Abstract
Background
Previous studies have disclosed up‐regulation of MIR‐378 in acute myeloid leukemia (AML), and might consequently affect the outcome of the patients. Correspondingly, hypomethylation of MIR‐378 was also identified in AML, particularly for FAB‐M2 subtype with t(8;21) chromosomal translocation. Nevertheless, the methylation status of MIR‐378 has not been illustrated in myelodysplastic syndrome (MDS). Herein we designed to understand the methylation pattern of MIR‐378 involved in MDS and clinical interrelation thereof.
Methods
Real‐time quantitative methylation‐specific PCR (RQ‐MSP) was performed to evaluate the methylation degree of MIR‐378 5’‐flanking region on bone marrow mononuclear cells collected from 95 de novo MDS patients. Five gene mutations (IDH1, IDH2, DNMT3A, U2AF1, and SF3B1) were detected by high‐resolution melting analysis to further evaluate the clinical relevance of hypomethylation of MIR‐378.
Results
Unmethylated level of MIR‐378 5’‐flanking region was significantly higher in MDS patients than that in controls (p = .034). Hypomethylated MIR‐378 was identified in 20 of 95 (21%) cases with MDS. Male patients appeared to be more frequent to harbor MIR‐378 hypomethylation compared to female patients (15/55, 27.3% vs. 4/40, 10.0%, p = .04). There was no significant difference in age, white blood cell counts, platelet counts, hemoglobin concentration, and karyotypes between the patients with and without MIR‐378‐hypomethylation. Distinct distribution of five gene mutations was not observed in the two groups as well. However, MIR‐378‐hypomethylated patients had significantly shorter overall survival than those without MIR‐378 hypomethylation (p = .036). Moreover, among patients <60 years, hypomethylation of MIR‐378 was confirmed to be an independent adverse prognostic factor by both Kaplan–Meier and Multivariate Cox analyses.
Conclusion
Hypomethylation of MIR‐378 5’‐flanking region is an adverse prognosticator in MDS, particularly in patients <60 years.
Keywords: hypomethylation, MIR‐378, myelodysplastic syndrome, prognosis
Hypomethylation level of MIR‐378 was significantly higher in myelodysplastic syndrome (MDS) patients than that in controls (p = .034). MIR‐378‐hypomethylated patients had significantly shorter overall survival than those without MIR‐378 hypomethylation (p = .036). Both Kaplan–Meier and Multivariate Cox analyses confirmed that hypomethylation of MIR‐378 5’‐flanking region is an adverse prognosticator in MDS, particularly in patients <60 years.

1. INTRODUCTION
Myelodysplastic syndrome (MDS) is a hematopoietic malignance characterized by dyshematopoiesis with a high tendency to transformation of acute myeloid leukemia (AML) (Valent et al., 2007). Cytogenetic as well as genetic alterations have been widely illustrated and considered to play a vital role in the processes of MDS (Adès, Itzykson, & Fenaux, 2014; Lee, Podoltsev, Gore, & Zeidan, 2015). Meanwhile, epigenetic variations, particularly for the aberrant DNA methylation, were also deemed to participate in the development of MDS (Issa, 2013; Itzykson & Fenaux, 2014). In spite of promising clinical application of hypomethylating agents (HMAs) presented recently, however, primary drug resistance and recurrence or disease progress during the treatment are the main obstacles (Carella, 2015), the outcome of patients with MDS generally remains dismal. The knowledge about pathogenesis of MDS is still insufficient. Until recently, a group of small noncoding RNA molecules (19–24 nts), microRNAs, have emerged as crucial roles in nearly all kinds of biologic functions such as development, differentiation, apoptosis, and proliferation (Schmittgen, 2008; Winter, Jung, Keller, Gregory, & Diederichs, 2009). Ectopic expression of miRNAs was implicated in a wide scale of diseases including cancers (Barbarotto, Schmittgen, & Calin, 2008; Calin & Croce, 2006). Increasing number of studies have even supposed that deregulation of various microRNAs contributed to pathogenesis of MDS and transformation into AML (Pons et al., 2009; Votavova et al., 2011), furthermore, large number of microRNAs could be served as useful biomarkers to predict the outcome (Bhise et al., 2016; Cattaneo et al., 2015; Xu, Guo, Zhang, Chen, & Hu, 2016). Strikingly, researchers have reported distinctive influence of MIR‐378 in many cancers, such as lung cancer (Chen et al., 2012), colorectal carcinoma (Faltejskova et al., 2012), gastric cancer (Deng et al., 2013). Our previous study preliminarily observed overexpressed MIR‐378 was often related to AML and exerted adverse effects on prognosis (Qian et al., 2013). Correspondingly, hypomethylated MIR‐378 was revealed to be associated with AML, especially in FAB‐M2 subtype (Xiao‐Wen et al., 2015). However, there is limited information about MIR‐378 methylation status in MDS. This work was to determine the methylation status of the 5’‐flanking region of MIR‐378, which contains 212 CpG dinucleotides, and further to analyse the clinical role in MDS.
2. MATERIALS AND METHODS
2.1. Clinical samples
This work was approved by both the Ethics Committees of the Affiliated People's Hospital of Jiangsu University and Kunshan Third People's Hospital. A total of 95 primary MDS patients, treated at the Affiliated People’ Hospital of Jiangsu University or Kunshan Third People's Hospital were included in this study. All bone marrow (BM) samples were obtained at initial diagnosis. Written informed consents were signed by each participant before the BM samples were collected. BM samples from 16 health donors were served as controls. The diagnosis of each case was established according to World Health Organization (WHO) and French‐American‐British (FAB) classification (Bennett et al., 2010; Vardiman et al., 2009). The risk classification for each patient was on the basis of International Prognosis Scoring System (IPSS) for MDS (Greenberg et al., 1997). The clinical features as well as laboratory tests of whole MDS patients were displayed in Table 1.
Table 1.
Correlation of MIR‐378 hypomethylation with clinical features in MDS
| Clinical parameters | MIR‐378 hypomethylation | |||
|---|---|---|---|---|
| No (n = 75) | Yes (n = 20) | Total (n = 95) | p | |
| Age (years)a | 60 (20–86) | 63.5 (27–85) | 60 (20–86) | .560 |
| Sex (male/female) | 39/36 | 16/4 | 55/40 | .040 |
| WBC (×109/L)a | 2.8 (0.9–26.6) | 2.4 (1.3–19.5) | 2.7 (0.9–26.6) | .204 |
| HB (g/L)a | 68 (26–128) | 58 (29–104) | 63.5 (26–118) | .088 |
| PLT (×109/L)a | 63 (0–754) | 60 (9–1176) | 61.5 (0–1176) | .964 |
| Cytogenetics | .607 | |||
| Good | 54 | 15 | 69 | |
| Intermediate | 9 | 1 | 10 | |
| Poor | 5 | 3 | 8 | |
| No data | 7 | 1 | 8 | |
| WHO classifications | .486 | |||
| RARS | 8 | 4 | 12 | |
| RCMD (RS) | 32 | 5 | 37 | |
| 5q‐ | 2 | 0 | 2 | |
| RAEB‐1 | 16 | 4 | 20 | |
| RAEB‐2 | 16 | 7 | 23 | |
| MDS‐U | 1 | 0 | 1 | |
| FAB classifications | .222 | |||
| RA | 32 | 7 | 39 | |
| RARS | 15 | 2 | 17 | |
| RAEB | 26 | 11 | 37 | |
| RAEBt | 2 | 0 | 2 | |
| IPSS | .541 | |||
| Low | 5 | 3 | 8 | |
| Int‐1 | 45 | 9 | 54 | |
| Int‐2 | 13 | 4 | 17 | |
| High | 8 | 2 | 10 | |
| No data | 4 | 2 | 6 | |
| Gene mutations | ||||
| U2AF1 (+/−) | 5/66 | 2/15 | 7/81 | .616 |
| IDH1/2 (+/−) | 4/68 | 0/17 | 4/85 | 1.000 |
| DNMT3A (+/−) | 2/70 | 1/16 | 3/86 | .475 |
| SF3B1 (+/−) | 5/66 | 1/16 | 6/82 | 1.000 |
Abbreviations: FAB, French‐American‐British; HB, hemoglobin; IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndrome; PLT, platelet count; RA, refractory anemia; RAEB, RA with excess of blasts; RARS, RA with ringed sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RCMD‐RS, RCMD with ringed sideroblasts; WBC, white blood cells; WHO, World Health Organization.
Median (range).
2.2. Therapeutic regimen
Treatment options including supportive care, low‐ or high‐intensity therapies, were given generally according to the risk classification and the wishes of patients. Briefly, symptomatic Low/Int‐1 patients were managed with hematopoietic growth factors, thalidomide or immunosuppressive therapy, and supportive treatment. The treatment for Int‐2/High patients included decitabine or chemotherapy with CAG (aclacinomycin, cytarabine, and G‐CSF) or HAG (homoharringtonine, cytarabine, and G‐CSF) protocol.
2.3. DNA isolation, bisulfite modification
The BM mononuclear cells (BMNCs) were obtained by using of Ficoll‐Hypaque gradient. Genomic DNA extraction from the BMNCs was conducted via genomic DNA purification kit (Gentra), and then CpGenome™ DNA Modification Kit (Chemicon) was used to treat the production of genomic DNA according to the manufacturer's protocol.
2.4. Methylation‐specific PCR
The methylation‐specific PCR (MSP) reactions for methylated MIR‐378 and unmethylated MIR‐378 were performed on Step One Plus (Applied Biosystems) using two sets of methylated (M) and unmethylated (U) specific primers (Table 2). For the control, we chose ALU repetitive sequence as was described before (Qian et al., 2011). The amplification reaction system consisted of 10 μM SYBR Premix Ex Taq II, 0.4 μM of primers, 0.4 μL 50× ROX (Takara), and 20 ng of bisulfite‐modified DNA. Amplification for methylated specific primers was conducted in the conditions of 95°C for 30 s, cycled at 95°C for 5 s, 62°C for 30 s, and 72°C for 30, and a fluorescence collection step at 75°C for 30 s (40 cycles), followed by a melting program at 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s. In real‐time quantitative MSP (RQ‐MSP) for unmethylated specific primers of MIR‐378, the amplification was carried out in the following program: 95°C for 30 s, then 40 cycles of 95°C for 5 s, 62°C for 30 s, 72°C for 30 s, and 75°C for 30 s (fluorescence collection step), followed by a melting program at 95°C for 15 s, 60°C for 60 s, 95°C for 15 s, and 60°C for 15 s.
Table 2.
The sequences of primers used in RQ‐MSP and BSP
| Forward (5’→3’) | Reverse (5’→3’) | Product (bp) | |
|---|---|---|---|
| RQ‐MSP | |||
| M | GGATGAGTTTTGAGTCGTTT | CCATACAAACCGCTCACTCC | 129 |
| U | TAGGATGAGTTTTGAGTTGT | ATAATCCCATACAAACCACT | 137 |
| BSP | AGGATTTTTTGGTGATTTTTG | TCACCCTCTAACTACATAATCCC | 200 |
Abbreviations: BSP, bisulfite‐sequencing PCR; RQ‐MSP, real‐time quantitative methylation‐specific PCR.
The normalized ratio (Nunmethylation‐ MIR‐378) was used to evaluate unmethylated level of MIR‐378. Nunmethylation‐ MIR‐378 was determined using the 2−ΔΔCT method comparing the ALU gene level. This means that a higher value of Nunmethylation‐ MIR‐378 represents a lower methylation status or hypomethylation of MIR‐378. The products of MSP were resolved in the concentration of 2% agarose gels to visualize under UV illumination.
2.5. Bisulfite‐sequencing PCR
In our experiment, Bisulfite‐sequencing PCR (BSP) was used to further determine DNA methylation density of MIR‐378 in samples from three MIR‐378‐hypermethylated and three MIR‐378 hypomethylated MDS patients according to the results of RQ‐MSP. The PCR was conducted with the reaction system containing 6.25 μM of dNTP mixture, 10× PCR buffer (0.25 mM KCl), 0.75 U of Hot start DNA polymerase (Takara), 0.5 μM of primers, and 20 ng of modified DNA on iCycler Thermal Cycler (Eppendorf). The bisulfite‐treated DNA was amplified with sequencing primers in Table 2. The conditions of amplification were 98°C for 10 s, then cycled at 98°C for 10 s, 56°C for 30 s, 72°C for 30 s, and entered to extension at 72°C for 7 min. After the program of purification, the PCR products were cloned into pMD®19‐T Vector (Takara), and 10 clones of each BSP product were sequenced (http://www.bgitechsolutions.cn/bbs). The percentage of methylation was calculated from the number of methylated CpG divided by total CpG loci of all 10 clones.
2.6. Mutations analysis
High‐resolution melting analysis (HRMA) (Lin et al., 2011, 2012; Qian et al., 2012; Yang et al., 2013) was conducted to analyze IDH1, IDH2, DNMT3A, SF3B1, and U2AF1 gene mutations. Direct DNA sequencing was employed to verify the positive results.
2.7. Statistical analysis
The SPSS software 22.0 package was used for Statistical analysis. Mann–Whitney/Wilcoxon test was performed to compare two groups with continuous variables. For the comparison of categorical variables between patient teams, the statistical method utilized was Pearson Chi‐square analysis or Fisher exact test. Kaplan–Meier method was employed for survival analysis, and the identification of the independent risk factors affecting the survival was carried out by Cox regression model. A two‐tailed p < .05 was considered statistically significant.
3. RESULTS
3.1. Hypomethylation of MIR‐378 5’‐flanking region in MDS patients
The human MIR‐378 gene locates in chromosome 5q32, according to bioinformatics analysis, there are three CpG islands within −2.0 kb consisted in MIR‐378 5’‐flanking region (Xiao‐Wen et al., 2015). As was disclosed by RQ‐MSP, the unmethylated level of MIR‐378 5’‐flanking region was substantially higher in MDS patients (median 0.021, range 0–23.990) than that in controls (median 0.005, range 0–1.000) (p = .034, Figure 1). The products of RQ‐MSP for MIR‐378 methylation and MIR‐378 unmethylation were analyzed by agarose gel electrophoresis and the representative electrophoretogram is displayed in Figure 2. Bisulfite sequencing was further performed to examine three patients with MIR‐378 hypomethylation and three patients with MIR‐378 hypermethylation on the basis of RQ‐MSP, and the results are shown in Figure 3. There was a significantly negative correlation between the results of RQ‐MSP and BSP (R = −.886, p = .019). Thus, our BSP analysis confirmed the results of RQ‐MSP.
Figure 1.
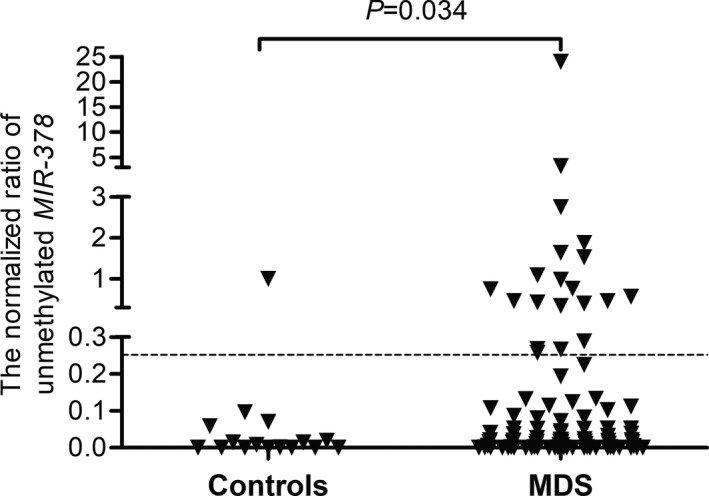
Relative Levels of MIR‐378 unmethylation in MDS and control. The cutoff value of the mean plus 1 SD (0.252) in normal controls was displayed with the dotted line. MDS, myelodysplastic syndrome
Figure 2.
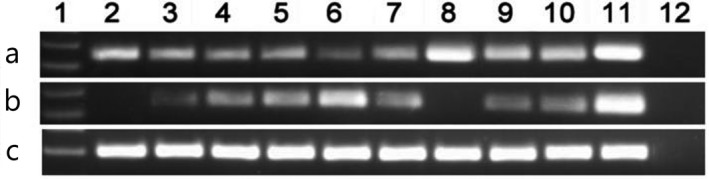
Electrophoresis results of RQ‐MSP products in MDS patients. (a) MIR‐378 methylation; (b) MIR‐378 unmethylation; (c) ALU. 1: Gene Ruler™ 100bp DNA ladder; 2–3: normal controls; 4–10: MDS samples; 11: cloned plasmid; 12: negative control. MDS, myelodysplastic syndrome; RQ‐MSP, real‐time quantitative methylation‐specific PCR
Figure 3.
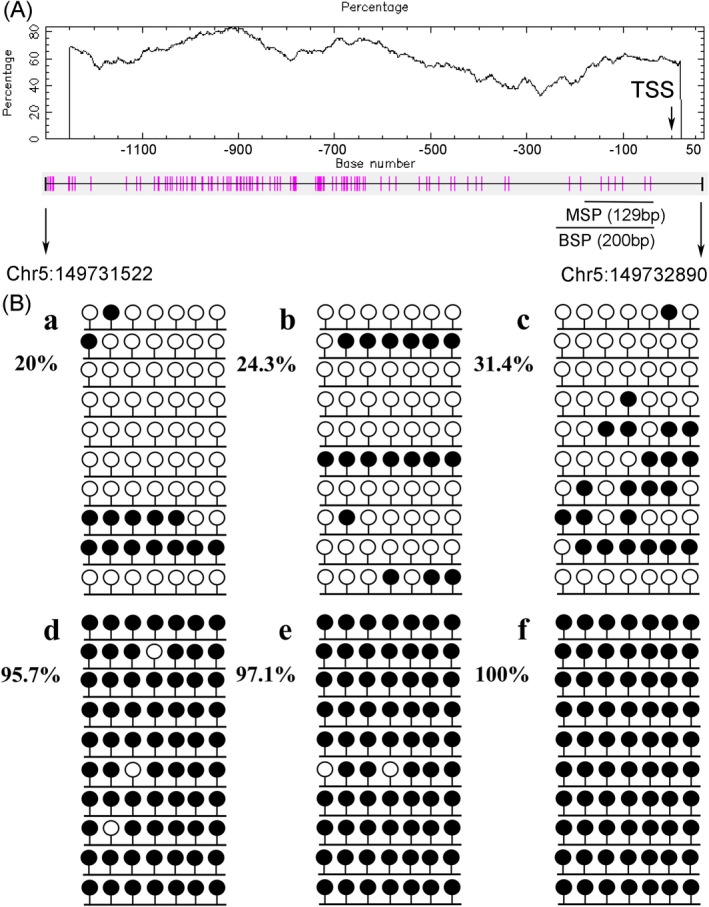
The CpG site of MIR‐378 5’‐flanking region and methylation density of MIR‐378 5’‐flanking region identified in three hypomethylated and three hypermethylated MDS patients by bisulfite sequencing. (A) diagrammatic sketches of the CpG site of MIR‐378 5’‐flanking region. The vertical lines represent the cytosine resides of CpGs. The black and gray arrows indicate the locations of the primers for BSP and RQ‐MSP respectively. (B) methylation density. Black lollipop: methylated CpG dinucleotide; Blank lollipop: unmethylated CpG dinucleotide. (a–c) Three hypomethylated samples; (d–f) three hypermethylated samples. The results of RQ‐MSP and bisulfite‐sequencing were significantly negative correlated (R = −.886, p = .019). BSP, bisulfite‐sequencing PCR; MDS, myelodysplastic syndrome; RQ‐MSP, real‐time quantitative methylation‐specific PCR
3.2. Association between MIR‐378 5’‐flanking region methylation and clinical characteristics
For clinical relevance analysis, the MDS patients were divided into two groups, the one with hypomethylated MIR‐378 and the other without hypomethylated MIR‐378, based on the value of the mean plus 1 SD (0.252) in normal controls. Accordingly, hypomethylated MIR‐378 was identified in 20 of 95 (21.1%) cases, however, there is no statistical significance as compared to healthy donors (6.3% [1/16], p = .188). No significant difference was observed in age, white blood cell counts, platelets, hemoglobin, as well as karyotypic groups between the two groups (p > .05). The incidence of hypomethylation of MIR‐378 in male (15/55, 27.3%) was significantly higher than that in female (4/40, 10.0%) (p = .040). There is no significant difference in distribution of MIR‐378 hypomethylaton in the FAB classifications, WHO classifications, or IPSS subgroups. It was also failed to find distinction of the five gene mutations between the two groups (p > .05) (Table 1).
3.3. Correlation between MIR‐378 5’‐flanking region methylation and prognosis
A total of 77 cases with available follow‐up data were included in our survival analysis. The median survival of all patients was about 24 months. Hypomethylated patients presented significantly worse survival compared to hypermethylated patients (median 8 vs. 30 months, respectively, p = .036, Figure 4a). Subgroup analysis revealed that patients with MIR‐378 hypomethylation presented slightly worse OS (median 7 months) than those without MIR‐378 hypomethylation (median not obtained) in cytogenetically normal patients (p = .152). Multivariate analysis including sex (male vs. female), IPSS (Low/Int‐1/Int‐2/High), MIR‐378 hypomethylation (yes vs. no), five gene mutations (mt vs. wt) failed to demonstrat independent prognostic factor of MIR‐378 hypomethylation in whole MDS patients (Table 3). However, among the age <60 years, patients with MIR‐378 hypomethylation had significantly shorter OS (median 9 months) than those without MIR‐378 hypomethylation (median not obtained) (p = .027, Figure 4b). The negative prognostic impact was also confirmed by multivariate analysis (Table 4).
Figure 4.
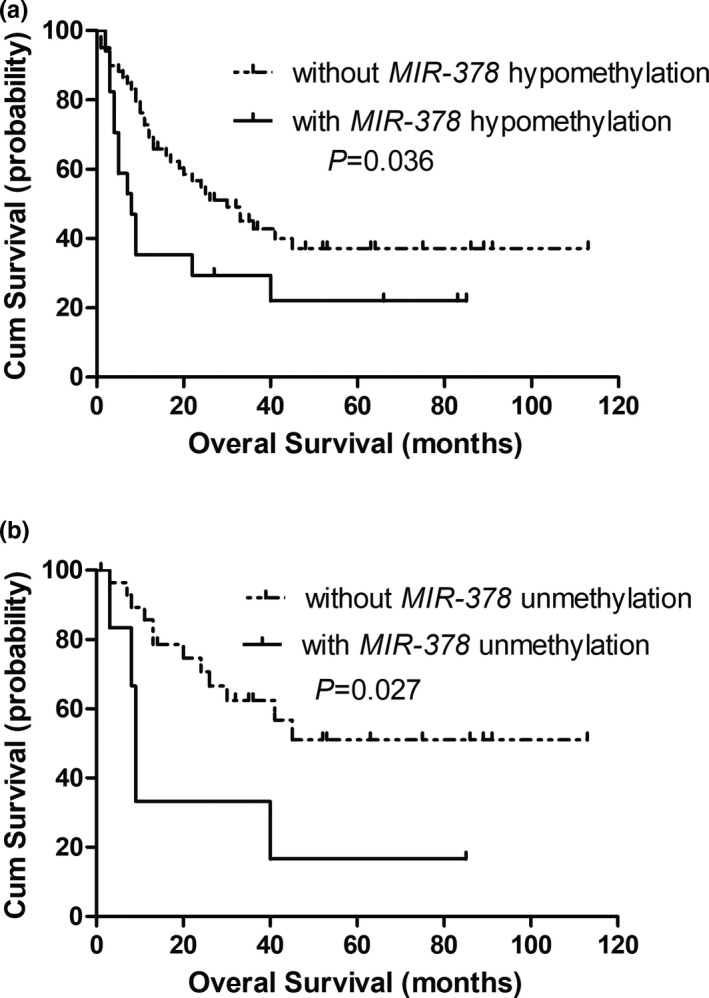
Overall survival of MDS patients. (a) All patients; (b) MDS patients <60 years. MDS, myelodysplastic syndrome
Table 3.
Multivariate analyses of prognostic factors for overall survival in MDS patients
| OS multivariate analysis | |||
|---|---|---|---|
| HRa | 95% CI | p value | |
| Sex | 0.385 | 0.189–0.786 | .009 |
| IPSS stratification | 1.525 | 1.076–2.161 | .018 |
| MIR‐378 hypomethylation | 1.709 | 0.830–3.519 | .146 |
| IDH1/2 mutation | 7.827 | 0.685–89.402 | .098 |
| DNMT3A mutation | 2.530 | 0.758–8.443 | .131 |
| U2AF1 mutation | 1.410 | 0.364–5.456 | .619 |
| SF3B1 mutation | 3.240 | 0.898–11.693 | .073 |
Abbreviations: CI, confidential interval; HR, hazard ratio; IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndrome; OS, overall survival.
HR >1 indicates an increased risk of an event for the first category listed.
Table 4.
Multivariate analyses of prognostic factors for overall survival in young (age <60 years) MDS patients
| OS multivariate analysis | |||
|---|---|---|---|
| HRa | 95% CI | p value | |
| Sex | 1.664 | 0.398–6.954 | .485 |
| IPSS stratification | 4.181 | 1.908–9.159 | .000 |
| MIR‐378 hypomethylation | 6.619 | 1.694–25.864 | .007 |
| IDH1/2 mutation | 7.827 | 0.685–89.402 | .098 |
| DNMT3A mutation | 17.391 | 1.551–194.943 | .021 |
| U2AF1 mutation | 1.410 | 0.364–5.456 | .619 |
| SF3B1 mutation | 31.725 | 2.604–386.451 | .007 |
Abbreviations: CI, confidential interval; HR, hazard ratio; IPSS, International Prognostic Scoring System; MDS, myelodysplastic syndrome; OS, overall survival.
HR >1 indicates an increased risk of an event for the first category listed.
4. DISCUSSION
Increasing numbers of microRNAs have been evidenced to participate in tumor initiation, evolution, drug‐resistance, and disease recurrence (Eitan et al., 2009; Esquela‐Kerscher & Slack, 2006; Fattore, Sacconi, Mancini, & Ciliberto, 2017; Ipn, Ng, Baharuddin, & Zakaria, 2017; Nicoloso, Spizzo, Shimizu, Rossi, & Calin, 2009; Ventura & Jacks, 2009), moreover, several of them have been confirmed to be useful tumor diagnostic and prognostic biomarkers. It is reported that MIR‐378 had the abilities to enhance some types of cancer cell proliferation, reduce the cell apoptosis in vitro, as well as promote neoplasm growth, angiogenesis, and metastasis in vivo (Chen et al., 2012; Lee, Deng, Wang, & Yang, 2007; Ma et al., 2014; Skrzypek et al., 2013; Yu et al., 2014). Clinically, a meta‐analysis presented recently has demonstrated the prospective diagnostic value of MIR‐378 in human cancers (Li, Shen, Li, Chen, & Chen, 2016). In AML, we firstly reported aberrant overexpression of MIR‐378 presented in FAB‐M2 subtype, particularly in cytogenetical classification of t(8;21) chromosomal translocation (Qian et al., 2013). Our RQ‐MSP analysis correspondently shown high incidence of hypomethylation of MIR‐378 presented in FAB‐M2 subtype, as well as for the cohort with t(8;21) chromosomal aberration (Xiao‐Wen et al., 2015). In vitro experiment, THP‐1 cells, an AML cell line, treated with a HMA 5‐aza‐dC, exhibited up‐expression of MIR‐378 in a dose‐dependent manner (Xiao‐Wen et al., 2015), providing the evidence of methylation regulation of MIR‐378 in AML cell line (Xiao‐Wen et al., 2015). Meanwhile, aberrant expression of MIR‐378 has even been reported in MDS by other studies, and the distinct expression depends on the different context of cytogenetics or different stage of MDS (Erdogan et al., 2011; Merkerova et al., 2011). Here in, we highlighted MIR‐378 methylation status in MDS, and the result demonstrated the generally hypomethylated MIR‐378 in MDS, those were compared to normal controls. Hypomethylation of MIR‐378 could be detected in 21% cases in our experiment. However, the distribution hypomethylation was not distinguishing either in cytogenetic classifications or in IPSS‐risk subgroups. Interestingly, our data indicated that MIR‐378 hypomethylation occurred more frequently in male patients. This phenomenon may be attributed to the prevalence of smoking in male people in China. It is crucial that the change of DNA methylation is another contribution by tobacco smoke in cancer (Alghanim, Wu, & Mccord, 2018; Tsaprouni et al., 2014). Whether MIR‐378 hypomethylation is correlated with smoking needs epidemiological investigation. In the present study a panel of gene mutations (IDH1, IDH2, DNMT3A, SF3B1,and U2AF1), genetically reported to be the important regulator in DNA methylation and RNA spliceosome and highly associated with MDS by recent studies (Adès et al., 2014; Gill, Leung, & Kwong, 2016; Lee et al., 2015; Lin et al., 2011, 2012), was analyzed by HRMA to further explore the correlation of hypomethylated MIR‐378 and genetic changes in MDS. However, no significant difference was found in the frequencies of five gene mutations (IDH1, IDH2, DNMT3A, SF3B1, and U2AF1) between patients with and without MIR‐378 hypomethylation.
Up‐expression rather than hypomethylation of MIR‐378 has been disclosed to be an adverse prognosticator in AML by our former studies (Qian et al., 2013; Xiao‐Wen et al., 2015). Notably, in this study we demonstrated that hypomethylation of MIR‐378 predicted poor outcome in MDS, particularly in patients <60 years. In our multivariate analysis, IPSS stratification, DNMT3A and SF3B1 mutations could significantly predict the outcome of younger patients (<60 years), in addition to these, hypomethylation of MIR‐378 also acts as a risk factor independently, providing helpful information for the prognosis of MDS. Further studies are needed to confirm the results before it can be used routinely as a potential marker for risk stratification in the patients with MDS.
Briefly, our study revealed that hypomethylaiton of MIR‐378 was a common epigenetic phenomenon in MDS, and predicted poor outcome of more young patients (<60 years).
CONFLICT OF INTEREST
None declared.
AUTHORS CONTRIBUTION
Jiang Lin and Jun Qian designed the study; Xiang‐mei Wen, Ying‐ying Zhang and Ji‐chun Ma performed experiments; De‐hong Wu, Xiao‐wen analyzed the data, created figures, and assisted in manuscript writing; Dong‐ming Yao and Jing‐dong Zhou analyzed data; Hong Guo, Peng‐fei Wu, Xing‐li Zhang, and Hong‐chun Qiu contributed patients data.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (81270630, 81970118), the Medical Innovation Team of Jiangsu Province (CXTDB2017002), 333 Project of Jiangsu Province (BRA2016131), Six Talent Peaks Project in Jiangsu Province (2015‐WSN‐115), Zhenjiang Clinical Research Center of Hematology (SS2018009), the Social Development Foundation of Zhenjiang (SH2017040, SH2018044), the Social Development Foundation of Kunshan (KS1624), “Postgraduate Research & Practice Innovation Program of Jiangsu Province” (KYCX17_1821), Clinical Medical Science, the Development Foundation of Jiangsu University (JLY20160011), and the Key Medical Talent Program of Zhenjiang City.
Wu D‐H, Zhu X‐W, Wen X‐M, et al. Hypomethylation of MIR‐378 5’‐flanking region predicts poor survival in young patients with myelodysplastic syndrome. Mol Genet Genomic Med. 2020;8:e1067 10.1002/mgg3.1067
De‐hong Wu, Xiao‐wen Zhu, and Xiang‐mei Wen authors contributed equally to this work.
Funding information
This study was supported by the National Natural Science Foundation of China (81270630, 81970118), the Medical Innovation Team of Jiangsu Province (CXTDB2017002), 333 Project of Jiangsu Province (BRA2016131), Six Talent Peaks Project in Jiangsu Province (2015‐WSN‐115), th eZhenjiang Clinical Research Center of Hematology (SS2018009), the Social Development Foundation of Zhenjiang (SH2017040, SH2018044), the Social Development Foundation of Kunshan (KS1624), “Postgraduate Research & Practice Innovation Program of Jiangsu Province” (KYCX17_1821), Clinical Medical Science, the Development Foundation of Jiangsu University (JLY20160011), the Key Medical Talent Program of Zhenjiang City.
Contributor Information
Jiang Lin, Email: linjiangmail@sina.com.
Jun Qian, Email: qianjun0007@hotmail.com.
REFERENCES
- Adès, L. , Itzykson, R. , & Fenaux, P. (2014). Myelodysplastic syndromes. Lancet, 383(9936), 2239–2252. 10.1016/S0140-6736(13)61901-7 [DOI] [PubMed] [Google Scholar]
- Alghanim, H. , Wu, W. , & Mccord, B. (2018). DNA methylation assay based on pyrosequencing for determination of smoking status. Electrophoresis, 39, 2806‐2814. 10.1002/elps.201800098 [DOI] [PubMed] [Google Scholar]
- Barbarotto, E. , Schmittgen, T. D. , & Calin, G. A. (2008). MicroRNAs and cancer: Profile, profile, profile. International Journal of Cancer, 122(5), 969–977. 10.1002/ijc.23343 [DOI] [PubMed] [Google Scholar]
- Bennett, J. M. , Catovsky, D. , Daniel, M. T. , Flandrin, G. , Galton, D. A. , Gralnick, H. R. , & Sultan, C. (2010). Proposals for the classification of the myelodysplastic syndromes. British Journal of Haematology, 51(2), 189–199. 10.1111/j.1365-2141.1982.tb08475.x [DOI] [PubMed] [Google Scholar]
- Bhise, N. S. , Chauhan, L. , Shin, M. , Cao, X. , Pounds, S. , Lamba, V. , & Lamba, J. K. (2016). MicroRNA‐mRNA pairs associated with outcome in AML: From in vitro cell‐based studies to AML patients. Frontiers in Pharmacology, 6(e2141), 324 10.3389/fphar.2015.00324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G. A. , & Croce, C. M. (2006). MicroRNA signatures in human cancers. Nature Reviews Cancer, 6(11), 857–866. 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- Carella, A. M. (2015). Clinical outcome after failure of hypomethylating therapy for myelodysplastic syndromes. European Journal of Haematology, 94(6), 546–553. 10.1111/ejh.12521 [DOI] [PubMed] [Google Scholar]
- Cattaneo, M. , Pelosi, E. , Castelli, G. , Cerio, A. M. , D′angiò, A. , Porretti, L. , … Biunno, I. (2015). A miRNA signature in human cord blood stem and progenitor cells as potential biomarker of specific acute myeloid leukemia subtypes. Journal of Cellular Physiology, 230(8), 1770 10.1002/jcp.24876 [DOI] [PubMed] [Google Scholar]
- Chen, L. T. , Xu, S. D. , Xu, H. , Zhang, J. F. , Ning, J. F. , & Wang, S. F. (2012). MicroRNA‐378 is associated with non‐small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Medical Oncology, 29(3), 1673–1680. 10.1007/s12032-011-0083-x [DOI] [PubMed] [Google Scholar]
- Deng, H. , Guo, Y. , Song, H. , Xiao, B. , Sun, W. , Liu, Z. , … Guo, J. (2013). MicroRNA‐195 and microRNA‐378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene, 518(2), 351–359. 10.1016/j.gene.2012.12.103 [DOI] [PubMed] [Google Scholar]
- Eitan, R. , Kushnir, M. , Lithwick‐Yanai, G. , David, M. B. , Hoshen, M. , Glezerman, M. , … Levavi, H. (2009). Tumor microRNA expression patterns associated with resistance to platinum based chemotherapy and survival in ovarian cancer patients. Gynecologic Oncology, 114(2), 253–259. 10.1016/j.ygyno.2009.04.024 [DOI] [PubMed] [Google Scholar]
- Erdogan, B. , Facey, C. , Qualtieri, J. , Tedesco, J. , Rinker, E. , Isett, R. B. , … Kim, A. S. (2011). Diagnostic microRNAs in myelodysplastic syndrome. Experimental Hematology, 39(9), 915–926.e912. 10.1016/j.exphem.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Esquela‐Kerscher, A. , & Slack, F. J. . (2006). Oncomirs‐microRNAs with a role in cancer. Nature Reviews Cancer, 6(4), 259–269. 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- Faltejskova, P. , Svoboda, M. , Srutova, K. , Mlcochova, J. , Besse, A. , Nekvindova, J. , … Slaby, O. (2012). Identification and functional screening of microRNAs highly deregulated in colorectal cancer. Journal of Cellular & Molecular Medicine, 16(11), 2655–2666. 10.1111/j.1582-4934.2012.01579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattore, L. , Sacconi, A. , Mancini, R. , & Ciliberto, G. (2017). MicroRNA‐driven deregulation of cytokine expression helps development of drug resistance in metastatic melanoma. Cytokine & Growth Factor Reviews, 36, 39–48. 10.1016/j.cytogfr.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Gill, H. , Leung, A. Y. H. , & Kwong, Y. L. (2016). Molecular and cellular mechanisms of myelodysplastic syndrome: Implications on targeted therapy. International Journal of Molecular Sciences, 17(4), 440 10.3390/ijms17040440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, P. , Cox, C. , LeBeau, M. M. , Fenaux, P. , Morel, P. , Sanz, G. , … Bennett, J. (1997). International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood, 89(6), 2079–2088. 10.1182/blood.V89.6.2079 [DOI] [PubMed] [Google Scholar]
- Ipn, B. , Ng, C. C. , Baharuddin, P. , & Zakaria, Z. (2017). MicroRNA expression patterns and target prediction in multiple myeloma development and malignancy. Genes & Genomics, 39(5), 1–8. 10.1007/s13258-017-0518-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa, J. P. (2013). The myelodysplastic syndrome as a prototypical epigenetic disease. Blood, 121(19), 3811–3817. 10.1182/blood-2013-02-451757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzykson, R. , & Fenaux, P. (2014). Epigenetics of myelodysplastic syndromes. Leukemia, 28(3), 497–506. 10.1038/leu.2013.343 [DOI] [PubMed] [Google Scholar]
- Lee, D. Y. , Deng, Z. , Wang, C. H. , & Yang, B. B. (2007). MicroRNA‐378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus‐1 expression. Proceedings of the National Academy of Sciences of the United States of America, 104(51), 20350 10.1073/pnas.0706901104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. J. , Podoltsev, N. , Gore, S. D. , & Zeidan, A. M. (2015). The evolving field of prognostication and risk stratification in MDS: Recent developments and future directions. Blood Reviews, 30(1), 1–10. 10.1016/j.blre.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Li, Z. Z. , Shen, L. F. , Li, Y. Y. , Chen, P. , & Chen, L. Z. (2016). Clinical utility of microRNA‐378 as early diagnostic biomarker of human cancers: A meta‐analysis of diagnostic test. Oncotarget, 7(36), 58569–58578. 10.18632/oncotarget.10707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. , Yao, D.‐M. , Qian, J. , Chen, Q. , Qian, W. , Li, Y. , … Xu, W.‐R. (2011). Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE, 6(10), e26906 10.1371/journal.pone.0026906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. , Yao, D.‐M. , Qian, J. , Chen, Q. , Qian, W. , Li, Y. , … Xu, W.‐R. (2012). IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Annals of Hematology, 91(4), 519–525. 10.1007/s00277-011-1352-7 [DOI] [PubMed] [Google Scholar]
- Ma, J. , Lin, J. , Qian, J. , Qian, W. , Yin, J. , Yang, B. , … Deng, Z. (2014). MiR‐378 promotes the migration of liver cancer cells by down‐regulating Fus expression. Cellular Physiology & Biochemistry, 34(6), 2266–2274. 10.1159/000369669 [DOI] [PubMed] [Google Scholar]
- Merkerova, M. D. , Krejcik, Z. , Votavova, H. , Belickova, M. , Vasikova, A. , & Cermak, J. (2011). Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. European Journal of Human Genetics, 19(3), 313 10.1038/ejhg.2010.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloso, M. S. , Spizzo, R. , Shimizu, M. , Rossi, S. , & Calin, G. A. (2009). MicroRNAs–the micro steering wheel of tumour metastases. Nature Reviews Cancer, 9(4), 293–302. 10.1038/nrc2619 [DOI] [PubMed] [Google Scholar]
- Pons, A. , Nomdedeu, B. , Navarro, A. , Gaya, A. , Gel, B. , Diaz, T. , … Monzo, M. (2009). Hematopoiesis‐related microRNA expression in myelodysplastic syndromes. Leukaemia & Lymphoma, 50(11), 1854–1859. 10.3109/10428190903147645 [DOI] [PubMed] [Google Scholar]
- Qian, J. , Lin, J. , Qian, W. , Ma, J.‐C. , Qian, S.‐X. , Li, Y. , … Deng, Z.‐Q. (2013). Overexpression of miR‐378 is frequent and may affect treatment outcomes in patients with acute myeloid leukemia. Leukemia Research, 37(7), 765–768. 10.1016/j.leukres.2013.03.014 [DOI] [PubMed] [Google Scholar]
- Qian, J. , Yao, D.‐M. , Lin, J. , Qian, W. , Wang, C.‐Z. , Chai, H.‐Y. , … Chen, X.‐X. (2012). U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS ONE, 7(9), e45760 10.1371/journal.pone.0045760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, J. , Zhu, Z. H. , Lin, J. , Ming, Y. D. , Li, Y. , Yang, J. , & Wang, C. Z. (2011). Hypomethylation of PRAME promoter is associated with poor prognosis in myelodysplastic syndrome. British Journal of Haematology, 154(1), 153–155. 10.1111/j.1365-2141.2011.08585.x [DOI] [PubMed] [Google Scholar]
- Schmittgen, T. D. (2008). Regulation of microRNA processing in development, differentiation and cancer. Journal of Cellular & Molecular Medicine, 12(5b), 1811–1819. 10.1111/j.1582-4934.2008.00483.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek, K. , Tertil, M. , Golda, S. , Ciesla, M. , Weglarczyk, K. , Collet, G. , … Dulak, J. (2013). Interplay between heme oxygenase‐1 and miR‐378 affects non‐small cell lung carcinoma growth, vascularization, and metastasis. Antioxidants & Redox Signaling, 19(7), 644–660. 10.1089/ars.2013.5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaprouni, L. G. , Yang, T.‐P. , Bell, J. , Dick, K. J. , Kanoni, S. , Nisbet, J. , … Deloukas, P. (2014). Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics, 9(10), 1382–1396. 10.4161/15592294.2014.969637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, P. , Horny, H.‐P. , Bennett, J. M. , Fonatsch, C. , Germing, U. , Greenberg, P. , … Wells, D. A. (2007). Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leukemia Research, 31(6), 727–736. 10.1016/j.leukres.2006.11.009 [DOI] [PubMed] [Google Scholar]
- Vardiman, J. W. , Thiele, J. , Arber, D. A. , Brunning, R. D. , Borowitz, M. J. , Porwit, A. , … Bloomfield, C. D. (2009). The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood, 114(5), 937–951. 10.1182/blood-2009-03-209262 [DOI] [PubMed] [Google Scholar]
- Ventura, A. , & Jacks, T. (2009). MicroRNAs and cancer: Short RNAs go a long way. Cell, 136(4), 586–591. 10.1016/j.cell.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votavova, H. , Grmanova, M. , Dostalova, M. M. , Belickova, M. , Vasikova, A. , Neuwirtova, R. , & Cermak, J. (2011). Differential expression of microRNAs in CD34+ cells of 5q‐ syndrome. Journal of Hematology & Oncology, 4(1), 1 10.1186/1756-8722-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, J. , Jung, S. , Keller, S. , Gregory, R. I. , & Diederichs, S. (2009). Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nature Cell Biology, 11(3), 228 10.1038/ncb0309-228 [DOI] [PubMed] [Google Scholar]
- Xiao‐Wen, Z. , Xiang‐Mei, W. , Ying‐Ying, Z. , Lei, Y. , Hong, G. , Jing, Y. , … Jiang, L. (2015). The 5' flanking region of miR‐378 is hypomethylated in acute myeloid leukemia. International Journal of Clinical and Experimental Pathology, 8(5), 4321–4331. [PMC free article] [PubMed] [Google Scholar]
- Xu, L. H. , Guo, Y. , Zhang, X. L. , Chen, J. J. , & Hu, S. Y. (2016). Blood‐based circulating microRNAs are potential diagnostic biomarkers for leukemia: Result from a meta‐analysis. Cellular Physiology and Biochemistry, 38(3), 939 10.1159/000443046 [DOI] [PubMed] [Google Scholar]
- Yang, J. , Qian, J. , Lin, J. , Yang, X. F. , Qian, W. , Chen, Q. , … Xiao, G. F. (2013). Development of a high‐resolution melting analysis for the detection of the SF3B1 mutations. Genetic Testing and Molecular Biomarkers, 17(4), 342–347. 10.1089/gtmb.2012.0364 [DOI] [PubMed] [Google Scholar]
- Yu, B.‐L. , Peng, X.‐H. , Zhao, F.‐P. , Liu, X. , Lu, J. , Wang, L. U. , … Li, X.‐P. (2014). MicroRNA‐378 functions as an onco‐miR in nasopharyngeal carcinoma by repressing TOB2 expression. International Journal of Oncology, 44(4), 1215 10.3892/ijo.2014.2283 [DOI] [PubMed] [Google Scholar]


