Abstract
Invasive candidiasis (IC) is the most common nosocomial infection and a leading cause of mycoses-related deaths. High-systemic toxicity and emergence of antifungal-resistant species warrant the development of newer preventive approaches against IC. Here, we have adopted an immunotherapeutic peptide vaccine-based approach, to enhance the body’s immune response against invasive candida infections. Using computational tools, we screened the entire candida proteome (6030 proteins) and identified the most immunodominant HLA class I, HLA class II and B- cell epitopes. By further immunoinformatic analyses for enhanced vaccine efficacy, we selected the 18- most promising epitopes, which were joined together using molecular linkers to create a multivalent recombinant protein against Candida albicans (mvPC). To increase mvPC’s immunogenicity, we added a synthetic adjuvant (RS09) to the mvPC design. The selected mvPC epitopes are homologous against all currently available annotated reference sequences of 22 C. albicans strains, thus offering a higher coverage and greater protective response. A major advantage of the current vaccine approach is mvPC’s multivalent nature (recognizing multiple-epitopes), which is likely to provide enhanced protection against complex candida antigens. Here, we describe the computational analyses leading to mvPC design.
Subject terms: Protein analysis, Peptide vaccines
Introduction
Invasive candidiasis (IC) is one of the most common public health problems and is a major therapeutic challenge1. Increasing frequency of the pre-disposing risk factors has led to a remarkable 20-fold higher incidence of IC in just two decades2. However, unlike other fungal infections, IC is usually due to an endogenous candida overgrowth at mucosal surfaces (mucosal candidiasis)3. Candida albicans (C. albicans) is the most common etiological agent of IC and is found in ~ 60% of clinical isolates of candidiasis4. In healthy individuals, C. albicans co-exists with the host in a harmless commensal (yeast) form without causing disease5. However, certain underlying conditions (e.g., major surgery, broad-spectrum antibacterial therapy, immunodeficiency diseases [AIDS, diabetes, cancer chemotherapy, etc.], or even premature birth) can cause candida to become pathogenic6. The pathogenesis of candida requires a switch from its commensal yeast form to a pathogenic fungal (hyphal) form5. The presence of hyphae enables candida to cause widespread damage to the underlying tissues. If the infection remains unchecked, candida can gain access to the host vasculature5. Once candida enters the bloodstream, it can spread throughout the body, causing life-threatening IC with a high mortality rate of 60%7. The current treatment with a limited set of available drugs leads to high drug-induced systemic toxicity and is associated with an increased emergence of anti-fungal-resistant candida species2. Therefore, despite the progress in medicine in general, IC is a leading cause of mycoses-related deaths8.
Immunosuppression is the leading cause of fatal, invasive candida infections. In healthy individuals, the ability of candida to alter its morphology is largely kept in check by an effective immune response9. The significance of an effective immune response in preventing IC, suggests that mimicking a natural immune response to candida may be an effective strategy to control its burden. In line with this, vaccination approaches against IC include PEV710 and NDV311, which have completed Phase I studies and are in further stages of immunogenicity and toxicity testing. PEV7 is a virosomal vaccine to protect women suffering from chronic vaginal yeast infections (vulvovaginal candidiasis or VVC). It consists of a truncated recombinant secreted aspartic protease 2 (Sap2), currently in clinical testing by Pevion BiotechAG10. NDV3 targets the recombinant N-terminal region of the hyphal protein agglutinin-like sequence three protein (rAls3p-N) and is being developed by NovaDigm Therapeutics12. Another vaccination approach from Novartis Pharmaceuticals (Efungumab) based on monoclonal antibody targeting the heat shock protein 90 (Hsp90) progressed through to a Phase III clinical trial but was abandoned in later stages of development due to safety concerns13. Finally, a prophylactic and therapeutic IgM-monoclonal antibody (MAb B6.1) by LigoCyte Pharmaceuticals that targeted (1 → 2)-β-mannotriose also failed during development14. Thus, there are no FDA-approved candida vaccines for human use.
The failure of current vaccination approaches in eliciting an effective anti-candida immune response is attributed to a variety of reasons. One of the major drawbacks is candida’s ability to evolve and ultimately escape the host immune surveillance15. Therefore, simultaneous targeting against multiple candida epitopes (multivalence) is expected to provide improved outcomes. So far, the multivalent-vaccine approach has not been adopted against C. albicans. Due to the complex nature of candida antigens and its ability to escape host-immune surveillance, a multi-epitope vaccine will likely be more beneficial in inducing a stronger and broader immune responses15,16. While most vaccines (to date) focused only on one antigen, our approach involves the simultaneous targeting of multiple candida antigens which are molecularly linked to form a single recombinant protein. Our strategy involves experimental validation of single peptide antigens whereby positive peptide epitopes can be combined to design a multivalent recombinant protein vaccine against C. albicans. The findings presented in the current study detail in silico epitope mapping and provide future directions for vaccine design against C. albicans.
Methods
Antigenicity prediction
C. albicans sc5314 (the most common clinical isolate)17 was used as a reference strain to retrieve its entire proteomic sequences (consisting of 6030 proteins) from the NCBI protein database. Next, each of these proteins fasta sequences was run on VaxiJen server, which utilizes an alignment-independent method based on principal amino acid properties18. Based on the published literature19, an antigenicity probability >0.9 was considered acceptable for subunit vaccines. The proteins were filtered based on their antigenicity score (>0.9) and subcellular localization (extracellular, plasma membrane or nuclear), using a web server (CELLO2GO20) for protein subcellular localization prediction, which shortlisted the number of proteins to 36 (Supplementary file 1, yellow and green highlight). Since our goal is to stop candida’s switch to the pathogenic fungal form (without affecting its commensalism), we selected five of the 36 proteins (Als4p, Als3p, Fav2p, Als2p, Eap1p) (Supplementary file 1, green highlight) with known functions in hyphae formation. Further, we included three more hyphal proteins (Hyr1p, Hwp1p, Sap2p) (Supplementary file 1, blue highlight) from published studies21. The antigenicity score of these proteins was slightly below our cutoff (0.9), but due to their role in candida hyphae formation, we selected them for further development (Table 1).
Table 1.
Most-antigenic proteins in C. albicans proteome.
| Accession Number | Vaxijen Analyses | Subcellular Localization |
|---|---|---|
| XP_710425.2 | Als4p [Candida albicans SC5314] Overall Antigen Prediction = 1.2231 (Probable ANTIGEN) | *Exc/PM/Nu |
| XP_710435.2 | Als3p [Candida albicans SC5314] Overall Antigen Prediction = 0.9490 (Probable ANTIGEN) | Exc/PM/Nu |
| XP_711172.1 | Fav2p [Candida albicans SC5314] Overall Antigen Prediction = 0.9661 (Probable ANTIGEN) | Exc/Nu |
| XP_712646.2 | Als2p [Candida albicans SC5314] Overall Antigen Prediction = 1.2323 (Probable ANTIGEN) | Exc/PM/Nu |
| XP_714572.2 | Eap1p [Candida albicans SC5314] Overall Antigen Prediction = 1.0124 (Probable ANTIGEN) | Exc/Nu |
| XP_722183.2 | Hyr1p [Candida albicans SC5314] Overall Antigen Prediction = 0.8286 (Probable ANTIGEN) | PM |
| XP_709961.2 | Hwp1p [Candida albicans SC5314] Overall Antigen Prediction = 0.7474 (Probable ANTIGEN) | PM |
| XP_711047.1 | Sap2p [Candida albicans SC5314] Overall Antigen Prediction = 0.7035 (Probable ANTIGEN) | Exc |
*Extracellular/Plasma membrane/Nuclear.
Epitope mapping
Using a NetCTL server22, we screened for the most antigenic HLA class I epitopes. HLA class I alleles are sub-grouped into 12 superfamilies (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, B62). We screened each of the eight-hyphal proteins against each of the HLA class I superfamily (a total of 8 × 12 = 96 queries). The threshold values used were (HLAI binding [epitope identification] >0.75, weight on proteasomal C- terminal cleavage = 0.15; and weight on TAP [transport efficiency] = 0.05)22 (data not shown). For HLA class II epitope binding, we used the IEDB prediction server23, with low percentile rank and IC50 value as the selection criteria. Based on the spatial structure of the epitopes, the B-cell epitope structure can be categorized as continuous (linear) and discontinuous (conformational) epitopes24. For identification of linear B-cell epitopes, we used the BCPreds server25, while discontinuous B-cell epitope analyses were done using the IEDB prediction server23. B-cell epitopes were 20mer in length. Supplementary file 2 lists 240 selected HLA class I, HLA class II and B-cell epitopes in each of the 8 selected hyphal proteins. Next, to eliminate the possibility of overlapping epitopes we ran the 240 selected epitopes on the IEDB cluster analysis algorithm and filtered the epitopes for sequence identity (Supplementary file 2). This reduced the number of epitopes to a total of 214. Binding predictions for 10 conserved HLA class II T- cell epitopes (Table 2) and phenotype frequency of HLA allele type was analyzed using IEDB prediction server. IEDB recommended consensus percentile rank of the top 10% was taken for making selections (Supplementary file 3).
Table 2.
Selected HLA class I (9mer), HLA class II (15mer) and B- cell (20mer) epitopes.
| EPITOPE | PROTEIN |
|---|---|
| SNGLNDWNYPISSES | XP_710425.2 Als4p |
| LVTYQNVPAGYRPFV | XP_710425.2 Als4p |
| MLLQFLLLSLCVSVA | XP_712646.2 Als2p |
| RILLSRILPSLSQAV | XP_710425.2 Als4p |
| MLQQYTLLLIYLSVA | XP_710435.2 Als3p |
| PSLNKVSTLFVAPQC | XP_710435.2 Als3p |
| HVGITKGLNDWNYPV | XP_710435.2 Als3p |
| MFLKNIFIALAIALL | XP_711047.1 Sap2p, XP_718053.2 Sap1p, XP_723210.1 Sap3p |
| NNMRLTFGAAIIGIA | XP_709961.2 Hwp1p |
| ISTFEGAGNNMRLTF | XP_709961.2 Hwp1p |
| PYDKCQLLF | XP_711047.1 Sap2p |
| TVTAPPGGTDTVIIREPPNP | XP_710425.2 Als4p, XP_712646.2 Als2p, XP_718077.1 Als1p |
| KTNEAGGSYDNVPVTLKKQG | XP_711047.1 Sap2p |
| NSPDAATGQIIFGGVDNAKY | XP_711047.1 Sap2p |
| VVKTPKAFPVTNGQEGKTSK | XP_711047.1 Sap2p |
| EFAASLQGDDGQPYDKCQLL | XP_711047.1 Sap2p |
| ADFCKQKGTYDPSGSSASQD | XP_711047.1 Sap2p |
| LDGSASTTATVTPSLTDLQA | XP_711172.1 Fav2p |
Conservation analyses
Further, we did the conservation analysis of 214 epitopes with 22 C. albicans strains with an annotated sequence available on NCBI. The selection of epitopes in the conserved regions of the C. albicans sequence will ensure higher coverage and protection against candidiasis. Using this analysis tool, we found 18 epitopes showing a 100% conservancy (Table 2). Vaccine designing was done using these 18 epitopes.
Peptide fusion
To ease any potential issues with the delivery of 18 single peptides, we constructed a fusion protein using molecular linker peptides. For example, intra Tc and Th epitopes were joined by AAY and GPGPG linker, respectively (Fig. 1). We also incorporated a TLR4 agonist RS09 (APPHALS) at the N- terminal end of the final vaccine construct. Molecular linking of 18- single candida epitopes will facilitate better in vivo antigen processing and presentation. The choice of linker peptides and the adjuvant was based on the study published by Pandey et al.19. RS09 mimics lipopolysaccharide (LPS), a natural TLR4 ligand26. Thus, the presence of RS09 enables co-stimulation of TCRs, driving a more robust immune activation. Use of synthetic adjuvants (RS09) is a safer approach and considered an advancement over traditional vaccination approaches, such as using Freund’s adjuvant27. The Tc epitope and adjuvant were joined by the EAAAK linker sequences (Fig. 1). Next, we performed the secondary structure analyses of the final mvPC vaccine construct using the RaptorX server28,29. Our initial analyses showed ~10% protein disorder. To further enhance protein stability, we identified the region of disorders and removed the peptide (GPGPGKTNEAGGSYDNVPVTLKKQG) (not shown in Fig. 1). Further rearrangements led to a final 349aa-long mvPC vaccine with 1% protein disorder (Fig. 2).
Figure 1.
mvPC design showing peptide fusion by molecular linkers (red) and synthetic RS09 adjuvant (green).
Figure 2.
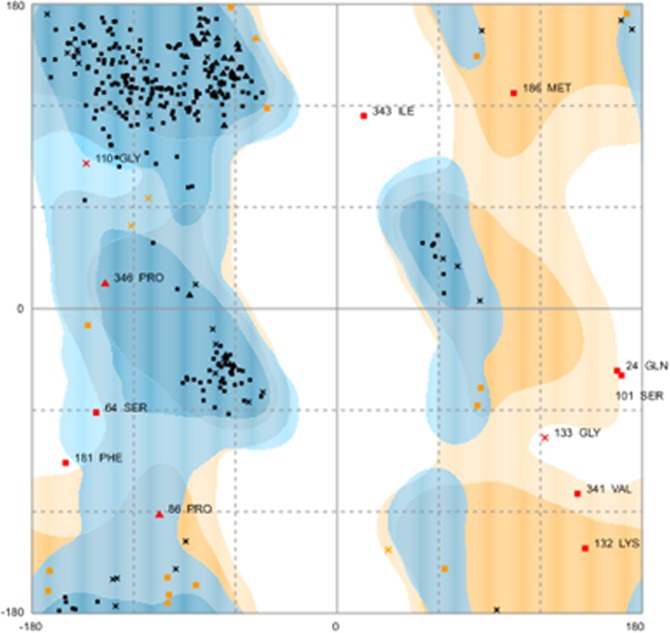
Ramachandran plot showing mvPC stability - Number of Residues in favored region 317 (91.4%); allowed region 18 (5.2%); and outlier region 12 (3.5%).
Sequence validation
Since fusing several epitopes can change the 3D- spatial arrangement of epitopes, we decided to validate T- cell (HLA class I and HLA class II) and B- cell (linear and discontinuous epitopes) in the final protein sequence. We found 100% conservation of 9mer (HLA class I), 15mer (HLA class II) and 20mer (B-cell) epitopes. We also found that the selected epitopes show 80–95% conservancy with other candida species (C. dubliniensis, C. parapsilosis, and C. orthopsilosis) (data not shown).
Biophysiochemical analyses
The biophysiochemical characteristics of the final mvPC vaccine were evaluated on the ProtParam server30,31. A predicted molecular weight of 36.3 kDa favored the antigenicity of the vaccine construct. mvPC’s isoelectric point (pI) is 6.14, suggesting that it is near neutral pH. The estimated half-life in mammalian reticulocytes is predicted to be ~4.4 h. The instability index (II) of the final mvPC vaccine is computed to be 33.79. Based on these analyses, mvPC vaccine is stable.
Antigenicity and allergenicity analyses
The efficacy of any vaccine is primarily determined by its antigenicity (i.e., potential to trigger an immune response). The antigenicity of the final mvPC vaccine was tested using ANTIGENpro32 and VaxiJen33,34 and was found to be 0.87 and 0.80, respectively. These values of antigenicity of the final mvPC vaccine are acceptable and comparable to other published subunit vaccines19. Elicitation of an allergic response will be unacceptable to vaccine administration; therefore, to rule out any potential allergic response, allergenicity was tested using the AllerTOP35 server, and the mvPC vaccine was found to be non-allergic. We also performed proteasome cleavage analysis of final mvPC construct using NetChop3.1 and MHCII-NP on IEDB server. The final mvPC was cleaved by proteasomes to generate the predicted T- cell epitopes identified in Table 2 (Supplementary file 4).
mvPC tertiary structure analyses
To visualize the tertiary structure of the final mvPC vaccine, we used a template-based tertiary structure prediction algorithm, RaptorX28,29. Given the input sequence, RaptorX predicts its secondary and tertiary structures, as well as solvent accessibility and disordered regions. Using RaptorX, mvPC was found to be a single-domain stable protein. RaptorX used 2y7lA (top-ranked template) for mvPC structure prediction. A p-value of 5.67e-10 confirms the confidence in the mvPC tertiary structure prediction. Overall uGDT was found to be 106 (uGDT >50 is considered a good prediction). In the final structure, 7 (2%) positions were predicted to be disordered. Secondary structures in protein are 6%H, 40%E, and 53%C, while solvent access probability is 29%E, 39%M, and 30%B.
The Rampage server was used to identify the tertiary structure stability prediction of the final mvPC construct. The number of residues in the favored region was 310 (89.3%); the number of residues in the allowed region was 19 (5.5%); and the number of residues in the outlier region was 18 (5.2%) (data not shown). We further refined the final 3D- structure using the Galaxy refine server and found improvement in the favored region. The number of residues in the favored region was 317 (91.4%); the number of residues in the allowed region was 18 (5.2%); and the number of residues in the outlier region was 12 (3.5%) (Fig. 2). We finally decided to work with the refined model and generated a 3D- structure using Galaxy refine server (Fig. 3). We performed aggregation analysis of unrefined 3D- structure of mvPC using Aggrescan3.0 in dynamic mode. The average A3D score of input structure is 0.036 and minimum energy of a model_7 in dynamic mode is −0.0742 (Supplementary file 4).
Figure 3.
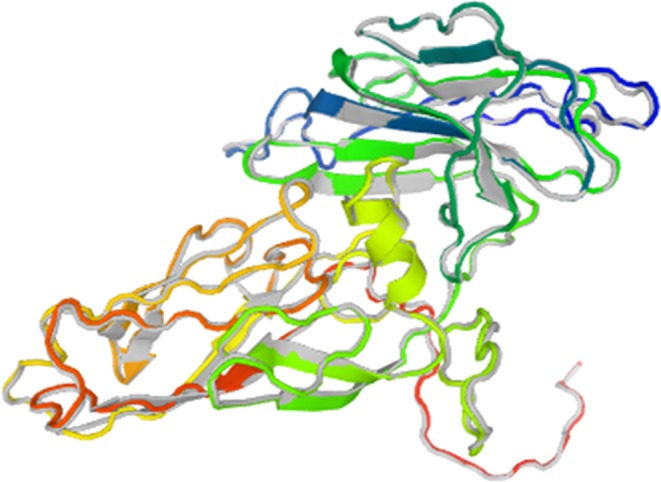
mvPC 3D- structure generated using Galaxy refine server.
Results and Discussion
High drug-toxicity and emergence of drug-resistant candida species has led to a wide-consensus on the development of immunotherapeutic approaches against IC36–39. However, the development of an effective vaccine molecule using conventional approaches involves inoculation of live-attenuated or inactivated pathogen, requiring laborious biochemical, immunological, and microbiological methods to identify the antigenic components40. Moreover, these approaches are limited in application. For example, a large number of patients with immunosuppressive conditions (such as cancer, solid organ or hematopoietic stem cell transplant, HIV, acquired or congenital immunodeficiency)6,41 are at a higher-risk for systemic candida infection and require alternate approaches to immunization.
Recent advances in genomic and proteomic approaches have revealed the power of computational tools to design effective and safe new-generation vaccines42,43. The success of immunoinformatic approaches in vaccinology was first demonstrated in 2013 with the licensing of Bexsero and Trumenba, protein-based vaccines against serotype B meningococcal vaccines44,45. Since then, these approaches have been used successfully in the design of subunit vaccines against some of the most infectious and difficult pathogens, including, Streptococcus pneumoniae (pneumonia)46, Escherichia coli47, Clostridium difficile48, Chlamydia trachomatis49, human cytomegalovirus (HCMV), respiratory syncytial virus (RSV), HIV, influenza and dengue viruses50, which demonstrate the significance of immunoinformatic approaches in facilitating the vaccine development process.
The present study represents the first proteome-wide immunoinformatic approach to identify the immunodominant epitopes and design a multivalent subunit vaccine against C. albicans. Using web-based servers, we screened the entire candida proteome (consisting of 6030 proteins), to identify the most immunodominant candida antigens. Of note, our immunogenicity analyses focused on identifying epitopes in the hyphal proteins, which help candida adhere to and invade epithelial cells, resulting in severe damage to the host cells5. Thus, targeting hyphae will preserve candida’s yeast form without affecting host-fungal commensalism. Eight antigenic proteins with known functions in hyphal formation (Als4p, Als3p, Fav2p, Als2p, Eap1p, Hyr1p, Hwp1p, Sap2p) were selected further for epitope mapping (Table 1). Immunogenicity testing led to the selection of 18 unique epitopes [10 CD4+ T- helper or Th (15mer), 7 B- cell (20mer) and 1 CD8+ T- cytotoxic or Tc (9mer)]. The rationale for selecting these epitopes lies in the significance of Th cells in recognizing HLA class II proteins and in turn activating both B- cells to secrete antibodies and activating Tc cells to kill infected target cells51. Antibody response to C. albicans is ideal to target carbohydrate moieties on the fungal cell wall, as well as some secreted proteins (secreted aspartyl proteinase or SAP52) and has been shown to be effective in providing resistance against IC53. Although Tc play a minor role in natural immunological defense against candida and have not been studied much, previous studies show that Tc are effective in controlling fungal infection post-vaccination54–56. Therefore, a combination of strong humoral and cell-mediated immune responses is likely to confer an effective immune response against pathogenic candida. Conservancy analysis also show that the selected epitopes (in addition to the eight hyphal proteins), are also present in other candida proteins showing sequence homology (Sap1p, Sap 3p and Als1p, Table 2). Further, while designing the mvPC vaccine we have not only identified immunodominant epitopes in the antigenic proteins of the reference strain but also checked their conservancy across all known 22 strains of C. albicans whose proteome sequences are available in NCBI. Our selected antigens are in the highly-conserved regions of the candida proteome, offering protection against the 22 currently known candida strains. Selection of conserved candida epitopes also minimizes the chances of mutation in all the conserved regions simultaneously, limiting the emergence of new-resistant candida species. The selected epitopes also show conservation (a.k.a immunoprotection) with other pathogenic candida species (C. dubliniensis, C. parapsilosis, and C. orthopsilosis). Lastly, selected epitopes show HLA binding affinity in all 27 reference alleles of HLA class II with a number of epitopes demonstrating binding ability with multiple HLA subtypes. The presence of RS09 (via activation of the TLR4 signalling pathway) is expected to boost anti-candida immune response by skewing Th response towards the Th17 subtype which plays a major role in mounting an immune response in clearance of pathogenic C. albicans57,58. RS09 has previously been shown to provide better adjuvanticity with fewer side effects in HIV-159.
It is important to note here that the efficacy of peptide vaccine is also largely dependent on the HLA type of the individual. A functional response will only be generated in individuals with a particular HLA type capable of binding a particular peptide epitope. Studies are underway to assess immunogenicity using single peptides in HLA-matched PBMC samples. Experimental testing of individual single peptides prior to linking them in the recombinant mvPC protein will eliminate the non-immunogenic intervening sequences. Collectively, the mvPC design demonstrates our goal to induce an effective immune response using a minimal well-defined antigen.
Supplementary information
Acknowledgements
The authors acknowledge the Creighton University School of Dentistry (Omaha, NE) and the University of Nebraska Medical Center (Omaha, NE) for supporting the conduct of this research. This work was supported in part by a grant from the Park Dental Foundation (S.M.R.S.).
Author contributions
S.T. and V.K. conceptualized the design of work and performed the data analyses and interpretation. S.T. wrote the first draft of the manuscript. V.K., S.S., M.W., B.L. and L.L. discussed the results, did a critical revision of the article and contributed to the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shikha Tarang and Varun Kesherwani.
Supplementary information
is available for this paper at 10.1038/s41598-020-57906-x.
References
- 1.Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers. 2018;4:18026. doi: 10.1038/nrdp.2018.26. [DOI] [PubMed] [Google Scholar]
- 2.Pierce CG, Lopez-Ribot JL. Candidiasis drug discovery and development: New approaches targeting virulence for discovering and identifying new drugs. Expert Opin Drug Discov. 2013;8:1117–1126. doi: 10.1517/17460441.2013.807245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vazquez JA, Sobel JD. Mucosal candidiasis. Infect Dis Clin North Am. 2002;16:793–820. doi: 10.1016/S0891-5520(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 4.Mora Carpio, A. L. & Climaco, A. Candidiasis, fungemia. (StatPearls Publishing LLC, 2019).
- 5.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yapar N. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag. 2014;10:95–105. doi: 10.2147/TCRM.S40160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano R, Sakamoto Y, Kudo K, Ohnishi M. Retrospective analysis of mortality and candida isolates of 75 patients with candidemia: A single hospital experience. Infect Drug Resist. 2015;8:199–205. doi: 10.2147/IDR.S80677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaller MA, Diekema DJ. Rare and emerging opportunistic fungal pathogens: Concern for resistance beyond candida albicans and aspergillus fumigatus. J Clin Microbiol. 2004;42:4419–4431. doi: 10.1128/JCM.42.10.4419-4431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Silva Dantas A, et al. Cell biology of candida albicans-host interactions. Curr Opin Microbiol. 2016;34:111–118. doi: 10.1016/j.mib.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Bernardis F, et al. A virosomal vaccine against candidal vaginitis: Immunogenicity, efficacy and safety profile in animal models. Vaccine. 2012;30:4490–4498. doi: 10.1016/j.vaccine.2012.04.069. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt CS, et al. NDV-3, a recombinant alum-adjuvanted vaccine for candida and staphylococcus aureus, is safe and immunogenic in healthy adults. Vaccine. 2012;30:7594–7600. doi: 10.1016/j.vaccine.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim AS, et al. NDV-3 protects mice from vulvovaginal candidiasis through T- and B-cell immune response. Vaccine. 2013;31:5549–5556. doi: 10.1016/j.vaccine.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pachl J, et al. A randomized, blinded, multicenter trial of lipid-associated amphotericin B alone versus in combination with an antibody-based inhibitor of heat shock protein 90 in patients with invasive candidiasis. Clin Infect Dis. 2006;42:1404–1413. doi: 10.1086/503428. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Jang EC, Han Y. Combination immunotherapy of MAb B6.1 with fluconazole augments therapeutic effect to disseminated candidiasis. Arch Pharm Res. 2011;34:399–405. doi: 10.1007/s12272-011-0307-9. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez-Lopez C, Lorenz MC. Fungal immune evasion in a model host-pathogen interaction: Candida albicans versus macrophages. PLoS Pathog. 2013;9:e1003741. doi: 10.1371/journal.ppat.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassone A. Development of vaccines for candida albicans: Fighting a skilled transformer. Nat Rev Microbiol. 2013;11:884–891. doi: 10.1038/nrmicro3156. [DOI] [PubMed] [Google Scholar]
- 17.Sampaio P, et al. Virulence attenuation of candida albicans genetic variants isolated from a patient with a recurrent bloodstream infection. PLoS One. 2010;5:e10155. doi: 10.1371/journal.pone.0010155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doytchinova IA, Flower DR. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandey RK, Bhatt TK, Prajapati VK. Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating anopheles salivary protein. Sci Rep. 2018;8:1125. doi: 10.1038/s41598-018-19456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu CS, et al. CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS One. 2014;9:e99368. doi: 10.1371/journal.pone.0099368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaffin WL, Lopez-Ribot JL, Casanova M, Gozalbo D, Martinez JP. Cell wall and secreted proteins of candida albicans: Identification, function, and expression. Microbiol Mol Biol Rev. 1998;62:130–180. doi: 10.1128/MMBR.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen MV, et al. An integrative approach to CTL epitope prediction: A combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35:2295–2303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 23.Andreatta M, et al. An automated benchmarking platform for MHC class II binding prediction methods. Bioinformatics. 2018;34:1522–1528. doi: 10.1093/bioinformatics/btx820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potocnakova L, Bhide M, Pulzova LB. An introduction to B-cell epitope mapping and in silico epitope prediction. J Immunol Res. 2016;2016:6760830. doi: 10.1155/2016/6760830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Manzalawy Y, Dobbs D, Honavar V. Predicting flexible length linear B-cell epitopes. Comput Syst Bioinformatics Conf. 2008;7:121–132. doi: 10.1142/9781848162648_0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shanmugam A, et al. Synthetic toll like receptor-4 (TLR-4) agonist peptides as a novel class of adjuvants. PLoS One. 2012;7:e30839. doi: 10.1371/journal.pone.0030839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coler RN, et al. A synthetic adjuvant to enhance and expand immune responses to influenza vaccines. PLoS One. 2010;5:e13677. doi: 10.1371/journal.pone.0013677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallberg M, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallberg M, Margaryan G, Wang S, Ma J, Xu J. RaptorX server: A resource for template-based protein structure modeling. Methods Mol Biol. 2014;1137:17–27. doi: 10.1007/978-1-4939-0366-5_2. [DOI] [PubMed] [Google Scholar]
- 30.Garg VK, et al. MFPPI - multi FASTA ProtParam interface. Bioinformation. 2016;12:74–77. doi: 10.6026/97320630012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafarga T, O’Connor P, Hayes M. Identification of novel dipeptidyl peptidase-IV and angiotensin-I-converting enzyme inhibitory peptides from meat proteins using in silico analysis. Peptides. 2014;59:53–62. doi: 10.1016/j.peptides.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Magnan CN, et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26:2936–2943. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaiswal V, Chauhan RS, Rout C. Common antigens prediction in bacterial bioweapons: A perspective for vaccine design. Infect Genet Evol. 2014;21:315–319. doi: 10.1016/j.meegid.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 34.Delfani S, et al. In silico analysis for identifying potential vaccine candidates against staphylococcus aureus. Clin Exp Vaccine Res. 2015;4:99–106. doi: 10.7774/cevr.2015.4.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v.2–a server for in silico prediction of allergens. J Mol Model. 2014;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 36.Tso GHW, Reales-Calderon JA, Pavelka N. The elusive anti-candida vaccine: Lessons from the past and opportunities for the future. Front Immunol. 2018;9:897. doi: 10.3389/fimmu.2018.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van de Veerdonk FL, Netea MG, Joosten LA, van der Meer JW, Kullberg BJ. Novel strategies for the prevention and treatment of candida infections: The potential of immunotherapy. FEMS Microbiol Rev. 2010;34:1063–1075. doi: 10.1111/j.1574-6976.2010.00232.x. [DOI] [PubMed] [Google Scholar]
- 38.Pabst HF, Swanson R. Immunotherapy in candidiasis. Lancet. 1972;1:1022. doi: 10.1016/s0140-6736(72)91200-7. [DOI] [PubMed] [Google Scholar]
- 39.Wang XJ, et al. Vaccines in the treatment of invasive candidiasis. Virulence. 2015;6:309–315. doi: 10.1080/21505594.2014.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnon R, Ben-Yedidia T. Old and new vaccine approaches. Int Immunopharmacol. 2003;3:1195–1204. doi: 10.1016/S1567-5769(03)00016-X. [DOI] [PubMed] [Google Scholar]
- 41.Lamoth F, Lockhart SR, Berkow EL, Calandra T. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother. 2018;73:4–13. doi: 10.1093/jac/dkx444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine. 2001;19:2688–2691. doi: 10.1016/S0264-410X(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 43.Sette A, Rappuoli R. Reverse vaccinology: Developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy PC, Sharyan A, Sheikhi Moghaddam L. Meningococcal vaccines: Current status and emerging strategies. Vaccines (Basel). 2018;6:10. doi: 10.3390/vaccines6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pizza M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 46.Lauer P, et al. Genome analysis reveals pili in group B streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 47.Berlanda Scorza F, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic escherichia coli DeltatolR IHE3034 mutant. Mol Cell Proteomics. 2008;7:473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Stabler RA, et al. Comparative genome and phenotypic analysis of clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montigiani S, et al. Genomic approach for analysis of surface proteins in chlamydia pneumoniae. Infect Immun. 2002;70:368–379. doi: 10.1128/IAI.70.1.368-379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bidmos FA, Siris S, Gladstone CA, Langford PR. Bacterial vaccine antigen discovery in the reverse vaccinology 2.0 era: Progress and challenges. Front Immunol. 2018;9:2315. doi: 10.3389/fimmu.2018.02315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palucka K, Banchereau J. Linking innate and adaptive immunity. Nat Med. 1999;5:868–870. doi: 10.1038/11303. [DOI] [PubMed] [Google Scholar]
- 52.Richardson JP, Moyes DL. Adaptive immune responses to candida albicans infection. Virulence. 2015;6:327–337. doi: 10.1080/21505594.2015.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang MX, et al. Human recombinant antimannan immunoglobulin G1 antibody confers resistance to hematogenously disseminated candidiasis in mice. Infect Immun. 2006;74:362–369. doi: 10.1128/IAI.74.1.362-369.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumaresan PR, da Silva TA, Kontoyiannis DP. Methods of controlling invasive fungal infections using CD8(+) T cells. Front Immunol. 2018;8:1939. doi: 10.3389/fimmu.2017.01939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nanjappa SG, Heninger E, Wuthrich M, Sullivan T, Klein B. Protective antifungal memory CD8(+) T cells are maintained in the absence of CD4(+) T cell help and cognate antigen in mice. J Clin Invest. 2012;122:987–999. doi: 10.1172/JCI58762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wuthrich M, Filutowicz HI, Warner T, Deepe GS, Jr., Klein BS. Vaccine immunity to pathogenic fungi overcomes the requirement for CD4 help in exogenous antigen presentation to CD8+ T cells: Implications for vaccine development in immune-deficient hosts. J Exp Med. 2003;197:1405–1416. doi: 10.1084/jem.20030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds JM, Martinez GJ, Chung Y, Dong C. Toll-like receptor 4 signaling in T cells promotes autoimmune inflammation. Proc Natl Acad Sci USA. 2012;109:13064–13069. doi: 10.1073/pnas.1120585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davila E, Kolls J. A “toll” for Th17 cell expansion. J Leukoc Biol. 2010;88:5–7. doi: 10.1189/jlb.0110057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M, Jiang Y, Gong T, Zhang Z, Sun X. Intranasal vaccination against HIV-1 with adenoviral vector-based nanocomplex using synthetic TLR-4 agonist peptide as adjuvant. Mol Pharm. 2016;13:885–894. doi: 10.1021/acs.molpharmaceut.5b00802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.