988). Effect sizes were analyzed using the fixed‐effects model.
The distribution of effect sizes was examined using tests of heterogeneity. Significant heterogeneity indicates that differences across effect sizes are likely due to sources other than sampling error.
3. RESULTS
Of 599 patients, consanguineous marriages were seen in 397 (66.2%) in comparison to 189 (31.6%) nonconsanguineous marriages and 13 patients with unknown consanguinity (2.2%). Some values were observed to be either missing or unknown for some parameters and therefore sums are not equal to 599 for some variables.
3.1. Relationship between consanguinity and demographic factors
Mothers from consanguineous marriages at HMC were more likely to be unemployed (unemployed: N: 96/135, 71.1% vs. employed: N: 21/41, 51.2%) (χ 2(1, N = 176) = 5.58, p < .05; OR = 2.34; 95% CI: 1.15, 4.81) and to have lower education (B = −0.052, R 2 = .022, p < .05) compared to mothers from nonconsanguineous marriages.
For SC, mothers from consanguineous marriages were more likely to be unemployed (unemployed: N: 167/237, 70.5% vs. employed: N: 83/137, 60.6%) (χ 2(1, N = 374) = 3.82, p < .05; OR = 1.55; 95% CI: 0.99, 2.42) compared to mothers from nonconsanguineous marriages. The results on education were not significant (B = −0.030, R 2 = .008, p = .091).
For the total sample, mothers from consanguineous marriages were more likely to be unemployed (unemployed: N: 263/372, 71.7%; employed: N: 104/178, 58.4%) (χ 2(1, N = 550) = 8.17, p < .01; OR = 1.72; 95% CI: 1.18, 2.49) and to have lower education (B = −0.035, R 2 = .010, p < .05) compared to mothers from nonconsanguineous marriages.
No significant relationship between father's occupation status and level of education in one hand and consanguinity on the other hand were found.
3.2. Effect of consanguinity on genetic disorder among cases treated at HMC
At HMC, among the consanguineous marriages, 66 (45.5%) have Autosomal Recessive conditions, 5 (3.4%) have Autosomal Dominant, 2 (1.4%) have X‐linked, 2 (1.4%) have chromosomal disorders, and 76 (52.4%) have undiagnosed conditions. Consanguineous marriages had significantly higher risk of Autosomal Recessive disorder compared to nonconsanguineous marriages (Table 2).
Table 2.
Effect of consanguinity on genetic disorder (HMC Data, n = 194)
|
Consanguineous marriages N (%) (n = 145) |
Nonconsanguineous marriages N (%) (n = 49) |
Odds ratio (OR, 95% CI) N (%) |
p‐valuea | |
|---|---|---|---|---|
| Autosomal Recessive | 66 (45.5%) | 11 (22.4%) | 2.89 (1.37, 6.09) | .005 |
| Autosomal Dominant | 5 (3.4%) | 4 (8.2%) | 2.49 (0.64, 9.67) | .188 |
| X‐linked | 2 (1.4%) | 1 (2.0%) | 1.49 (0.13, 16.79) | .747 |
| Chromosomal disorder | 2 (1.4%) | 4 (8.2%) | 6.36 (1.13, 35.85) | .036 |
| Undiagnosed | 76 (52.4%) | 27 (55.1%) | 1.11 (0.58, 2.14) | .744 |
| Gender: Male | 84 (57.9) | 24 (49.0) | 1.43 (0.75, 2.75) | .277 |
| Age ≤14 years | 125 (86.8%) | 42 (85.7%) | 1.10 (0.43, 2.79) | .931 |
Nonconsanguineous marriages were considered as a reference category in the logistic regression analysis.
Abbreviation: HMC, Hamad Medical Corporation.
Logistic regression analysis.
In contrast, no significant differences were found for undiagnosed disease, X‐linked and Autosomal Dominant between nonconsanguineous and consanguineous marriages. On the other hand, nonconsanguineous marriages were more likely to have chromosomal abnormality compared to consanguineous marriages (Table 2).
A univariate logistic regression was applied to assess the association between consanguinity and genetic conditions. The consanguineous marriages have 2.9 times increased risk of developing Autosomal Recessive disorders. On the other hand, nonconsanguineous marriages were significantly related to chromosomal abnormality with increased risk of 6.36 (Table 2 and Figure 1).
Figure 1.
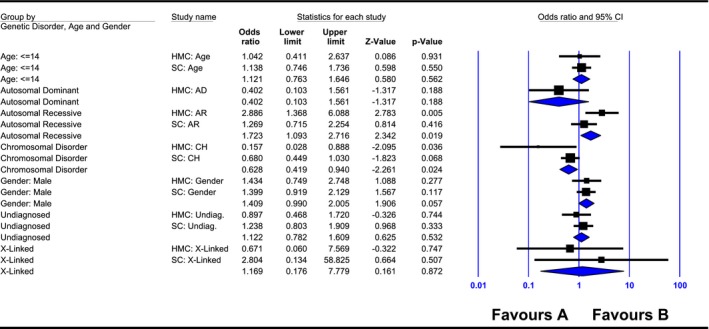
Meta‐analysis for the specific odds ratio (95% confidence interval “CI”) of each study (institutions: Hamad Medical Corporation [HMC] and Shafallah Center [SC]) and the total pooled odds ratio for each genetic disorder, age, and gender with regards to their relationship to consanguinity
There were no significant differences between young (<14 years) and adults (>14 years) within consanguineous and nonconsanguineous families for each genetic disorder (Table 3).
Table 3.
Consanguinity, and genetic disorder by age groups (HMC data)
|
Consanguineous marriages N (%) |
p‐valuea |
Nonconsanguineous marriages N (%) |
p‐valuea | |||
|---|---|---|---|---|---|---|
| Age ≤14 years | Age >14 years | Age ≤14 years | Age >14 years | |||
| Autosomal Recessive | 53 (42.4) | 12 (63.2) | .074 | 9 (21.4) | 2 (28.6) | .500 |
| Autosomal Dominant | 5 (4.0) | 0 (0) | .488 | 3 (7.1) | 1 (14.3) | .472 |
| X‐linked | 2 (1.6) | 0 (0) | .753 | 1 (2.4) | 0 (0) | .857 |
| Chromosomal disorder | 2 (1.6) | 0 (0) | .753 | 3 (7.1) | 1(14.3) | .472 |
| Undiagnosed | 69 (55.2) | 7 (36.8) | .106 | 24 (57.1) | 3 (42.9) | .382 |
Overall‐ p = .247 (χ 2 = 5.42), Overall‐ p = .808 (χ 2 = 0.97).
Abbreviation: HMC, Hamad Medical Corporation.
Pearson Chi‐squared and Fisher Exact tests.
Multinominal logistic regression was performed to investigate whether specific level of relatedness (first cousin maternal; first cousin paternal, first cousin unknown, double first cousin, second cousin, same family tribe) in comparison to nonconsanguinity predicts mode of inheritance of genetic condition or not. It was found that Autosomal Recessive disorder was more likely to be predicted by paternal first cousin (OR: 3.70; 95% CI: 1.44–9.52; p = .007), double first cousin (OR: 6.17; 95% CI: 1.25–30.32; p = .025), and general first cousin (OR: 4.44; 95% CI: 1.49–13.23; p = .007) in comparison to nonconsanguinity. The rest of the relatedness levels were not significant for Autosomal Recessive disorder.
None of the other mode of inheritance of genetic conditions was predicted by specific relatedness level.
3.3. Effect of consanguinity on genetic disorder among cases treated at SC
At SC, among the consanguineous marriages there were 44 (17.5%) with Autosomal Recessive conditions, 2 (0.8%) with X‐linked, 109 (43.3%) chromosomal disorders, and 97 (38.5%) with undiagnosed conditions (Table 4).
Table 4.
Effect of consanguinity on genetic disorder (SC Data) (n = 392)
|
Consanguineous marriages N (%) (n = 252) |
Nonconsanguineous marriages N (%) (n = 140) |
Odds ratio (OR, 95% CI) N (%) |
p‐valuea | |
|---|---|---|---|---|
| Autosomal Recessive | 44 (17.5) | 20 (14.3) | 1.27 (0.71, 2.25) | .416 |
| Autosomal Dominant | — | — | — | — |
| X‐linked | 2 (0.8) | 0 (0) | 2.80 (0.13, 58.83) | .507 |
| Chromosomal disorder | 109 (43.3) | 74 (52.9) | 1.47 (0.97, 2.22) | .068 |
| Undiagnosed | 97 (38.5) | 47 (33.6) | 1.24 (0.80, 1.91) | .333 |
| Gender: Male | 159 (63.1) | 77 (55.0) | 1.40 (0.92, 2.13) | .117 |
| Age ≤14 years | 105 (41.8) | 54 (39.1) | 1.12 (0.73, 1.71) | .604 |
Nonconsanguineous marriages were considered as a reference category in the logistic regression analysis.
Abbreviation: SC, Shafallah Center.
Logistic regression analysis.
No significant differences between consanguineous and nonconsanguineous marriages were observed for all genetic disorders.
Among consanguineous and nonconsanguineous marriages, the percentage of chromosomal disorders was significantly lower among adults as compared to the pediatric subjects. On the other hand, adults have higher undiagnosed disorder compared to the pediatric group among consanguineous marriages (Table 5).
Table 5.
Consanguinity, and genetic disorder by age groups (SC Data)
|
Consanguineous marriages N (%) |
p‐valuea |
Nonconsanguineous marriages N (%) |
p‐valuea | |||
|---|---|---|---|---|---|---|
| Age ≤14 years | Age >14 years | Age ≤14 years | Age >14 years | |||
| Autosomal Recessive | 20 (19.0) | 23 (15.8) | .302 | 3 (5.6) | 16 (19.0) | .020 |
| Autosomal Dominant | — | — | — | — | — | — |
| X‐linked | 2 (1.9) | 0 (0) | .174 | — | — | — |
| Chromosomal disorder | 54 (51.4) | 55 (37.7) | .021 | 36 (66.7) | 37 (44.0) | .007 |
| Undiagnosed | 29 (27.6) | 68 (46.6) | .002 | 15 (27.8) | 32 (38.1) | .143 |
Overall‐ p = .009 (χ 2 = 11.51), Overall‐ p = .018 (χ 2 = 8.02).
Abbreviation: SC, Shafallah Center.
Pearson Chi‐squared and Fisher Exact tests
In addition, among nonconsanguineous marriages the percentage of Autosomal Recessive was significantly higher among adults as compared to the pediatric group (Table 5).
With regards to relatedness effect on genetic disorders, the multinominal logistic regression analysis showed that undiagnosed disorder was more likely to be predicted by first cousin (OR: 4.40; 95% CI: 1.29–15.05; p = .018) in contrast to nonconsanguineous marriages. The rest of the relatedness levels were not significant for undiagnosed disorders. None of the other mode of inheritance of genetic conditions was predicted by specific relatedness level.
3.4. Effect of consanguinity on various genetic disorders in the total cohort cases
We calculated the overall pooled odds ratios for each genetic disorder across both institutions (HMC and SC, Figure 1). Overall odds ratio for Autosomal Recessive disorders was significantly associated with consanguinity. Consanguineous marriages had approximately two times increased risk of developing Autosomal Recessive disorders compared to nonconsanguineous marriages.
On the other hand, nonconsanguineous marriages had a significant higher risk of chromosomal disorders compared to consanguineous marriages.
No significant differences were found for undiagnosed disease, X‐linked and Autosomal Dominant, age, and gender between nonconsanguineous and consanguineous marriages. The heterogeneity was not significant within each genetic disorder (Table 6 and Figure 1).
Table 6.
Heterogeneity for the total sample (HMC and SC)
| Group | Number studies | Heterogeneity | |||
|---|---|---|---|---|---|
| Q‐value | df (Q) | p‐value | τ‐squared | ||
| Fixed effect analysis | |||||
| Age: <14 | 2 | 0.029 | 1 | .866 | 0.000 |
| Autosomal Recessive | 2 | 2.923 | 1 | .087 | 0.222 |
| Autosomal Dominant | 1 | 0.000 | 0 | 1.000 | 0.000 |
| Chromosomal | 2 | 2.599 | 1 | .107 | 0.659 |
| Gender: Male | 2 | 0.004 | 1 | .949 | 0.000 |
| Undiagnosed | 2 | 0.652 | 1 | .419 | 0.000 |
| X‐linked | 2 | 0.519 | 1 | .471 | 0.000 |
| Total within | 6.725 | 6 | .347 | ||
| Total between | 15.162 | 6 | .019 | ||
| Overall | 13 | 21.887 | 12 | .039 | 0.086 |
Abbreviation: HMC, Hamad Medical Corporation; SC, Shafallah Center.
4. DISCUSSION
Our results clearly demonstrate that consanguinity is indeed a major risk factor in the occurrence of Autosomal Recessive diseases. The relatedness analysis showed that the Autosomal Recessive disorder group was more likely to be predicted by paternal first cousin, double first cousin, and first cousin in the HMC data.
In contrast, chromosomal abnormality was significantly related to nonconsanguinity in HMC but not SC. Chromosomal abnormality was also more common among pediatric patients compared to adults in consanguineous and nonconsanguineous marriages in SC but not HMC, which could be attributed generally to chromosomal abnormalities mostly diagnosed in this pediatric age group (≤14 years).
Adults, on the other hand, were more likely to be undiagnosed (in consanguineous marriages) and to have Autosomal Recessive (in nonconsanguineous marriages) at SC compared to pediatric patients. This can be explained by the fact that the phenotype is usually atypical or milder in the age group above 14 years, thus, reaching a definitive diagnosis is more challenging.
These differences between the centers could be attributed mainly due to the fact that the two centers (HMC and SC) received different types of cases. HMC is the main tertiary care center for diagnosis and management for genetic disorders and the data were also collected through direct interviews with patients and through their medical record files. On the other hand, SC is mainly for rehabilitation that also includes cases with nongenetic disorders and the data were collected only by accessing the patients’ record file. Thus, there are obviously differences in the referral patterns and the nature of the patients among the two centers.
The effect of consanguinity on various genetic disorders in the total cohort cases confirmed our findings.
Many previous studies in the region have established the link between marriage practices and genetic disorders. In a recent study in Saudi Arabia, researchers indicated that the percentage of birth defects was as high as 54.5% in a consanguineous group (Majeed‐Saidan et al., 2015). Another study from Tunisia reported consanguineous marriages to have a relatively higher risk of producing offspring with genetic disorders, caused by the expression of rare recessive genes inherited from common ancestors, than that of the general population (Khlat & Khoury, 1991; Teebi, 1994). On the other hand, in an Egyptian study, it was reported that consanguinity has no significant effect in Autosomal Dominant disorders (Shawky, Elsayed, Ibrahim, & Seifeldin, 2012). Another study from Jordan, reported that approximately 30% of sporadic undiagnosed cases of mental retardation, congenital anomalies, and dysmorphism may have an Autosomal Recessive etiology with risks of recurrence in future pregnancies (Hamamy et al., 2007). It has also been noted that the appearance of rare syndromes such as Joubert syndrome is highly concentrated in particular tribes and families in the United Arab Emirates (Al‐Gazali et al., 1997).
There are no studies of this type concerned specifically with the Qatari society, where our observations strongly suggest that there is an increased risk of genetic disorders if parents are consanguineous. The objective of this study is to understand the role of consanguinity in specific categories of genetic disorders in the Qatari population. Qatar represents many aspects of the Arab Gulf countries in relation to its small population and historically high preference for consanguineous unions. Consanguineous marriages are also recognized as being associated with higher risk for Autosomal Recessive diseases than in the general population (Kumaramanickavel, Joseph, Vidhya, Arokiasamy, & Shridhara Shetty, 2002; Taillemite et al., 1985; World Health Organization, Regional Office for the Eastern Mediterranean, 1997) by favoring the expression of recessive deleterious alleles. This study mainly focusses on the relationship between marriage between relatives, and genetic diseases among Qataris. This study will help in reducing the health effects of marriage between relatives and recommend developing public health policies in Qatar in the long run.
The results also indicate that first cousin marriages, especially paternal ones, could be a risk factor for Autosomal Recessive disorder. We only speculate that this could be due to the fact that the Qatari society is a patriarchal one where paternal cousin marriages are more likely to happen and more predominant. This pattern could have started several generations back in the life of the current patients (e.g., their parents, grandparents, and great grandparents).
In addition, mothers from consanguineous marriages were more likely to be unemployed and have lower educational levels compared to mothers from nonconsanguineous marriages. We can speculate that unemployed females with lower educational level may lack the knowledge of the dangers and consequences of consanguineous marriages. They may also have limited social networks with most of their social life taking place within the same extended family. Thus, they are more likely to marry from the same family tribe compared to those who are employed and are educated to a lower level. In addition, it is possible that parental influence plays a bigger role among unemployed females with lower educational levels. Age also was a vital factor to explain some of the differences at one of the centers.
Thus, future interventions and policies should take into account these socioeconomic and demographic factors.
The results of the study can help to inform public knowledge and shape opinion in Qatar regarding genetic disorders and disabilities resulting from consanguineous marriage. As the results of this study show, it is imperative to advocate for a change to the cultural and social framework that reproduces and normalizes consanguineous marriages in the region. Policy makers should endorse social, educational, and public health initiatives to mitigate the impact of genetic disease in the Qatari society.
CONFLICT OF INTEREST
None declared.
AUTHORS' CONTRIBUTIONS
T.B.‐O. and K.Al.‐G.: designed the study and obtained funds, T.B.‐O. recruited patients into the study, M.E. interviewed patients and data acquisition, T.Y., collected the data, N.A.‐D. and M.S. supervised data analysis, N.A.‐D., P.C., and M.S. performed statistical analysis. T.B.‐O., M.E., T.Y., and M.S. and N.A.‐D. contributed to manuscript drafting. All authors critically reviewed and agreed on the final version of the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all patients for being included in the study.
CONSENT FOR PUBLICATION
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article. All enquiries should be directed to Nader Al‐Dewik: naldewik@hamad.qa.
ACKNOWLEDGMENTS
This study was financially supported by Qatar National Research Fund (QNRF) ‐ National Priorities Research Program (NPRP) grant no [4‐086‐5‐007]. We would like to thank QNRF for their support.
The statements made herein are solely the responsibility of the authors.
Ben‐Omran T, Al Ghanim K, Yavarna T, et al. Effects of consanguinity in a cohort of subjects with certain genetic disorders in Qatar. Mol Genet Genomic Med. 2020;8:e1051 10.1002/mgg3.1051
REFERENCES
- Abdulrazzaq, Y. M. , Bener, A. , Al‐Gazali, L. I. , Al‐Khayat, A. I. , Micallef, R. , & Gaber, T. (1997). A study of possible deleterious effects of consanguinity. Clinical Genetics, 51(3), 167–173. 10.1111/j.1399-0004.1997.tb02447.x [DOI] [PubMed] [Google Scholar]
- Al‐Gazali, L. I. , Bener, A. , Abdulrazzaq, Y. M. , Micallef, R. , Al‐khayat, A. I. , & Gaber, T. (1997). Consanguineous marriages in the United Arab Emirates. Journal of Biosocial Science, 29(4), 491–497. 10.1017/S0021932097004914 [DOI] [PubMed] [Google Scholar]
- Al‐Gazali, L. I. , Sztriha, L. , Dawodu, A. , Bakir, M. , Varghese, M. , Varady, E. , … Padmanabhan, R. (1999). Pattern of central nervous system anomalies in a population with a high rate of consanguineous marriages. Clinical Genetics, 55(2), 95–102. 10.1034/j.1399-0004.1999.550205.x [DOI] [PubMed] [Google Scholar]
- Bener, A. , & Alali, K. (2006). Consanguineous marriage in a newly developed country: The Qatari population. Journal of Biosocial Science, 38, 239–246. 10.1017/S0021932004007060 [DOI] [PubMed] [Google Scholar]
- Bener, A. , & Mohammad, R. R. (2017). Global distribution of consanguinity and their impact on complex diseases: Genetic disorders from an endogamous population. Egyptian Journal of Medical Human Genetics, 18(4), 315–320. 10.1016/j.ejmhg.2017.01.002 [DOI] [Google Scholar]
- Bittles, A. H. , & Black, M. L. (2010). Consanguinity, human evolution, and complex diseases. Proceedings of the National Academy of Sciences of the United States of America, 107(Suppl 1), 1779–1786. 10.1073/pnas.0906079106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein, M. , Hedges, L. , Higgins, J. , & Rothstein, H. (2005). Comprehensive meta-analysis (version 2) [Computer Software]. Englewood, NJ: Biostat. [Google Scholar]
- Catalogue for Transmission Genetics in Arabs . (2017). CTGA at a glance. Retrieved from http://cags.org.ae/ctga/graph/ [Google Scholar]
- Cohen, J. (1988). Statistical power analyses for the behavioral sciences (2nd edn). Hillsdale, NY: Erlbaum. [Google Scholar]
- El Mouzan, M. I. , Al Salloum, A. A. , Al Herbish, A. S. , Qurachi, M. M. , & Al Omar, A. A. (2008). Consanguinity and major genetic disorders in Saudi children: A community‐based cross‐sectional study. Annals of Saudi Medicine, 28(3), 169–173. 10.4103/0256-4947.51726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Shafei, A. , Rao, P. , & Sandhu, A. K. (1986). Congenital malformations and consanguinity. Australian and New Zealand Journal of Obstetrics and Gynaecology, 26(3), 168–172. 10.1111/j.1479-828X.1986.tb01559.x [DOI] [PubMed] [Google Scholar]
- Hamamy, H. (2012). Consanguineous marriages: Preconception consultation in primary health care settings. Journal of Community Genetics, 3(3), 185–192. 10.1007/s12687-011-0072-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamy, H. , Antonarakis, S. E. , Cavalli‐Sforza, L. L. , Temtamy, S. , Romeo, G. , Kate, L. P. T. , … Bittles, A. H. (2011). Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop Report. Genetics in Medicine, 13(9), 841–847. 10.1097/GIM.0b013e318217477f [DOI] [PubMed] [Google Scholar]
- Hamamy, H. , Masri, A. , Al‐Hadidy, A. , & Ajlouni, K. (2007). Consanguinity and genetic disorders. Profile from Jordan. Saudi Medical Journal, 28, 1015–1017. [PubMed] [Google Scholar]
- Hoodfar, E. , & Teebi, A. S. (1996). Genetic referrals of Middle Eastern origin in a western city: Inbreeding and disease profile. Journal of Medical Genetics, 33(3), 212–215. 10.1136/jmg.33.3.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkeni, E. , Monastiri, K. , Saket, B. , Guediche, M. N. , & Ben Cheikh, H. (2007). Interplay of socio‐economic factors, consanguinity, fertility, and offspring mortality in Monastir Tunisia. Croatian Medical Journal, 48(5), 701–707. [PMC free article] [PubMed] [Google Scholar]
- Khlat, M. , & Khoury, M. (1991). Inbreeding and diseases: Demographic, genetic, and epidemiologic perspectives. Epidemiologic Reviews, 13, 28–41. 10.1093/oxfordjournals.epirev.a036072 [DOI] [PubMed] [Google Scholar]
- Kumaramanickavel, G. , Joseph, B. , Vidhya, A. , Arokiasamy, T. , & Shridhara Shetty, N. (2002). Consanguinity and ocular genetic diseases in South India: Analysis of a five‐year study. Community Genetics, 5(3), 182–185. 10.1159/000066334 [DOI] [PubMed] [Google Scholar]
- Majeed‐Saidan, M. A. , Ammari, A. N. , AlHashem, A. M. , Al Rakaf, M. S. , Shoukri, M. M. , Garne, E. , & Kurdi, A. M. (2015). Effect of consanguinity on birth defects in Saudi women: Results from a nested case‐control study. Birth Defects Res A Clin Mol Teratol, 103(2), 100–104. 10.1002/bdra.23331 [DOI] [PubMed] [Google Scholar]
- Ross, L. E. (1997). Mate selection preferences among African American college students. Journal of Black Studies, 27(4), 554–569. 10.1177/002193479702700407 [DOI] [Google Scholar]
- Shawky, R. M. , Elsayed, N. S. , Ibrahim, D. S. , & Seifeldin, N. S. (2012). Profile of genetic disorders prevalent in northeast region of Cairo, Egypt. Egyptian Journal of Medical Human Genetics, 13(1), 45–62. 10.1016/j.ejmhg.2011.10.002 [DOI] [Google Scholar]
- Tadmouri, G. , Nair, P. , Obeid, T. , Al Ali, M. , Al Khaja, N. , & Hamamy, H. (2009). Consanguinity and reproductive health among Arabs. Reproductive Health, 6(1), 17 10.1186/1742-4755-6-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillemite, J. L. , Daffos, F. , Joye, N. , Forestier, F. , Portnoi, M. F. , & Capella‐Pavlovsky, M. (1985). Fetal karyotyping of fetal blood obtained from the umbilical vein using ultrasound. Journal de Gynécologie, Obstétrique et Biologie de la Reproduction (Paris), 14(3), 315–319. [PubMed] [Google Scholar]
- Teebi, A. S. (1994). Autosomal recessive disorders among Arabs: An overview from Kuwait. Journal of Medical Genetics, 31(3), 224–233. 10.1136/jmg.31.3.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teebi, A. S. , & El‐Shanti, H. I. (2006). Consanguinity: Implications for practice, research, and policy. Lancet, 367(9515), 970–971. 10.1016/S0140-6736(06)68406-7 [DOI] [PubMed] [Google Scholar]
- World Health Organization, Regional Office for the Eastern Mediterranean . (1997). Community control of genetic and congenital disorders. Retrieved from https://apps.who.int/iris/handle/10665/119571 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. All enquiries should be directed to Nader Al‐Dewik: naldewik@hamad.qa.


