Abstract
α-tomatine and dehydrotomatine are steroidal glycoalkaloids (SGAs) that accumulate in the mature green fruits, leaves, and flowers of tomatoes (Solanum lycopersicum) and function as defensive compounds against pathogens and predators. The aglycones of α-tomatine and dehydrotomatine are tomatidine and dehydrotomatidine (5,6-dehydrogenated tomatidine), and tomatidine is derived from dehydrotomatidine via four reaction steps: C3 oxidation, isomerization, C5α reduction, and C3 reduction. Our previous studies (Lee et al. 2019) revealed that Sl3βHSD is involved in the three reactions except for C5α reduction, and in the present study, we aimed to elucidate the gene responsible for the C5α reduction step in the conversion of dehydrotomatidine to tomatidine. We characterized the two genes, SlS5αR1 and SlS5αR2, which show high homology with DET2, a brassinosteroid 5α reductase of Arabidopsis thaliana. The expression pattern of SlS5αR2 is similar to those of SGA biosynthetic genes, while SlS5αR1 is ubiquitously expressed, suggesting the involvement of SlS5αR2 in SGA biosynthesis. Biochemical analysis of the recombinant proteins revealed that both of SlS5αR1 and SlS5αR2 catalyze the reduction of tomatid-4-en-3-one at C5α to yield tomatid-3-one. Then, SlS5αR1- or SlS5αR2-knockout hairy roots were constructed using CRISPR/Cas9 mediated genome editing. In the SlS5αR2-knockout hairy roots, the α-tomatine level was significantly decreased and dehydrotomatine was accumulated. On the other hand, no change in the amount of α-tomatine was observed in the SlS5αR1-knockout hairy root. These results indicate that SlS5αR2 is responsible for the C5α reduction in α-tomatine biosynthesis and that SlS5αR1 does not significantly contribute to α-tomatine biosynthesis.
Keywords: α-tomatine, CRISPR/Cas9, hairy roots, Solanum lycopersicum, steroidal glycoalkaloid
Introduction
Steroidal glycoalkaloids (SGAs) are typically found in members of Solanum species, and are known as toxic substances in Solanum food crops (Harrison 1990; Helmut 1998; Petersen et al. 1993) such as tomato (Solanum lycopersicum), potato (Solanum tuberosum), and eggplant (Solanum melongena). Because of their toxic effects on fungi, bacteria, insects, and animals, SGAs are considered to play defensive roles against a wide range of pathogens and predators (Friedman 2002, 2006). Tomato contains α-tomatine and dehydrotomatine as major SGAs in green tissues such as leaves and immature fruits (Friedman 2002). However, during the fruits ripening of tomato fruits, α-tomatine accumulated in immature fruits is metabolized and converted to nontoxic SGA esculeoside A (Iijima et al. 2009). In potatoes, α-solanine and α-chaconine are the two major SGAs, and their contents are especially high in tuber sprouts (Friedman 2006). Eggplant mainly produces α-solasonine and α-solamargine (Wu et al. 2013). The enormous structural diversity of SGAs is generated by various combinations of steroidal aglycones and sugar residues.
The common precursor for the biosynthesis of SGAs is cholesterol (Sawai et al. 2014), which undergoes subsequent modification via oxidation at the C16, C22, and C26 positions, transamination at the C26, and glycosylation at the C3 hydroxy group (Friedman 2002; Ginzberg et al. 2009; Ohyama et al. 2013; Petersen et al. 1993). Recently, several SGA biosynthetic genes were identified in tomatoes and potatoes. For instance, sterol side chain reductase 2 (SSR2) was identified as a key enzyme in cholesterol and the resulting SGAs biosynthesis (Sawai et al. 2014), and two cytochrome P450 monooxygenases (CYPs) (PGA1/CYP72A208 and PGA2/CYP72A188) and a 2-oxoglutarate-dependent dioxygenase (DOX) (16DOX) are responsible for hydroxylation of cholesterol at the C26, C22 and C16 positions, respectively, to produce SGAs (Itkin et al. 2013; Nakayasu et al. 2017; Umemoto et al. 2016). Additionally, the involvement of some UDP-dependent glycosyltransferases (UGTs), including GAME1 in tomato, were found to be involved in glycosylation steps in biosynthesis of SGAs (Itkin et al. 2011). However, the genes involved in the later biosynthetic processes have not yet been elucidated.
As described above, tomato contains α-tomatine and dehydrotomatine as major SGAs (Friedman 2002). α-Tomatine and dehydrotomatine are biosynthesized via four steps of glycosylations from their respective aglycones, tomatidine and dehydrotomatidine. Tomatidine possesses a single bond between C5 and C6 (dihydro type), while dehydrotomatidine has a Δ5,6 double bond (dehydro type), and is biosynthesized from dehydrotomatidine by the four-step reaction process shown in Figure 1. We recently identified that the gene designated as Sl3βHSD1 is involved in the conversion of dehydrotomatidine to tomatidine (Lee et al. 2019). Sl3βHSD1 is a multifunctional enzyme that possesses the activities of 3β-hydroxysteroid dehydrogenase (3βHSD) and 3-ketosteroid isomerase (3KSI) when acting on dehydrotomatidine to form tomatid-4-en-3-one and also shows the activity of 3-ketosteroid reductase (3KSR) when acting on tomatid-3-one to produce tomatidine (Lee et al. 2019). Therefore, Sl3βHSD1 is involved in the three reaction steps among the four reactions from dehydrotomatidine to tomatidine, and however, it has not yet been identified as the gene involved in a 5α-reduction step from tomatid-4-en-3-one to tomatid-3-one.
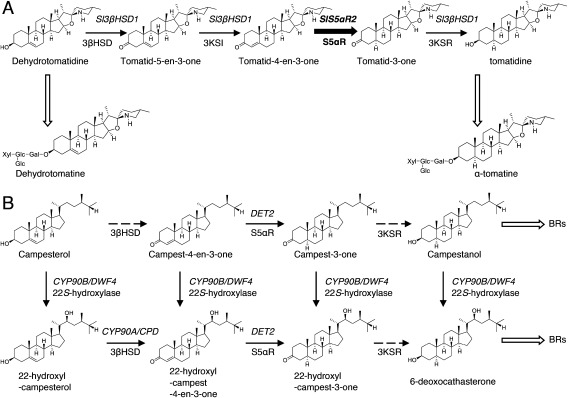
Figure 1. The putative biosynthetic pathways for (A) α-tomatine in tomatoes and (B) brassinosteroids (BRs) in plants. The thick solid arrow indicates the reaction step characterized in this work. Black thin solid arrows indicate the single reaction step characterized previously, and dashed arrows indicate the uncharacterized step. White thick arrows represent multiple reaction steps. The names of enzymes are indicated under the arrows. 3βHSD, 3β-hydroxysteroid dehydrogenase; 3KSI, 3-ketosteroid isomerase; S5αR, steroid 5α-reductase; 3KSR, 3-ketosteroid reductase. The genes previously characterized are shown in the upper part of the arrows. Sl3βHSD1 involved in the conversion of dehydrotomatidine to tomatidine in tomatoes; DWF4, dwarf4; DET2, de-etiolated-2; CPD, constitutive photomorphogenesis dwarf.
The 5α-reduction step is thought to be catalyzed by a steroid 5α-reductase (S5αR), which is involved in the NADPH-dependent reduction of the Δ4,5 double bond in various steroids and steroid hormones. In plants, Arabidopsis DET2 (AtDET2) has been identified as a biosynthetic gene of plant hormone brassinosteroids, and it encodes an S5αR protein that shares significant sequence identity with mammalian S5αRs (Noguchi et al. 1999). AtDET2 catalyzes the NADPH-dependent reduction of the Δ4,5 double bond in various steroids. The Arabidopsis det2 mutant shows a small dark-green dwarf phenotype, as a consequence of brassinosteroid deficiency via lack of the conversion of campest-4-en-3-one to campest-3-one (Noguchi et al. 1999). Tomato LeDET2, which is an ortholog of AtDET2 has been characterized as a functional steroid 5α-reductase (Rosati et al. 2005). Moreover, the presence of two isozymes was suggested by a crude enzyme assay performed on tomatoes (Rosati et al. 2005). We recently isolated and characterized SlS5αR1, and co-incubation of SlS5αR1 with Sl3βHSD1 and dehydrotomatidine in vitro demonstrated that the four reaction steps converting dehydrotomatidine to tomatidine were reconstituted in the in vitro assay (Lee et al. 2019). However, whether SlS5αR1 contributes to α-tomatine biosynthesis in vivo has remained unknown.
In the present study, we isolated a second DET2 homolog from tomato, designated as SlS5αR2. The expression pattern of SlS5αR2 was similar to those of known SGA biosynthetic genes in tomato. In vitro assay of recombinant SlS5αR2 using progesterone as a substrate revealed that SlS5αR2 encodes a functional steroid 5α-reductase. Additionally, simultaneous incubation of SlS5αR2 with Sl3βHSD1 resulted in the successive conversion of dehydrotomatidine to tomatidine in vitro, as observed in the case of recombinant SlS5αR1. Furthermore, CRISPR/Cas9-mediated knockout of either SlS5αR2 or SlS5αR1 was conducted in tomato hairy roots. While disruption of SlS5αR1 did not affect the endogenous SGA levels, SlS5αR2-knockout tomato hairy roots showed drastic reduction in the α-tomatine level and significant accumulation of dehydrotomatine. Based on all these findings, we conclude that SlS5αR2, but not SlS5αR1, is essential for the 5α-reduction step of α-tomatine biosynthesis in tomatoes.
Materials and methods
Plant materials
The tomato plant used in this study was S. lycopersicum cv Micro-Tom (TOMJPF00001), obtained from the NBRP (MEXT, Japan). The seeds were germinated and cultivated under a 16/8-h light/dark photoperiod in plant thermostatic chambers at 25°C until the plants produced ripening red fruits.
Chemicals
Progesterone were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan), and 5α-pregnane-3,20-dione was purchased from Santa Cruz Biotechnology (Satnta Cruz, CA, USA). Dehydrotomatidine was isolated in our lab from tomatidine purchased from Chromadex (Irvine, CA, USA) using high-performance liquid chromatography (HPLC). α-Tomatine was purchased from Sigma-Aldrich.
RNA extraction and reverse transcription
Total RNAs were extracted using the RNAeasy Plant Mini Kit (QIAGEN, Hilden, Germany) and the RNase-Free DNase Set (QIAGEN) according to the manufacturer’s instructions. The extracted total RNAs were used to synthesize the first strand of cDNA using the Transcriptor First Strand cDNA Synthesis Kit (TOYOBO, Osaka, Japan) for real-time quantitative polymerase chain reaction (PCR).
Real-time quantitative PCR analysis
Real-time quantitative PCR (qPCR) was performed with the LightCycler®Nano (Roche, Basel, Switzerland) using GeneAce SYBR® qPCR Mix α (Nippon Gene), with the following primers 1 and 2 for SlS5αR1, 3 and 4 for SlS5αR2, 5 and 6 for ubiquitin (Supplementary Table S2), using as template cDNAs from the various tomato tissues; flower, green fruit (G fruit), yellow fruit (Y fruit), orange fruit (O fruit), and red fruit (R fruit) as templates. Cycling was conducted at 95°C for 10 min, 45 cycles at 95°C for 30 s, and 60°C for 1 min for amplification, followed by holding at 95°C for 30 s and ramping up from 60 to 95°C at 0.1°C s−1 to perform a melting curve analysis. Each assay was repeated three times. The expression levels were normalized against the values obtained for the ubiquitin gene, which was used as an internal reference in tomato. Data acquisition and analysis were carried out using LightCycler®Nano software (Roche).
Cloning of SlS5αR1, SlS5αR1 and Sl3βHSD1
The DNA fragments containing the ORF of Solyc09g013070 (SlS5αR1) and Solyc10g086500 (SlS5αR2) were isolated using the cDNA template from the tomato leaves with primer sets 7 and 8 for SlS5αR1, and 9 and 10 for SlS5αR2 (Supplementary Table S2). The PCR product was purified using Wizard® SV Gel and the PCR Clean-Up System (Promega, WI, USA) and cloned into the pMD19 T-vector (TaKaRa, Shiga, Japan). The sequence of the cloned DNA was confirmed using the ABI 3130 genetic analyzer (Applied Biosystems, CA, USA) and analyzed using BioEdit, a biological sequence alignment editor (http://www.mbio.ncsu.edu/). Similarly, the open reading frame (ORF) of Sl3βHSD1 was isolated using a clone of the LEFL1039AB03 (identical to Solyc01g073640 in the tomato ITAG 2.4) in the Kazusa Full-length Tomato cDNA Database (http://www.pgb.kazusa.or.jp/kaftom/clone.html), purchased from Kazusa DNA Research Institute. The ORF of Sl3βHSD1 was amplified using PCR with the clone as a template and primer sets 11 and 12 (Supplementary Table S2).
Heterologous expression of the recombinant SlS5αR1, SlS5αR2 and Sl3βHSD1 proteins
The DNA fragments containing the ORF of the SlS5αR1 and SlS5αR2 gene were ligated into a pFastBac1 vector (Invitrogen, Carlsbad, CA), and were then introduced into Escherichia coli DH10Bac (Life Technologies) to generate the corresponding recombinant bacmid DNAs. Preparation of recombinant baculovirus DNAs that contained SlS5αR1 and SlS5αR2 ORFs and transfection of Spodoptera frugiperda 9 insect cells were carried out according to the manufacturer’s instructions (Life Technologies). Heterologous expression of SlS5αR1 and SlS5αR2 proteins in insect cells was conducted as described by Ohnishi et al. (2006). Similarly, the DNA fragments containing the Sl3βHSD1 gene ORF were ligated into pCold ProS2 vector (TaKaRa), and recombinant Sl3βHSD1 protein was prepared using bacterial expression system in E.coli strain BL21 (DE3) as described by Lee et al. (2019).
In vitro enzyme activity assay
To prepare tomatid-4-en-3-one, we conducted in vitro enzyme assay of recombinant Sl3βHSD1 protein, and the assay was performed using 100 µl of reaction mixture comprising 100 mM bis-tris-HCl (pH 7.2), 2.5 mM NAD+ as a coenzyme, 50 μM dehydrotomatidine as a substrate, and the purified Sl3βHSD1. The enzymatically synthesized tomatid-4-en-3-one was further metabolized by recombinant SlS5Rα1 to produce tomatid-3-one, and the reaction was carried out as described below.
Microsomal fractions of the insect cells expressing SlS5αR1 or SlS5αR2 were obtained from 100 ml of cultured cell suspension. Infected cells were washed with phosphate-buffered saline and suspended in buffer A, consisting of 20 mM potassium phosphate (pH 7.5), 20% (v/v) glycerol, 1 mM ethylenediaminetetraacetic acid, and 1 mM dithiothreitol. The cells were sonicated and cell debris was removed by centrifugation at 10,000 g for 15 min, and the resulting pellets were homogenized with buffer A to provide the microsomal fractions. The assay of SlS5αR1 and SlS5αR2 for steroid C5 reduction were performed using 100 µl reaction mixture comprising 100 mM potassium phosphate (pH 7.2), 1 mM NADPH or NADH as a coenzyme, the enzymatically synthesized tomatido-4en-3-one, and the microsomal fraction containing SlS5αR1 or SlS5αR2. The conversion of dehydrotomatidine to tomatidine was reconstituted in an in vitro assay by mixing the purified Sl3βHSD1 protein with SlS5αR1 or SlS5αR2-containing microsomes. The reaction mixture consisted of 100 mM bis-tris-HCl buffer (pH 7.2), purified Sl3βHSD1 protein, microsomes containing SlS5αR1 or SlS5αR2, 2.5 mM NAD+, 1 mM NADPH, and 25 μM dehydrotomatidine. All the reactions were performed at 30°C for 2 h and were terminated by the addition of 100 µl ethyl acetate. The reaction products were extracted three times with an equal volume of ethyl acetate and the organic phase was collected and evaporated. The residues were analyzed using gas chromatography-mass spectrometry (GC-MS) and/or liquid chromatography-mass spectrometry (LC-MS).
GC-MS analysis of the reaction product was conducted as described previously (Lee et al. 2019). Briefly, the residue was trimethylsilylated, and then subjected to a GC-MS-QP2010 Ultra (Shimadzu, Kyoto, Japan) with a DB-1MS (30 m×0.25 mm, 0.25 µm film thickness; J&W Scientific, CA, USA) capillary column. An MS scan mode with a mass range of m/z 50–700 were used. Progesterone and 5α-pregnane-3,20-dione were monitored at a selected ion monitoring (SIM) mode chromatogram at m/z 229 and 231, respectively. Tomatid-4-en-3-one and tomatid-3-one were analyzed via SIM mode at m/z 125. For LC-MS analysis, the residue was dissolved in 200 µl of methanol. LC-MS analysis of each reaction product was carried out as described above Lee et al. (2019), using an ACQUITY UPLC H-Class System (Waters, MA, USA) with an Acquity UPLC HSS-T3 column (1.8 µm, 2.1×100 mm2, Waters) at 40°C and an SQ Detector 2 (Waters). Data acquisition and analysis were performed using MassLynx 4.1 software (Waters). Reaction products were analyzed by SIM-mode chromatography at m/z 412, 414, and 416.
Construction of CRISPR/Cas9 vectors
The knockout tomato hairy roots for each of SlS5αR1 and SlS5αR2 were generated by targeted genome editing with the CRISPR/Cas9 system. We used the CRISPR/Cas9 binary vector pMgP237-2A-GFP to express multiplex gRNAs (Hashimoto et al. 2018; Nakayasu et al. 2018). To design a gRNA target with low off-target effect in SlS5αR1 and SlS5αR2, we conducted in silico analyses using the Web tool Design sgRNAs for CRISPRko (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and Cas-OT software (Xiao et al. 2014). Then we selected target sequences named SlS5R1_492-511 and SlS5R1_552-571 in the coding region of SlS5αR1, and SlS5R2_449-468 and SlS5R2_511-530 in the coding region of SlS5αR2, respectively. To enhance the efficiency of gRNA transcription from U6 promoter, one G was added to the 5ʹ- end of SlS5R1_492-511, SlS5R1_552-571 and SlS5R2_511-530. Two DNA fragments composed of the gRNA scaffold and tRNA scaffold between two target sequences, SlS5R1_492-511/ SlS5R1_552-571 and SlS5R2_449-468/SlS5R2_511-530, were generated by PCR using pMD-gtRNA containing gRNA and tRNA scaffolds as a template and primer sets containing restriction enzyme BsaI sites (13 and 14 for SlS5R1_492-511/ SlS5R1_552-571, 15 and 16 for SlS5R2_449-468/SlS5R2_511-530) (Supplementary Table S2). The units containing two gRNAs–tRNAs were then independently inserted into the BsaI site of pMgP237-2A-GFP using Golden Gate Cloning methods to construct the CRIPR/Cas9 vectors, labeled pMgP237-SlS5αR1ko and pMgP237-SlS5αR2ko, respectively.
Generation of SlS5RαR1- and SlS5RαR2-disrupted transgenic tomato hairy roots
The vectors pMgP237-SlS5αR1ko and pMgP237-SlS5αR2ko were independently introduced into Agrobacterium rhizogenes ATC15834 by electroporation. Transgenic tomato hairy roots were generated as previously described by Thagun et al. (2016). Genome DNA was extracted from each established hairy root and transformants were selected by genomic PCR to amplify a partial fragment of the T-DNA region integrated into the genome using primers 17 and 18 (Supplementary Table S2). To analyze the mutations or large deletions in the transgenic hairy roots, the region including the target sites of gRNAs was amplified by PCR using primers 19 and 20 for pMgP237-SlS5αR1ko transformants and primers 21 and 22 for pMgP237-SlS5αR2ko transformants (Supplementary Table S2). The PCR fragments obtained from each transformant were then subjected to agarose-gel electrophoresis or microchip electrophoresis using MultiNA (Shimadzu). Furthermore, each PCR fragment from four independent transgenic hairy root lines (pMgP237-SlS5αR1ko_#1 and #6, pMgP237-SlS5αR2ko_#7 and #12) was cleaned using the Wizard® SV Gel and PCR Clean-Up System (Promega, Japan) according to the manufacturer’s instructions, and cloned into pCR™4Blunt-TOPO® (Invitrogen). Sanger sequencing of each of the cloned DNAs was performed using a sequencing service (Eurofins Genomics).
Quantification of SGAs in tomato hairy roots
SGAs were extracted from 100 mg fresh weight of harvested transgenic lines, and each sample was prepared and analyzed by LC-MS. The LC-MS analysis was performed using the LC-MS/MS system; water with 0.1% (v/v) formic acid (A) and acetonitrile (B) were used as mobile phases, with an elution gradient of 10–55% B from 0 to 30 min and 55–75% B from 30 to 35 min (linear gradient). MS scan mode was used with a mass range of m/z 400–1,200. The quantities of α-tomatine and dehydrotomatine were calculated from the peak area using an α-tomatine calibration curve.
Phylogenetic analaysis
Amino acid sequences of AtDET2 homologs were retrieved from the Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and Sol Genomics networks (https://solgenomics.net/). A phylogenetic tree was constructed using the MEGA7 program and the maximum likelihood method with the following parameters: Poisson correction, pairwise deletion and bootstrap (1,000 replications; random seed).
Results
Selection of candidate S5αR genes involved in α-tomatine biosynthesis
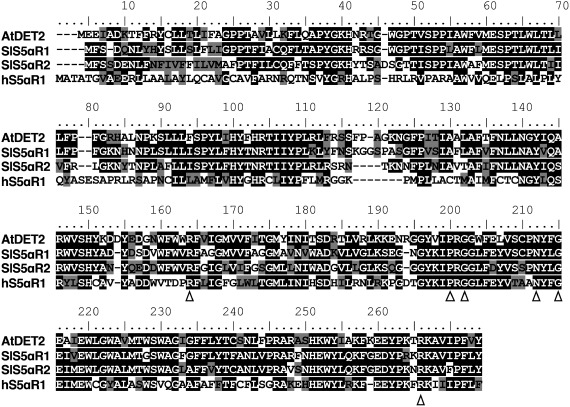
To survey the candidate gene encoding functional S5αR, BLASTX analysis against tomato transcripts database from Sol Genomics (http://solgenomics.nrt/) used AtDET2 as a query. This analysis allowed us to select two distinct cDNAs encoded by Solyc09g013070 and Solyc10g086500. Solyc09g013070 was identical to SlS5αR1, which has been already characterized to encode a functional steroid 5α-reductase (Lee et al. 2019), and Solyc10g086500 was identical to LeDET2, which was previously characterized to encode a functional steroid 5α-reductase (Rosati et al. 2005), and here we designated it as SlS5αR2. SlS5αR1 and SlS5αR2 show 63% and 57% amino acid identity to AtDET2, respectively, and share 67% amino acid identity to each other. Both SlS5αR1 and SlS5αR2 contain six (Arg-150/148, Pro-185/183, Gly-187/185, Asn-197/195, Gly-200/198 and Arg-251/249) of seven conserved amino acid residues that are part of a typical cofactor binding domain in mammalian 5α-reductase (Figure 2; Russell and Wilson 1994). Both SlS5αR1 and SlS5αR2 contain six potential transmembrane-spanning domains based on the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM), as well as AtDET2 predicted by the same program.
Figure 2. Multiple sequence alignment of SlS5αR1 and SlS5αR2 with AtDET2 from Arabidopsis thaliana and human hS5αR1. Black and gray shades indicate identical and similar amino acid residues, respectively. Conserved residues, which are required for steroid 5α-reduction activity, are marked by white triangles.
Expression analysis of SlS5αR1 and SlS5αR2 in various tissues of tomato
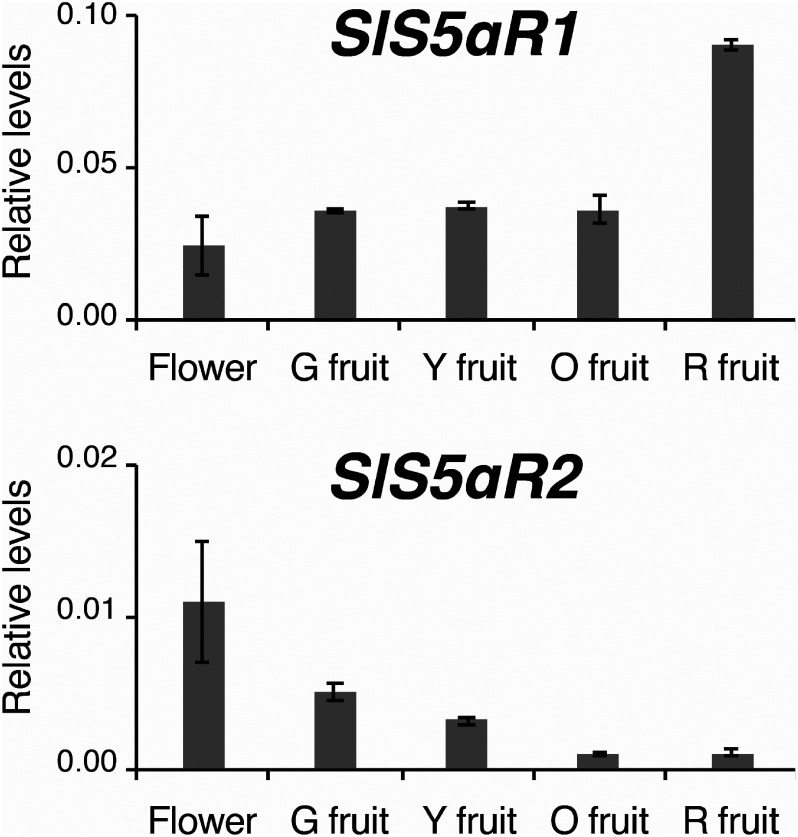
The expression of SlS5αR1 and SlS5αR2 was analyzed in the RNA-seq data of the Tomato Functional Genomics Database (http://ted.bti.cornell.edu/). SlS5αR2 showed an expression pattern similar to those of the known SGA biosynthetic genes in terms of higher expression in immature fruits and decreasing expression levels during ripening of fruits, whereas the transcript levels of SlS5αR1 remained high even in red ripe fruits where the expression of SGA genes was low (Supplementary Table S1). To confirm the expression of SlS5αR1 and SlS5αR2, real-time PCR was performed using the reverse-transcription products of total RNAs extracted from various tissues of S. lycopersicum cv. Micro-Tom. SlS5αR2 was highly expressed in flowers, where a large amount of α-tomatine is accumulated (Roddick 1974), while the expression level of SlS5αR2 was low in red mature fruits (Figure 3), in which α-tomatine is further metabolized to esculeocide A and the α-tomatine level is very low (Iijima et al. 2009). This expression pattern of SlS5αR2 is similar to those of the SGA biosynthetic genes Sl16DOX, GAME1, and Sl3βHSD1 (Lee et al. 2019; Nakayasu et al. 2017). On the other hand, SlS5αR1 was abundantly expressed in red mature fruits, and the expression pattern of SlS5αR1 is not in agreement with the known SGA biosynthetic genes (Figure 3).
Figure 3. Quantitative RT-PCR analysis of SlS5αR1 and SlS5αR2 expression in various tissues of tomatoes. G, Y, O, and R mean green, yellow, orange, and red, respectively. Transcript levels of SlS5αR1 and SlS5αR2 are shown relative to that of the ubiquitin gene as an internal reference. Error bars indicate standard error from the mean (n=3).
In vitro functional analysis of the C5α-reduction activity of SlS5αR2 on tomatid-4-en-3-one to tomatid-3-one
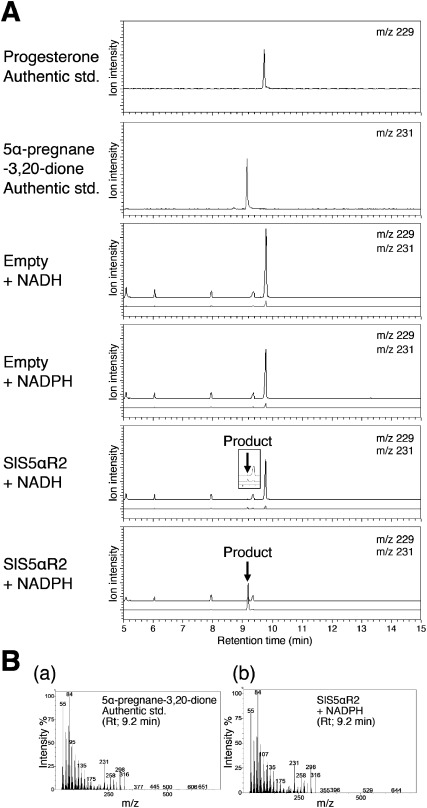
In our recent studies, the recombinant SlS5αR1 protein was expressed with a baculovirus-mediated expression system in Spodoptera frugiperda 9 insect cells and confirmed to catalyze C5α-reduction of tomatid-4-en-3-one to yield tomatid-3-one in vitro (Lee et al. 2019). Previously, it was reported that recombinant SlS5αR2 expressed in mammalian cells (COS-7) was active on typical substrates of mammalian S5αRs (progesterone, testosterone and androstenedione) (Rosati et al. 2005). In the present study, to compare the catalytic activities of SlS5αR1 and SlS5αR2, the recombinant SlS5αR2 was prepared with a baculovirus-mediated expression system and C5-reduction activities were analyzed. First, the functional expression of SlS5α2 was confirmed by in vitro enzyme assays using progesterone as a substrate in the presence of NADPH or NADH as a coenzyme, and the reaction products were analyzed by gas chromatography-mass spectrometry (GC-MS). A product with a retention time of 9.2 min and a major mass fragment ion at m/z 231 was detected. This product was identical to the authentic compound 5α-pregnane-3,20-dione in terms of both the retention time the mass spectrum (Figure 4). Although SlS5αR2 could catalyze the C5 reduction with either NADPH or NADH as a coenzyme, the reduction activity in the presence of NADPH was much higher than that in the presence of NADH. Thus, the functional expression of SlS5αR2 in the baculovirus-insect cell system was confirmed.
Figure 4. In vitro assay of the C5 reduction activity of SlS5αR1 with progesterone as the substrate. (A) Progesterone was monitored by selected ion monitoring (SIM) at m/z 229 and is shown in black, and the product 5α-pregnane-3,20-dione was monitored at m/z 231 and is shown in gray. (B) Mass spectrum of the (a) authentic 5α-pregnane-3,20-dione at a retention time of 9.2 min and (b) product peak of progesterone incubated with SlS5αR2 and NADPH.
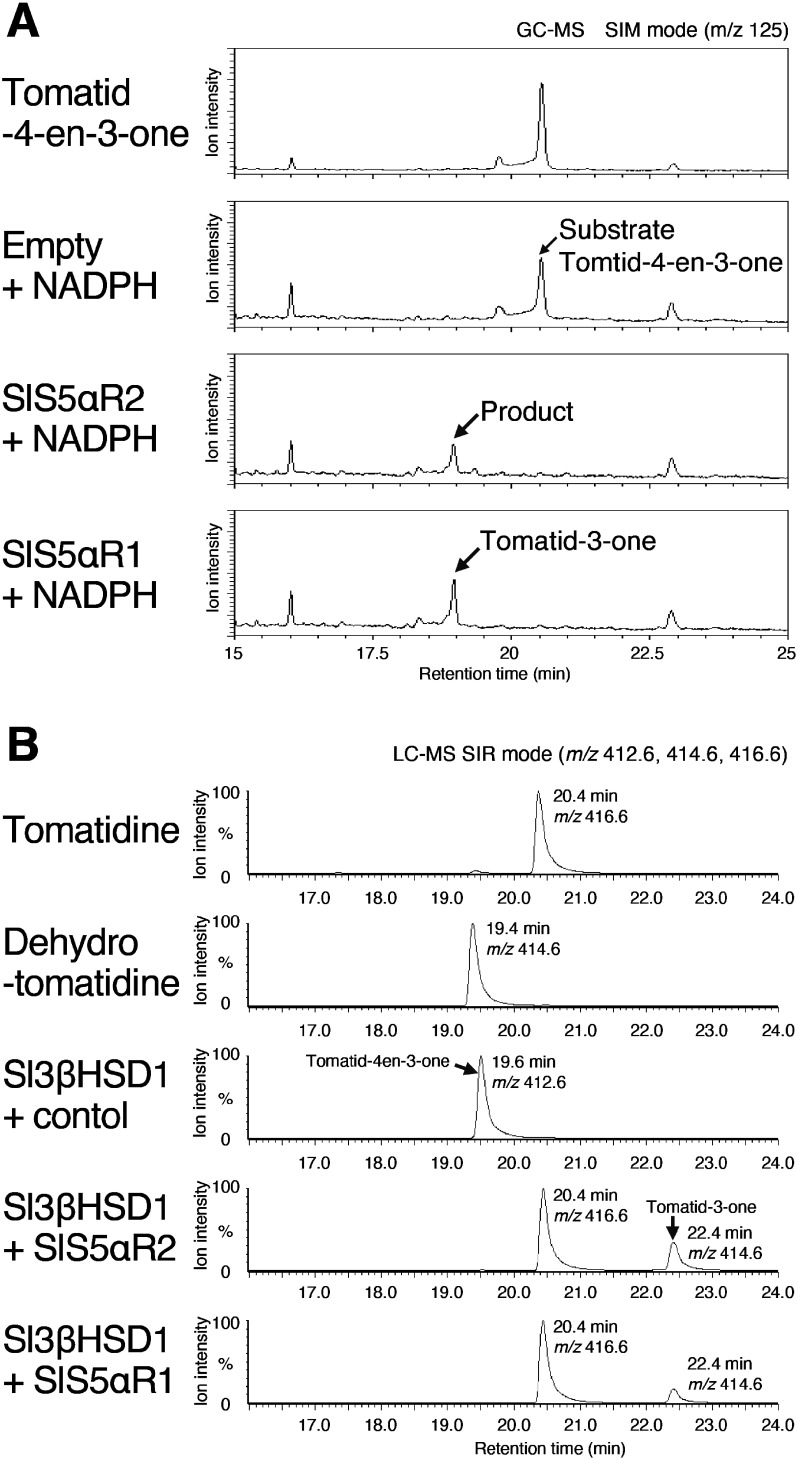
We evaluated the C5α-reduction activities on tomatid-4-en-3-one. Because authentic compounds of tomatid-4-en-3-one and tomatid-3-one are not commercially available, we obtained them by enzymatic synthesis using Sl3βHSD1 and SlS5αR1 (Lee et al. 2019). Tomatid-4-en-3-one was synthesized from dehydrotomatidine by enzymatic conversion with the recombinant Sl3βHSD1 using NAD+ as a coenzyme, and tomatid-3-one was synthesized from tomatid-4-en-3-one by enzymatic conversion with SlS5αR1 using NADPH as a coenzyme. Then, the recombinant SlS5αR2 was assayed with the enzymatically synthesized tomatid-4-en-3-one in the presence of NADPH, and the reaction products were analyzed using GC-MS (Figure 5A). Tomatid-4-en-3-one was metabolized to yield a product with a retention time of 18.9 min, and this product by SlS5αR2 was identical to tomatid-3-one, obtained by incubating tomatid-4-en-3-one with SlS5αR1 (Figure 5A). Thus, SlS5αR2 could catalyze the reduction of tomatid-4-en-3-one at C5α to yield tomatid-3-one.
Figure 5. In vitro assays of SlS5αR1 and SlS5αR2 with tomatid-4-en-3-one. (A) GC-MS analysis of the in vitro assays of SlS5αR1/2 and NADPH incubated with tomatide-4-en-3-one as a substrate. Substrate and products were monitored by SIM mode at m/z 125. (B) LC-MS analysis of the in vitro assays of SlS5αR1/2 and Sl3βHSD1 incubated with dehydrotomatidine as a substrate. Substrate and products were monitored by SIM mode at m/z 412, 414, and 416.
Next, we conducted a co-incubation assay of SlS5αR2 with Sl3βHSD1 using dehydrotomatidine as a substrate, and the reaction products were analyzed by LC-MS (Figure 5B). When incubating dehydrotomatidine with Sl3βHSD1, tomatid-4-en-3-one was detected with a retention time of 19.6 min and with a parent mass at m/z 412.6 (Figure 5B). Simultaneous incubation of dehydrotomatidine with SlS5αR2 and Sl3βHSD1 resulted in the detection of two major products with the retention times of 20.4 and 22.4 min, respectively (Figure 5B). The product with a retention time of 22.4 min gave a parent mass at m/z 414.6, which corresponded to [tomatid-3-one+H+]+ (Figure 5B). The product at a retention time of 20.4 min, which had a parent mass at m/z 416.6, was identical to the authentic tomatidine (Figure 5B). These results indicate that, during co-incubation with SlS5αR2 and Sl3βHSD1, dehydrotomatidine was at first converted to tomatid-4-en-3-one by Sl3βHSD1, and SlS5αR2 subsequently catalyzed the reduction of tomatid-4-en-3-one at C5α to form tomatid-3-one, and Sl3βHSD1 finally catalyzed the reduction of tomatid-3-one at C3 to yield tomatidine. Similar result was obtained in case of co-incubation of dehydrotomatidine with SlS5αR1 and Sl3βHSD1 (Figure 5B), and this result was consistent with our previous work (Lee et al. 2019). Taken together, either SlS5αR2 or SlS5αR1, in combination with Sl3βHSD1 and cofactors, could reconstitute the conversion of dehydrotomatidine to tomatidine in an in vitro assay.
CRISPR/Cas9-mediated genome editing of SlS5αR1 or SlS5αR2 in tomato hairy roots
To verify the contribution of SlS5αR1 and SlS5αR2 in α-tomatine biosynthesis in vivo, SlS5αR1 and SlS5αR2 were each independently disrupted using CRISPR/Cas9-mediated genome editing in tomato hairy roots. In this study, we used a CRISPR/Cas9 binary vector designated as pMgP237-2A-GFP that permits expression of multiplex gRNAs by a pre-tRNA processing mechanism (Hashimoto et al. 2018; Nakayasu et al. 2018). To design the gRNAs specific to SlS5αR1 and to SlS5αR2, we conducted in silico analysis using Design sgRNAs for CRISPRko and Cas-OT software (Xiao et al. 2014). Then, the selected target sequences were inserted into a pMgP237-2A-GFP backbone vector, and we constructed a targeting vector specific to SlS5αR1 and one specific to SlS5αR2, designated as pMgP237-SlS5αR1ko and pMgP237-SlS5αR2ko, respectively (Supplementary Figure S1). The hypocotyls of tomato cv. Micro-Tom were infected with A. rhizogenes harboring each of the two constructed vectors. Several lines of transgenic hairy roots were obtained, and the mutations in the region surrounding the target sites of gRNAs were analyzed.
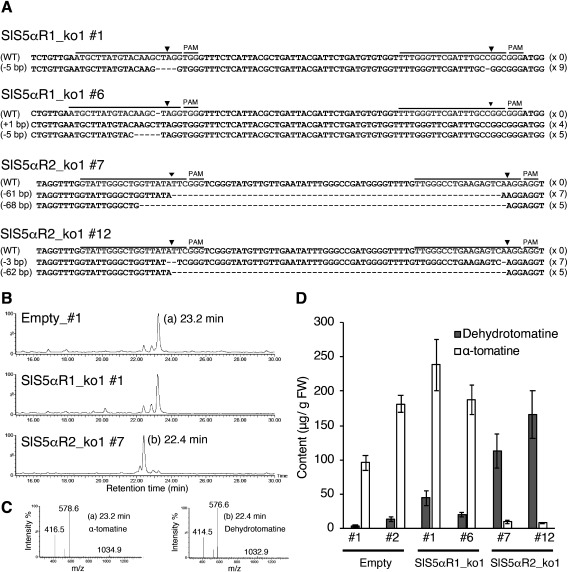
Among the pMgP237-SlS5αR1ko transgenic hairy roots, a formation of a heteroduplex, which indicates the mutation at the target site, was detected as shifted peaks in #1 and #6 (Supplementary Figure S2). Among the pMgP237-SlS5αR2ko transgenic hairy roots, deletion of a large segment or a formation of a heteroduplex was detected in #5, #7, #8, #10, #11, and #12 (Supplementary Figure S2). To confirm the mutations at the target sites, four lines for each of pMgP237-SlS5αR1ko (lines #1 and #6) and pMgP237-SlS5αR2ko (line #7 and #12) were selected, and sequencing analyses were conducted. The lines #1 and #6 for pMgP237-SlS5αR1ko had no intact SlS5αR1 sequences, and all of the sequences showed the translational frameshift mutations (Figure 6A). In the lines #7 and #12 for pMgP237-SlS5αR2ko, all the detected sequences showed some deletions in the SlS5αR2 gene (Figure 6A).
Figure 6. Characterization of SlS5αR1 or SlS5αR2-disrupted tomato hairy roots. (A) DNA sequences of gRNA target sites. Black and gray lines indicate gRNA target sites and PAM sequences, respectively. Black triangles indicate cleavage positions of target sites. Deletions or insertions of nucleotides are presented on the left sides of the sequences, and the numbers of detected sequences are shown on the left sides. (B) LC-MS analysis of SGAs in SlS5αR1_ko1_#1 and SlS5αR2 _ko2_#7. (C) Mass spectrum of the α-tomatine (peak at 23.2 min) and dehydrotomatine (peak at 22.4 min). (D) Quantification of α-tomatine and dehydrotomatine contents using LC-MS. Error bars indicate standard error (n=3).
Endogenous SGAs of these selected lines were extracted and analyzed by LC-MS. The level of α-tomatine in the pMgP237-SlS5αR1ko_#1 and #6 was similar to that in the vector control, and the amounts of dehydrotomatine were not affected (Figure 6B–D). On the other hand, the α-tomatine level was significantly decreased in the pMgP237-SlS5αR2ko_#7 and #12, and a corresponding increase in dehydrotomatine level was detected as compared with that in the vector control (Figure 6B–D). These results indicate that SlS5αR2 is involved in the conversion of dehydrotomatidine to tomatidine in tomato, while SlS5αR1 is not responsible for biosynthesis of SGAs.
Discussion
In the present study, we found that SlS5αR1 and SlS5αR2 catalyze the conversion of tomatid-4-en-3-one to tomatid-3-one in vitro (Figure 5A). In the SlS5αR2-knockout tomato hairy roots, α-tomatine was dramatically decreased and corresponding amounts of dehydrotomatine were accumulated, while the disruption of SlS5αR1 did not affect the α-tomatine and dehydrotomatine levels (Figure 6). These results clearly demonstrate that SlS5αR2, but not SlS5αR1, is involved in the C5α reduction step of the metabolic conversion of dehydrotomatidine to tomatidine in α-tomatine biosynthesis.
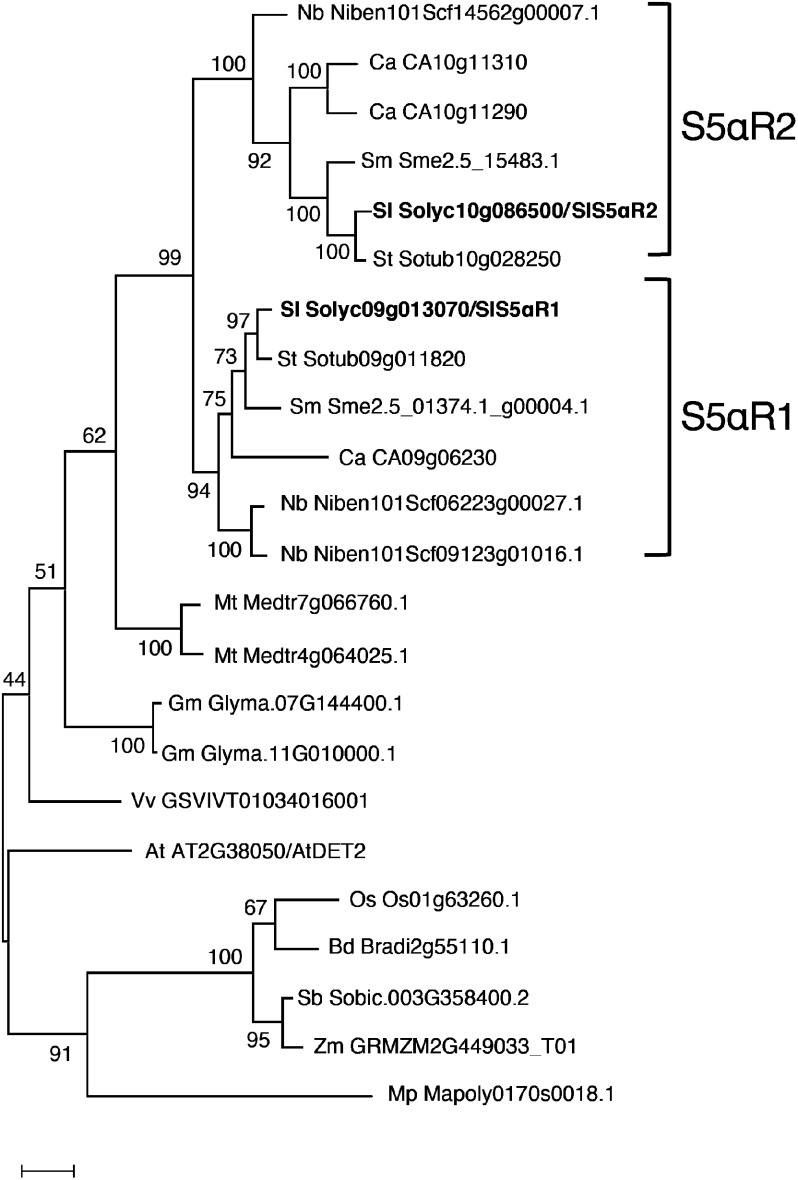
Arabidopsis DET2 (AtDET2) is the first gene isolated in plants coding functional steroid 5α-reductase, and the Arabidopsis det2-1 mutant shows a dwarf and de-etiolated phenotype because of a deficiency of bioactive brassinosteroids (Noguchi et al. 1999). In plants, the reduction of the steroid Δ4,5 double bond is an important reaction in the biosynthesis of the plant hormone BRs since all known bioactive BRs lack Δ4,5 double bond (Rosati et al. 2005). Steroid 5α-reductase is likely to be conserved in plant kingdom, and we found that some plant species have multiple S5αR genes in their genome. We found that all Solanaceae species analyzed have two or more S5αR genes in the genome, and phylogenetic analysis demonstrated that the S5αR proteins of solanaceous plants are clearly divided into two distinguishable clades (Figure 7), suggesting that duplication of the ancestral S5αR gene has occurred in solanaceous plants. SlS5αR1 shows higher homology to AtDET2, suggesting that SlS5αR1 would keep to function only in the biosynthesis of phytosterols and brassinosteroids. Then, the duplication gives rise to SlS5αR2, which is specialized in α-tomatine biosynthesis. The duplication and neofunctionalization have been reported for several enzymes involved in phytosterols, BRs and cholesterol biosynthesis (Christ et al. 2019; Sawai et al. 2014; Sonawane et al. 2017). For instance, potato and tomato have two DWF1 homologs named SSR1 and SSR2 (Sawai et al. 2014). SSR1 corresponds to DWF1 and has Δ24(28) reduction activity required for phytosterol biosynthesis, while SSR2 catalyzes Δ24(25) reduction of cycloartenol and desmosterol for cholesterol biosynthesis. Our in vitro assay revealed that both SlS5Rα1 and SlS5αR2 encode the functional steroid 5α-reductase and catalyze the conversion of 5α-reduction of tomatid-4-en-3-one to yield tomatid-3-one (Figure 5A). Furthermore, in combination with Sl3βHSD1, each of SlS5αR1 and SlS5αR2 could complete the four reaction steps converting dehydrotomatidine to tomatidine in the in vitro assays (Figure 5B). These observations suggest that the different physiological roles for SlS5αR1 and SlS5αR2 reflect factors other than neofunctionalization of catalytic activity.
Figure 7. Phylogenetic analysis of plant AtDET2 homologs. Species were represented tomato (Sl), potato (St), eggplant (Sm), Capsicum annuum (Ca), Nicotiana benthamiana (Nb), Brachypodium distachyon (Bd), soybean (Gm), Medicago truncatula (Mt), rice (Os), Sorghum bicolor (Sb), Vitis vinifera (Vv), and maize (Zm). A phylogenetic tree was generated using the maximum likelihood method in MEGA7. Bootstrap values based on 1,000 replicates are shown at the branching points.
Recently, Thagun et al. (2016) reported that Jasmonate-Responsive ERF 4 (JRF4) comprehensively regulates α-tomatine biosynthetic genes, including those involved in the upstream mevalonate pathway, in tomatoes. Quantitative RT-PCR analysis in various tissues of tomatoes showed different expression patterns between SlS5αR1 and SlS5αR2, and the expression pattern of SlS5αR2 is in good agreement with those of known SGA biosynthetic genes. Therefore, our results indicate that acquisition of the transcriptional regulation, which cooperates with other SGA biosynthetic genes, contributes to functional specialization of SlS5αR2 for SGA biosynthesis. However, our quantitative RT-PCR analyses and RNA-seq data of the Tomato Functional Genomics Database (http://ted.bti.cornell.edu/) demonstrated that SlS5αR1 is also expressed in certain amounts in SGA containing tissues, suggesting that there are mechanisms, in addition to transcriptional control, to explain the functional differentiation of SlS5αR1 and SlS5αR2 in vivo. For example, post-translational regulation such as distinct intracellular localization and/or protein–protein interaction in the metabolon can be one of the mechanisms, as discussed below.
SlS5αR2-knockout tomato hairy roots contained very little α-tomatine, and the amount of dehydrotomatine increased to a level comparable to the level of α-tomatine accumulated in control tomato hairy roots (Figure 6). Dehydrotomatine is biosynthesized via glycosylation from dehydrotomatidine, which is not a direct substrate of SlS5αR2. Dehydrotomatidine is metabolized via oxidation and isomerization by Sl3βHSD1 to yield tomatid-4-en-3-one, which is a direct substrate of SlS5Rα2 (Figure 1). Considering that isomerase reaction of mammalian 3βHSD has been reported to be irreversible (Thomas et al. 2005), high accumulation of dehydrotomatine suggests that Sl3βHSD1 does not function well in the SlS5αR2-knockout tomato hairy roots. This may be the reason why dehydrotomatine but not tomatid-4-en-3-one accumulated in the SlS5αR2-knockout tomato hairy roots. It has been reported that protein–protein interaction that allows the formation of protein complexes of sequential metabolic enzymes, termed metabolon, enhances direct channeling of substrates between the biosynthetic enzymes, providing increased control over metabolic pathway fluxes (Obata 2019). So far, to the best of our knowledge, there has been no report of metabolon formation in SGA biosynthesis, but the physical protein–protein interaction of SlS5αR2 with Sl3βHSD1, or other SGA biosynthetic enzymes, may influence the functional differentiation of SlS5αR1 and SlS5αR2. In order to address this hypothesis, we need to examine in detail the subcellular localization of SlS5αR2 and other SGA biosynthetic enzymes, as well as protein–protein interactions among SGA biosynthetic enzymes.
In conclusion, we showed here that SlS5αR2 is a key enzyme in the production of tomatidine from dehydrotomatidine in tomatoes, thereby contributing to structural diversity of SGAs in the Solanaceae family. SGA is well known as a toxic and antinutritional substance in a Solanaceae crop, but SGAs are considered to play protective roles against plant pathogens and predators for the plants. Through this study, it may be possible to clarify the significance of solanaceous plants, producing SGA with various structures.
Acknowledgments
We thank Mina Yamamoto for technical assistance.
This study was supported by the Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Fellows.
Abbreviations
- SGA
steroidal glycoalkaloid
- CYP
cytochrome P450 monooxygenase
- DOX
2-oxoglutarate dependent dioxygenase
- S5αR
steroid 5α-reductase
- UGT
UDP-dependent glycosyltransferase
- 3βHSD
3β-hydroxysteroid dehydrogenase
- 3KSI
3-ketosteroid isomerase
- 3KSR
3-ketosteroid reductase
Supplementary Data
References
- Christ B, Xu C, Xu M, Li FS, Wada N, Mitchell AJ, Han XL, Wen ML, Fujita M, Weng JK (2019) Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat Commun 10: 3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M (2002) Tomato glycoalkaloids: Role in the plant and in the diet. J Agric Food Chem 50: 5751–5780 [DOI] [PubMed] [Google Scholar]
- Friedman M (2006) Potato glycoalkaloids and metabolites: Roles in the plant and in the diet. J Agric Food Chem 54: 8655–8681 [DOI] [PubMed] [Google Scholar]
- Ginzberg I, Tokuhisa J, Veilleux R (2009) Potato steroidal glycoalkaloids: Biosynthesis and genetic manipulation. Potato Res 52: 1–15 [Google Scholar]
- Harrison DM (1990) Steroidal alkaloids. Nat Prod Rep 7: 139–147 [Google Scholar]
- Hashimoto R, Ueta R, Abe C, Osakabe Y, Osakabe K (2018) Efficient multiplex genome editing induces precise, and self-ligated type mutations in tomato plants. Front Plant Sci 9: 916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmut R (1998) Solanum steroid alkaloids: An update. Alkaloids: Chem Biol Perspect 12: 103–185 [Google Scholar]
- Iijima Y, Fujiwara Y, Tokita T, Ikeda T, Nohara T, Aoki K, Shibata D (2009) Involvement of ethylene in the accumulation of esculeoside A during fruit ripening of tomato (Solanum lycopersicum). J Agric Food Chem 57: 3247–3252 [DOI] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, et al. (2011) GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23: 4507–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Nakayasu M, Akiyama R, Kobayashi M, Miyachi H, Sugimoto Y, Umemoto N, Saito K, Muranaka T, Mizutani M (2019) Identification of a 3β-hydroxysteroid dehydrogenase/3-ketosteroid reductase involved in α-tomatine biosynthesis in tomato. Plant Cell Physiol 60: 1304–1315 [DOI] [PubMed] [Google Scholar]
- Nakayasu M, Akiyama R, Lee HJ, Osakabe K, Osakabe Y, Watanabe B, Sugimoto Y, Umemoto N, Saito K, Muranaka T, et al. (2018) Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol Biochem 131: 70–77 [DOI] [PubMed] [Google Scholar]
- Nakayasu M, Umemoto N, Ohyama K, Fujimoto Y, Lee HJ, Watanabe B, Muranaka T, Saito K, Sugimoto Y, Mizutani M (2017) A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol 175: 120–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J (1999) Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol 120: 833–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T (2019) Metabolons in plant primary and secondary metabolism. Phytochem Rev 18: 1483–1507 [Google Scholar]
- Ohnishi T, Watanabe B, Sakata K, Mizutani M (2006) CYP724B2 and CYP90B3 function in the early C-22 hydroxylation step of brassinosteroid biosynthetic pathway in tomato. Biosci Biotechnol Biochem 70: 2071–2080 [DOI] [PubMed] [Google Scholar]
- Ohyama K, Okawa A, Moriuchi Y, Fujimoto Y (2013) Biosynthesis of steroidal alkaloids in Solanaceae plants: Involvement of an aldehyde intermediate during C-26 amination. Phytochemistry 89: 26–31 [DOI] [PubMed] [Google Scholar]
- Petersen HW, Mølgaard P, Nyman U, Olsen CE (1993) Chemotaxonomy of the tuber-bearing Solanum species, subsection Potatoe (Solanaceae). Biochem Syst Ecol 21: 629–644 [Google Scholar]
- Roddick JG (1974) The steroidal glycoalkaloid α-tomatine. Phytochemistry 13: 9–25 [Google Scholar]
- Rosati F, Bardazzi I, De Blasi P, Simi L, Scarpi D, Guarna A, Serio M, Racchi ML, Danza G (2005) 5α-reductase activity in Lycopersicon esculentum: Cloning and functional characterization of LeDET2 and evidence of the presence of two isoenzymes. J Steroid Biochem Mol Biol 96: 287–299 [DOI] [PubMed] [Google Scholar]
- Russell DW, Wilson JD (1994) Steroid 5 alpha-reductase: Two genes/two enzymes. Annu Rev Biochem 63: 25–61 [DOI] [PubMed] [Google Scholar]
- Sawai S, Ohyama K, Yasumoto S, Seki H, Sakuma T, Yamamoto T, Takebayashi Y, Kojima M, Sakakibara H, Aoki T, et al. (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 26: 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane PD, Pollier J, Panda S, Szymanski J, Massalha H, Yona M, Unger T, Malitsky S, Arendt P, Pauwels L, et al. (2017) Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat Plants 3: 16205. [DOI] [PubMed] [Google Scholar]
- Thagun C, Imanishi S, Kudo T, Nakabayashi R, Ohyama K, Mori T, Kawamoto K, Nakamura Y, Katayama M, Nonaka S, et al. (2016) Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol 57: 962–975 [DOI] [PubMed] [Google Scholar]
- Thomas JL, Boswell EL, Scaccia LA, Pletnev V, Umland TC (2005) Identification of key amino acids responsible for the substantially higher affinities of human type 1 3β-hydroxysteroid dehydrogenase/isomerase (3β-HSD1) for substrates, coenzymes, and inhibitors relative to human 3β-HSD2. J Biol Chem 280: 21321–21328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto N, Nakayasu M, Ohyama K, Yotsu-Yamashita M, Mizutani M, Seki H, Saito K, Muranaka T (2016) Two cytochrome P450 monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol 171: 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SB, Meyer RS, Whitaker BD, Litt A, Kennelly EJ (2013) A new liquid chromatography–mass spectrometry-based strategy to integrate chemistry, morphology, and evolution of eggplant (Solanum) species. J Chromatogr A 1314: 154–172 [DOI] [PubMed] [Google Scholar]
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, Zhang B (2014) CasOT: A genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30: 1180–1182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.