Abstract
p53 and aldehyde dehydrogenase (ALDH) have been implicated in key tumorigenesis processes including cancer initiating cell (CIC) maintenance; however, the relationship between these two mediators remains poorly defined. In this study, ALDH isoform expression diversity was revealed in CICs with disparate p53 functional states: gain of function, high risk p53 mutation (p53HRmut) and wildtype p53 (p53WT) inactivated by the human papillomavirus 16 (HPV16) E6 oncogene. Interrogation of head and neck squamous cell carcinoma (HNSCC) cell lines and patient tumors showed that HPV16+/p53WT cases have higher ALDH variance score (AVS), a measure of tumor ALDH isoform expression diversity, compared to HPV−/p53HRmut cases (p = 0.03). AVS and several individual ALDH isoforms were associated with prognosis in HPV16+/p53WT HNSCC but not in HPV−/p53HRmut HNSCC. Knockdown of the dominant ALDH isoform in high AVS HNSCC depleted the CIC pool in vitro and in vivo. Our results demonstrate that p53 functional states are associated with distinct ALDH isoform transcriptomic signatures. Moreover, tumor ALDH profiling may provide insight on which ALDH isoform to target in high AVS HNSCC tumors to deplete the CIC population.
Subject terms: Cancer stem cells, Head and neck cancer
Introduction
A frequent genetic event in the tumorigenesis cascade is aberrant p53 function, either loss of wildtype p53 (p53WT) function or gain of function, high-risk p53 mutation (p53HRmut). p53WT controls normal stem cell homeostasis and evidence exists to support p53WT dysfunction in cancer initiating cell (CIC) expansion and maintenance1. Nanog, a core embryonic stem cell transcription factor, known to modulate normal stem cell and CIC pluripotency is negatively regulated by p53WT2–4. In addition, CD44 and CD133, two well-recognized markers for CICs, are repressed by p53WT through direct promoter occupancy5,6. A logical extension of these studies is that loss of p53WT function will likely result in CIC expansion through multiple mechanisms to drive cancer promotion and progression. In contrast to p53WT, literature on p53HRmut in this space is scant. A recent paper reported that p53HRmut is more efficient than p53WT deficiency to facilitate somatic cell reprogramming; however, these p53HRmut reprogrammed cells exhibit genetic instability and have a higher potential for malignant transformation with tumor initiating properties7.
Aldehyde dehydrogenase (ALDH) is a superfamily consisting of 19 evolutionarily conserved isoforms and its activity is used as a functional assay to identify normal stem cells and CICs. Based on initial work, it was generally believed that ALDH1A1 is indispensable for high ALDH activity to maintain the CIC pool8–10. However, depending on the cell line or anatomical site, there is now compelling evidence that other ALDH isoforms may contribute to or be primarily responsible for elevated ALDH activity in CICs11–15. A relationship between p53 and ALDH was revealed when p53HRmut was shown to act as a transcription factor to drive ALDH1A1 transcription to confer a novel gain of function effect on CIC maintenance and/or expansion16. However, the influence of p53 functional states on the ALDH gene family in the context CICs and cancer, in general, remains poorly understood.
Head and neck squamous cell carcinomas (HNSCCs) are cancers from various anatomical site in the head and neck region and can be subdivided based on two major etiologic factors: smoking/alcohol use and high-risk human papillomavirus (HPV), in particular HPV16. HPV− HNSCCs are driven by smoking and alcohol use, and often harbor p53 genomic alterations, mostly gain of function, missense mutations. In contrast, HPV + HNSCCs have p53WT that is inactivated by the HPV oncogene, E6. These two distinct etiologies offer an unique opportunity to explore the relationship between p53 and ALDH without the use of genetic engineering approaches. Using HPV− and HPV16+ cell lines and patient tumors, we showed that p53 functional states have differential ALDH isoform expression diversity at the CIC and bulk tumor levels. HPV16+/p53WT tumors tend to display a dominant ALDH isoform expression pattern with enrichment of a particular ALDH family member. Interestingly, targeting the dominant ALDH isoform in high AVS HNSCC cell lines resulted in robust depletion of the CIC population. Our work demonstrates that p53 functional states are associated with distinct ALDH isoform transcriptomic signatures and suggests that tumor ALDH profiling may identify the isoform responsible for CIC maintenance in high AVS HNSCC.
Results
ALDH isoform transcriptomic profiles in CICs from HPV16+ and HPV− HNSCC cell lines
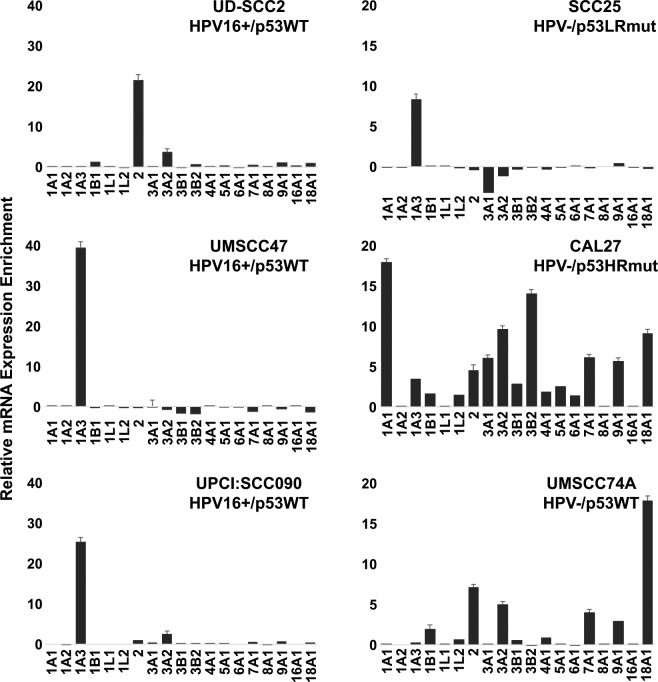
The HPV16+ HNSCC cell lines, UD-SCC2, UMSCC47, and UPCI-SCC090, have wildtype p53 that is inactivated by HPV16E617. The HPV− HSNCC cell lines have distinct p53 states; CAL27 has gain of function, high risk mutant p53 (H193L), SCC25 has loss of function p53 due to the deletion of two base pairs in codon 209, and UMSCC74A has wildtype p5318. ALDH activity measured using the ALDEFLUOR assay is a well-established methodology to identify and quantitate CICs in hematologic and solid malignancies. ALDHhigh CIC fraction varied across our panel of HNSCC cell lines (Supplemental Fig. 1). We collected ALDHhigh (top 5%) and ALDHlow (bottom 5%) populations to determine the ALDH isoform expression signature in CICs from these cell lines. As shown in Fig. 1a, ALDHhigh CICs from HPV16+/p53WT HNSCC displayed a dominant ALDH isoform expression signature; ALDH1A3 was enriched in UMSCC47 and UPCI-SCC090 CICs, whereas, ALDH2 was the main isoform enriched in UD-SCC2 CICs. Similarly, in SCC25, a HPV− HNSCC cell line with truncated, loss of function p53, ALDHhigh CICs were enriched for a single ALDH isoform, ALDH1A3. In contrast, high ALDH isoform expression diversity was shown in ALDHhigh CICs from HPV−/p53HRmut CAL27. HPV−/p53WT UMSCC74A CICs represent an intermediate group and had moderate ALDH isoform expression diversity.
Figure 1.
ALDH isoform transcriptomic profiles in CICs from HPV16+ and HPV− HNSCC cell lines. A panel of HPV16+ and HPV− HNSCC cell lines with distinct p53 functional states were sorted for ALDHhigh and ALDHlow subsets, and assessed for expression of individual ALDH isoforms. Data is presented as relative mRNA expression enrichment in the ALDHhigh CIC population (mean ± s.e.m.; n = 3).
HPV16+/p53WT HNSCC have higher ALDH variance score than HPV−/p53HRmut HNSCC
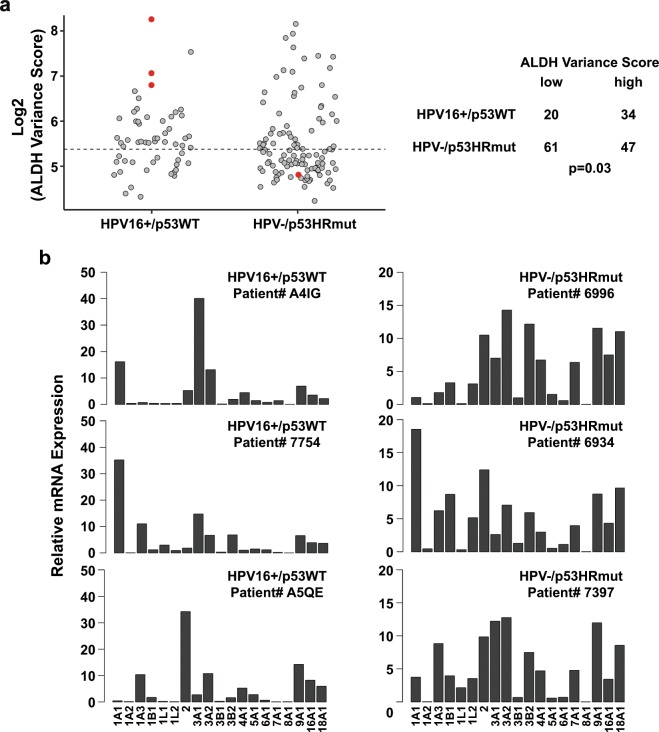
Our results using HNSCC cell lines suggest that HPV status and p53 functional states may drive distinct ALDH expression signatures. In order to extend these findings from cell lines to patient tumors, we developed a mathematical algorithm, ALDH variance score (AVS), as an approach to quantitate the ALDH isoform expression variance in individual samples. HPV16+/p53WT HNSCC cell lines had AVS >100; 109.6 for UD-SCC2, 313.1 for UMSCC47, and 137.4 for UPCI-SCC090. HPV− SCC25 expresses truncated, loss of function p53 and thus, has an inactivated p53 functional state analogous to HPV16-driven inactivation of p53WT. Similar to HPV16+/p53WT cell lines, SCC25 had a high AVS of 175.7. In comparison, HPV−/p53HRmut CAL27 HNSCC cell line had a low AVS of 28.4. Next, the AVS algorithm was applied to the HPV16+ and HPV− cohorts from the TCGA HNSCC dataset. The difference in AVS was unremarkable between HPV16+ and HPV− tumors (Supplemental Fig. 2a). Moreover, when tumors were binned based on p53 mutational status, AVS distribution was comparable across the p53WT, p53LRmut, and p53HRmut cohorts (Supplemental Fig. 2b). We focused our work on HPV16+/p53WT and HPV−/p53HRmut since these two distinct entities are frequently presented in the clinic. As shown in Fig. 2a, HPV16+/p53WT tumors are associated with high AVS, and HPV−/p53HRmut tumors tend to have low AVS (p = 0.03, Fisher’s exact test). Representative HPV16+/p53WT HNSCC tumors display a dominant ALDH isoform expression pattern with high AVS, whereas, representative HPV−/p53HRmut HNSCC tumors show greater ALDH isoform expression diversity with low AVS (Fig. 2b).
Figure 2.
HPV16+/p53WT HNSCCs have higher ALDH variance score than HPV−/p53HRmut HNSCCs. (a) ALDH variance score (AVS). AVS was calculated for HPV16+/p53WT and HPV−/p53HRmut HNSCC cell lines and primary tumors. Cell lines are represented by red circles and primary tumors from the TCGA HNSCC cohort are represented by grey circles. The dashed line represents the median AVS for the combined cohort. Median AVS for the combined cohort was used to stratify into two groups: low AVS and high AVS (p = 0.03, Fisher’s exact test). (b) ALDH isoform expression profiles from representative HPV16+/p53WT and HPV−/p53HRmut HNSCC patients.
ALDH variance score is associated with stemness index
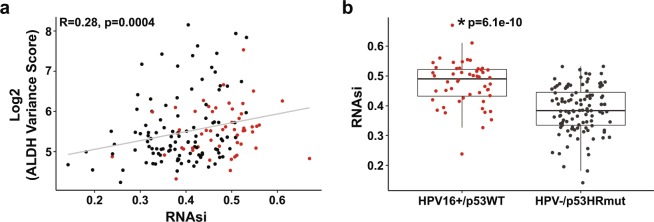
Direct quantitation of the CIC pool in individual TCGA HNSCC tumors is not feasible so we used the PanCancer Atlas stemness index (mRNAsi)19 as a tool to infer relative CIC frequency. High AVS is associated with a high mRNAsi (r = 0.28, p = 0.0004) in this TCGA HNSCC cohort (Fig. 3a). Moreover, mean mRNAsi was higher in HPV16+/p53WT tumors than in HPV−/p53HRmut HNSCC tumors (p = 6.1 × 10−10). This is consistent with a previous report showing that HPV16+/p53WT HNSCCs have a higher intrinsic CIC frequency than HPV−/p53HRmut HNSCC20 and thus, supports the potential utility of mRNAsi to infer relative CIC frequency from global transcriptome in patient tumors.
Figure 3.
ALDH variance score is associated with stemness index. (a) AVS and mRNAsi. High AVS is correlated with a high mRNAsi (r = 0.28, p = 0.0004, Spearman correlation). (b) mRNAsi in HPV16+/p53WT and HPV−/p53HRmut tumors. Data are presented as box plots (p = 6.1 × 10−10, Wilcoxon test).
ALDH variance score is associated with survival in HPV16+/p53WT HNSCC
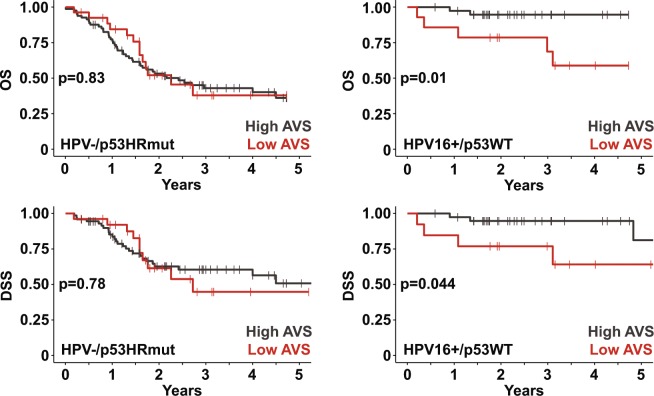
Since HPV16+/p53WT patients have superior overall survival (OS) and disease-specific survival (DSS) compared to HPV−/p53HRmut patients (Supplemental Fig. 3), the influence of AVS on prognosis was analyzed for each cohort separately. In Fig. 4, in the HPV16+/p53WT setting, high AVS patients had superior OS (log-rank, p = 0.01) and DSS (log-rank, p = 0.044) compared to low AVS patients. However, the prognostic utility of AVS was not observed in the HPV−/p53HRmut cohort. Next, we took a step further and investigated the clinical impact of individual ALDH isoforms. High ALDH1A3, ALDH3B1, ALDH7A1, and ALDH18A1 correlated with poor prognosis, whereas, elevated ALDH2, an isoform with tumor suppressive actions, was associated with better OS in HPV16+/p53WT patients (Table 1). None of the ALDH isoforms was prognostic in the HPV−/p53HRmut setting.
Figure 4.
ALDH variance score is a prognostic biomarker in HPV16+/p53WT HNSCC. Five-year Kaplan-Meier plots for overall and disease-specific survival in HPV16+/p53WT and HPV−/p53HRmut cohorts. Cohorts were stratified based on AVS: high AVS denotes top 75% and low AVS denotes bottom 25%. Log-rank test was used to compare the Kaplan-Meier plots.
Table 1.
Select ALDH isoforms are prognostic biomarkers in HPV16+/p53WT HNSCC.
| HPV16+/p53WT | HPV−/p53HRmut | |||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| ALDH1A1 | 0.94 (0.82, 1.08) | 0.376 | 0.99 (0.96, 1.02) | 0.643 |
| ALDH1A2 | 1.02 (0.81, 1.27) | 0.886 | 0.74 (0.22, 2.50) | 0.630 |
| ALDH1A3 | 1.17 (1.01, 1.36) | 0.033 | 1.01 (0.98, 1.05) | 0.438 |
| ALDH1B1 | 1.13 (0.91, 1.40) | 0.269 | 0.99 (0.91, 1.08) | 0.889 |
| ALDH1L1 | 1.07 (0.81, 1.42) | 0.617 | 1.38 (0.81, 2.36) | 0.234 |
| ALDH1L2 | 1.16 (0.35, 3.85) | 0.813 | 0.94 (0.79, 1.11) | 0.450 |
| ALDH2 | 0.85 (0.74, 0.98) | 0.023 | 0.99 (0.97, 1.03) | 0.937 |
| ALDH3A1 | 1.02 (0.97, 1.08) | 0.425 | 1.00 (0.99, 1.02) | 0.781 |
| ALDH3A2 | 0.85 (0.71, 1.00) | 0.056 | 0.97 (0.91, 1.04) | 0.381 |
| ALDH3B1 | 3.15 (1.25, 7.96) | 0.015 | 1.08 (0.75, 1.57) | 0.665 |
| ALDH3B2 | 1.07 (0.97, 1.18) | 0.205 | 0.99 (0.97, 1.02) | 0.712 |
| ALDH4A1 | 1.08 (0.88, 1.33) | 0.449 | 0.99 (0.92, 1.06) | 0.715 |
| ALDH5A1 | 0.97 (0.60, 1.59) | 0.915 | 1.15 (0.90, 1.47) | 0.267 |
| ALDH6A1 | 1.16 (0.25, 5.49) | 0.851 | 0.95 (0.53, 1.70) | 0.857 |
| ALDH7A1 | 1.25 (1.03, 1.50) | 0.021 | 1.02 (0.96, 1.09) | 0.576 |
| ALDH8A1 | 1.10 (0.80, 1.30) | 0.180 | 0.52 (0.09, 2.82) | 0.445 |
| ALDH9A1 | 0.90 (0.71, 1.14) | 0.390 | 1.04 (0.96, 1.12) | 0.351 |
| ALDH16A1 | 0.91 (0.64, 1.31) | 0.624 | 0.98 (0.90, 1.07) | 0.704 |
| ALDH18A1 | 1.33 (1.07, 1.65) | 0.011 | 0.99 (0.93, 1.06) | 0.889 |
Targeting the dominant ALDH isoform in high AVS HNSCC depletes the CIC pool
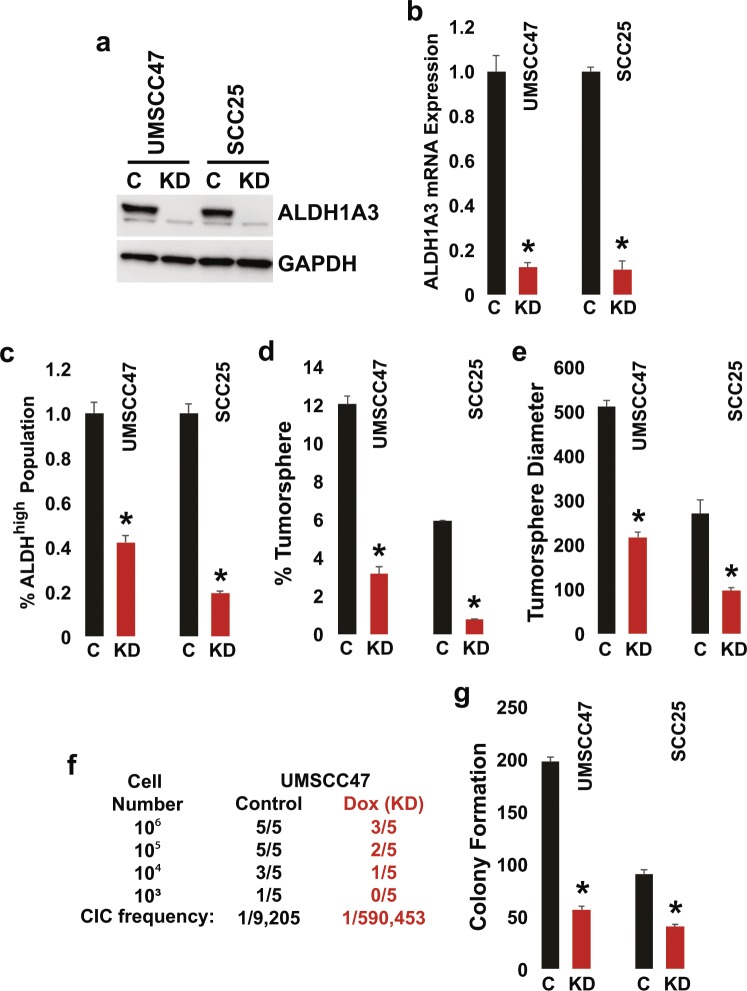
We hypothesized that high AVS is indicative of a homogenous CIC pool and thus, targeting the dominant ALDH isoform may result in contraction of the CIC population. Tetracycline-inducible ALDH1A3 shRNA polyclonal UMSCC47 and SCC25 cells were generated to determine if the dominant ALDH isoform is indispensable for CIC maintenance in representative HPV16+ and HPV− HNSCC with high AVS (Fig. 5). ALDH1A3 mRNA expression and protein levels were substantially reduced following induction with doxycycline. Knockdown of ALDH1A3 depleted the ALDHhigh CIC population by 57% in UMSCC47 and 79% in SCC25. Furthermore, tumorsphere formation efficiency and tumorsphere diameter were reduced; 71% and 58% inhibition in UMSCC47 and, 83% and 67% inhibition in SCC25, respectively. In vivo limiting dilution assay in NSG mice showed that ALDH1A3 knockdown dramatically depleted the CIC population by > 60-fold in UMSCC47. CIC frequency was reduced from 1/9,205 to 1/590,453 (p < 0.001).
Figure 5.
Targeting the dominant ALDH isoform in high AVS HNSCC depletes the CIC pool. UMSCC47 and SCC25 cells were transduced with the inducible pLV-RNAi/shRNA-ALDH1A3 and polyclonal cell populations were collected. Cells were stimulated with doxycycline at 1000 ng/ml for all the experiments. (a) ALDH1A3 protein levels. Cell lysates were immunoblotted with anti-ALDH1A3 and GAPDH antibodies. Representative image is cropped. (b) ALDH1A3 mRNA expression. ALDH1A3 and GAPDH expression was determined using qPCR with TaqMan primers. Data were normalized to GAPDH and are presented as mean ± s.e.m. (n = 3, *p < 0.05, two-tailed Student’s t-test). (c) ALDHhigh CIC population. Cells were analyzed by FACS and ALDHhigh CIC population was quantitated using the ALDEFLUOR assay. Data are presented as mean ± s.e.m. (n = 3, *p < 0.05, two-tailed Student’s t-test). (d,e) Tumorsphere formation efficiency and diameter. Cells were harvested, seeded on low-attachment plates in a defined, serum-free culture medium, and tumorspheres were allowed to grow. Tumorsphere formation efficiency was calculated as the number of tumorspheres formed divided by the original number of cells seeded. Data are presented as mean ± s.e.m. (n = 3, *p < 0.05, two-tailed Student’s t-test). (f) In vivo cancer initiating cell frequency. Indicated number of cells were implanted subcutaneously in the flanks of NSG mice. Tumor incidence (palpable tumor of any size) was noted over the course of the experiment. Cancer initiating cell (CIC) frequency was calculated using the L-Calc program. (g) Clonogenic survival. Cells were plated and allowed to grow in complete media for 10 days. Subsequently, colonies were fixed, stained with crystal violet, and counted. Data are presented as mean ± s.e.m. (n = 3, *p < 0.05, two-tailed Student’s t-test).
Discussion
There are hints in the literature that p53 functional states regulate ALDH to modulate the CIC pool. Reactivation of p53WT in HPV16+/p53WT HNSCC depleted the ALDHhigh CIC pool20. Knockout of p53HRmut in SW480 colorectal carcinoma cells resulted in CIC population contraction and reduction of ALDH1A1 expression16. Moreover, p53−/− RKO cells showed higher levels of ALDH1A3 compared to its isogenic p53+/+ counterpart16. These findings indicate that perturbations of p53 functional states have a consequence on CIC maintenance and regulation of certain ALDH isoforms. However, since these studies assessed only a select number of ALDH isoforms, the connection between p53 and ALDH in cancer remains poorly defined.
In this study, we assessed the expression profile of the entire ALDH gene family in HNSCC cell lines and primary tumors with defined HPV and p53 statuses. A dominant ALDH isoform expression signature was shown in HPV16+/p53WT CICs. In contrast, HPV−/p53HRmut CAL27 had CICs with considerable ALDH isoform expression diversity; seven isoforms were enriched by >5-fold. Using AVS as a measure of ALDH isoform expression diversity, analysis of the TCGA HNSCC dataset indicated that HPV16+/p53WT tumors have higher AVS compared to HPV−/p53HRmut tumors revealing that the differences in ALDH expression signature between p53 functional states may not be limited to the CIC subset but extend to the bulk tumor cell population as well. These findings led to the speculation that CIC frequency and/or genomic homogeneity is appreciably higher in HPV16+/p53WT tumors than in HPV−/p53HRmut tumors and thus, transcriptomes of HPV16+/p53WT tumors may better reflect the CIC population. This concept is supported by several pieces of evidence: (a) HPV16 preferentially infects basal cells in the squamous epithelium and these undifferentiated, isogenic cells are likely to be the cell of origin for HPV16+/p53WT tumors, (b) HPV16+/p53WT tumors have higher CIC frequency20 and mRNAsi (Fig. 3) than HPV−/p53HRmut tumors, and (c) HPV16+/p53WT tumors have lower aneuploidy score21 and mutant allele tumor heterogeneity (MATH)22 than HPV−/p53HRmut tumors (Supplemental Fig. 4).
The ALDH superfamily consists of 19 evolutionarily conserved isoforms recognized to oxidize aldehydes to carboxylic acids23. In addition to aldehyde metabolism, ALDHs are involved in a plethora of cellular processes which influence tumorigenesis, including retinoic acid (RA) synthesis and signaling, ultraviolet light absorption, hydroxyl radical scavenging, and antioxidant activity24,25. Multiple groups have investigated and shown select ALDH isoforms, in particular ALDH1 members, as prognostic biomarkers in a spectrum of solid malignancies26–28. We assessed the entire ALDH family and found a select number of isoforms, ALDH1A3, ALDH2, ALDH3B1, ALDH7A1, and ALDH18A1, to be associated with survival in HPV16+/p53WT HNSCC. ALDH2 has tumor suppressive actions and polymorphisms in this gene is associated with increased risk to a number of alcohol-related cancers29. The pro-tumorigenic role of ALDH1A3 in cancer is well-described and its utility as a prognostic biomarker is beginning to emerge11,14,30. Scant literature exists to link ALDH3B1, ALDH7A1, and ALDH18A1 to cancer, although, these studies provide initial evidence that these isoforms favor tumorigenesis31–33. Additional work is needed to improve our understanding of these poorly studied ALDH isoforms in tumorigenesis, stemness, and treatment relapse in HPV16-driven malignancies.
AVS was developed as a quantitative tool to capture ALDH isoform expression diversity from bulk tumor transcriptomic datasets. An initial concern was that AVS may just be capturing global genomic and transcriptomic differences. However, this potential issue was mitigated based on our analyses showing that AVS was not correlated with aneuploidy score or MATH (Supplemental Fig. 4) and moreover, assessment of the cytokeratin family, consisting of 37 genes, using the same methodology for AVS revealed that cytokeratin variance score was similar between HPV16+/p53WT and HPV−/p53HRmut tumors (Supplemental Fig. 5). Based on our findings that AVS is positively correlated with mRNAsi, we speculated that AVS may be a marker for CIC homogeneity such that high AVS is indicative of a tumor populated with homogenous CICs at high frequency. This notion is further supported by our data that the CIC pool was severely contracted as a consequence of targeting the dominant ALDH isoform in two high AVS HNSCC cell lines; one in the HPV16+ setting and the other in the HPV− setting. ALDH isoform specific inhibitors are actively being developed for oncologic indications, however, biomarkers are needed to select patients likely to respond to these targeted molecules. A potential clinical application for AVS is as a niche molecular biomarker to match high AVS HNSCC patients to specific ALDH isoform inhibitors.
Materials and Methods
Cell lines
SCC25 and CAL27 cell lines were purchased from ATCC (Manass, VA). UMSCC74A and UMSCC47 cell lines were obtained from Thomas Carey, University of Michigan. UPCI:SCC090 and UD-SCC2 cell lines were provided by Susanne Gollin, University of Pittsburgh and Henning Bier, Heinrich-Heine University, Dusseldorf, Germany. CAL27, UD-SCC2, UMSCC47, UMSCC74A, and UPCI:SCC090 were grown in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 100 µg/ml streptomycin, and 100 U/ml penicillin. SCC25 cells were grown in a 1:1 mixture of Ham’s F-12 and DMEM supplemented with 10% FBS, 0.4 µg/mL hydrocortisone, 100 µg/ml streptomycin, and 100 U/ml penicillin. Cell lines were maintained in a humidified atmosphere of 5% CO2 at 37 °C. Cell lines were authenticated using STR profile analysis and not tested for mycoplasma contamination.
Quantitative real-time PCR
Cells were extracted for total RNA using the TRIzol reagent (ThermoFisher Scientific, Waltham, MA) or TaqMan PreAmp Cells-to-CT kit (ThermoFisher Scientific). mRNA expression of ALDH isoforms was determined using the Applied Biosystems 7900HT Fast Real-Time PCR System with validated, pre-designed TaqMan primers (ThermoFisher Scientific). The expression of ALDH isoforms were normalized to glyceraldehyde 3-phosphate dehydrogenase (GADPH) using the ∆∆Ct method.
ALDH variance score
Clinical data from the TCGA HNSCC dataset were downloaded from the Genomics Data Commons data portal/TCGA Research Network: http://cancergenome.hih.gov/. HPV status for the TCGA HNSCC cohort were determined by PCR from the biospecimen center Nationwide Children’s Hospital, and downloaded from the auxiliary file of the clinical TCGA HNSCC dataset from the GDC data portal. All HPV16+ and HPV− cases from the TCGA HNSCC cohort were identified and analyzed. Mutational profiles of individual patients were obtained from the updated Pan-Cancer dataset. p53 missense mutations were classified as high risk using the evolutionary action score of p53 (EAp53) threshold of >7534. Pre-processed legacy RNA-seq data were downloaded from NCI Genomic Data Commons using TCGAbiolinks package in R and gene level GC-content normalization was performed35,36. RNA-Seq by Expectation Maximization (RSEM) of individual ALDH isoforms was converted to percent of total ALDH RSEM by dividing the RSEM for each ALDH isoform by the sum of RSEM from all ALDH isoforms. For each sample, the deviation of each ALDH isoform from the mean percentage was calculated and squared to avoid negative values. ALDH variance score (AVS) was then calculated as an average of the squared deviations using the formula:
Stemness index
The PanCancer Atlas published a set of stemness indices based on mRNA expression (mRNAsi) by machine-learning scoring the oncogenic dedifferentiation of TCGA tumor samples19. Briefly, the entire RNA expression gene set was evaluated and each gene was weighted based on stemness features. The stemness of each sample was scored accordingly and subsequently mapped to the [0, 1] range.
Inducible shRNA-ALDH1A3 expression system
Three targeting sequences to knockdown ALDH1A3 were designed, synthesized, and cloned into the inducible tetracycline-on pLV-RNAi (BioSettia, San Diego, CA) vector. Sequence 1: 5′ AAAAGGTCAAGTTCACCAAGATATTGGATCCAATATCTTGGTGAACTTGACC 3′; Sequence 2: 5′ AAAAGCAGAGAACTAGGTGAATATTGGATCCAATATTCACCTAGTTCTCTGC 3′ Sequence 3: 5′AAAAGCAGGTCTACTCTGAGTTTGTTTGGATCCAAACAAACTCAGAGTAGACCTGC 3′. Cells were transduced with pLV-RNAi/shRNA-ALDH1A3 and polyclonal cell populations were collected. Preliminary studies demonstrated that sequence 1 resulted in the most efficient knockdown of ALDH1A3. All experiments in this study were performed using sequence 1 and cells were stimulated with doxycycline at 1 µg/ml.
Immunoblot
Whole cell lysates were mixed with Laemmli loading buffer, boiled, separated by SDS–PAGE, and transferred to a nitrocellulose membrane. Subsequently, immunoblot analyses were performed using antibodies specific to ALDH1A3 (ab129815; abcam) or GADPH (AB2302; MilliporeSigma) and processed using the Pierce Fast Western Blot Kit (ThermoFisher Scientific).
ALDEFLUOR assay
Cells were suspended in ALDEFLUOR assay (Stem Cell Technologies, British Columbia, Canada) buffer containing ALDH substrate (bidipy-aminoacetaldehyde, 1 mM per 1 × 106 cells) and incubated for 45 mins at 37 °C. For each experiment, an aliquot of cells was exposed to diethylaminobenzaldehyde (50 mM), an ALDH inhibitor, to serve as the negative control. The sorting gate was established using the negative control as the baseline. Cell suspensions were centrifuged at 300 g for 5 mins at 4 °C and re-suspended in 0.5 mL ALDEFLUOR assay buffer for analysis. Fluorescence-activated cell sorting analyses were performed using BD FACS Calibur (BD Lifesciences, Franklin Lakes, NJ) at The Ohio State University Comprehensive Cancer Center Analytical Cytometry Core.
Tumorsphere formation assay
Cells were collected and seeded in a serum-free defined medium consisting of keratinocyte serum-free medium supplemented with epidermal growth factor, basic fibroblast growth factor, insulin and hydrocortisone in low-attachment plates (Corning Incorporated, Corning, NY). Tumorsphere formation efficiency was calculated by dividing the number of tumorspheres (≥50 µm in diameter) formed in 7 d by the initial number of cells seeded. Tumorsphere diameter was measured using the NIS-Elements software (Nikon Instruments, Melville, NY).
In vivo tumor incidence
Inducible shRNA-ALDH1A3 UMSCC47 cells were suspended in 50:50 DMEM:Matrigel and implanted subcutaneously into the flank of 6–8 week old, female NOD/SCID mice. Subsequently, for each cell dilution, mice were randomly assigned to two treatment arms; control diet (n = 5) or doxycycline (200 mg/kg)-containing diet (n = 5) ad libitum. Tumor incidence was monitored for 49 days following tumor cell implantation. Cancer initiating cell frequency was calculated using the L-Calc program (STEMCELL Technologies Inc., Vancouver, Canada). Sample size estimate was not performed for this experiment. All animals were included in our analysis. Investigative team was not blinded to the group allocation during the experiment and when assessing the endpoint of tumor incidence. Animal experiments were conducted in compliance with ethical regulations and under an approved protocol from The Ohio State University.
Statistical analyses
All analyses were performed in R. Fisher’s exact, Spearman, Student’s t, or Wilcoxon tests were used when appropriate to evaluate the association between categorical variables. Five-year OS and DSS plots were generated by the Kaplan-Meier method and log-rank test was used to compare the plots. Cox proportional hazards model was used for univariate analysis for individual ALDH isoforms as a continuous variable. All tests were two-tailed and p-values < 0.05 were considered significant.
Supplementary information
Acknowledgements
Supported in part by National Institutes of Health grants R01CA193590, R01DE023555, and R01GM117921, and University Hospitals Seidman Cancer Center
Author contributions
S.G., X.X., W.Q.O. and Q.P. conceived and designed the study. S.G., X.X., W.Q.O., K.C.R., J.G.Y., T.N.T. and Q.P. acquired, analyzed, or interpreted the data. S.G., W.Q.O. and Q.P. drafted and revised the manuscript. All authors approved the submitted version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shanying Gui and Xiujie Xie.
Supplementary information
is available for this paper at 10.1038/s41598-020-57758-5.
References
- 1.Bonizzi G, Cicalese A, Insinga A, Pelicci PG. The emerging role of p53 in stem cells. Trends Mol. Med. 2012;18:6–12. doi: 10.1016/j.molmed.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Lin T, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 3.Po A, et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 2010;29:2646–2658. doi: 10.1038/emboj.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zbinden M, et al. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park EK, et al. Transcriptional repression of cancer stem cell marker CD133 by tumor suppressor p53. Cell Death Dis. 2015;6:e1964. doi: 10.1038/cddis.2015.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godar S, et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarig R, et al. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J. Exp. Med. 2010;207:2127–2140. doi: 10.1084/jem.20100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YC, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem. Biophys. Res. Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 9.Clay MR, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head. Neck. 2010;32:1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao C, et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin. Cancer Res. 2014;20:4154–4166. doi: 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim IG, Lee JH, Kim SY, Kim JY, Cho EW. Fibulin-3 negatively regulates ALDH1 via c-MET suppression and increases gamma-radiation-induced sensitivity in some pancreatic cancer cell lines. Biochem. Biophys. Res. Commun. 2014;454:369–375. doi: 10.1016/j.bbrc.2014.10.084. [DOI] [PubMed] [Google Scholar]
- 13.Luo Y, et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cell. 2012;30:2100–2113. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcato P, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cell. 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 15.Kurth I, et al. Cancer stem cell related markers of radioresistance in head and neck squamous cell carcinoma. Oncotarget. 2015;6:34494–34509. doi: 10.18632/oncotarget.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solomon H, et al. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene. 2018;37:1669–1684. doi: 10.1038/s41388-017-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimple RJ, et al. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013;73:4791–4800. doi: 10.1158/0008-5472.CAN-13-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min BM, et al. Inactivation of the p53 gene by either mutation or HPV infection is extremely frequent in human oral squamous cell carcinoma cell lines. Eur. J. Cancer B Oral. Oncol. 1994;30B:338–345. doi: 10.1016/0964-1955(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 19.Malta TM, et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell. 2018;173:338–354 e315. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M, et al. Elevated intrinsic cancer stem cell population in human papillomavirus-associated head and neck squamous cell carcinoma. Cancer. 2014;120:992–1001. doi: 10.1002/cncr.28538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoadley KA, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304 e296. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral. Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol. Rev. 2007;59:125–150. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- 24.Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 25.King G, Holmes R. Human corneal and lens aldehyde dehydrogenases. Purification and properties of human lens ALDH1 and differential expression as major soluble proteins in human lens (ALDH1) and cornea (ALDH3) Adv. Exp. Med. Biol. 1997;414:19–27. doi: 10.1007/978-1-4615-5871-2_4. [DOI] [PubMed] [Google Scholar]
- 26.Charafe-Jauffret E, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang F, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol. Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, et al. Prognostic value of ALDH1 and Nestin in advanced cancer: a systematic meta-analysis with trial sequential analysis. Ther. Adv. Med. Oncol. 2019;11:1758835919830831. doi: 10.1177/1758835919830831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seitz HK, Stickel F. Acetaldehyde as an underestimated risk factor for cancer development: role of genetics in ethanol metabolism. Genes. Nutr. 2010;5:121–128. doi: 10.1007/s12263-009-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J. Clin. Invest. 2019;130:2043–2055. doi: 10.1172/JCI126414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Hoogen C, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 32.Marchitti SA, Orlicky DJ, Brocker C, Vasiliou V. Aldehyde dehydrogenase 3B1 (ALDH3B1): immunohistochemical tissue distribution and cellular-specific localization in normal and cancerous human tissues. J. Histochem. Cytochem. 2010;58:765–783. doi: 10.1369/jhc.2009.955773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang JQ, et al. Dietary cholesterol promotes steatohepatitis related hepatocellular carcinoma through dysregulated metabolism and calcium signaling. Nat. Commun. 2018;9:4490. doi: 10.1038/s41467-018-06931-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neskey DM, et al. Evolutionary Action Score of TP53 Identifies High-Risk Mutations Associated with Decreased Survival and Increased Distant Metastases in Head and Neck Cancer. Cancer Res. 2015;75:1527–1536. doi: 10.1158/0008-5472.CAN-14-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colaprico A, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva TC, et al. TCGA Workflow: Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Res. 2016;5:1542. doi: 10.12688/f1000research.8923.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.