Abstract
In our previous studies, the production of four bioactive molecules by Streptomyces sp. PAL114 in complex ISP2 broth medium has been described. Three of these molecules belong to the angucycline family. In this study, two novel antibiotics belonging to the same family were produced by strain PAL114 on M2 synthetic medium containing L-tryptophan as precursor. These antibiotics, named mzabimycins A and B, were intracellular and produced only in the presence of L-tryptophan. After four days of culturing PAL114 in the M2 medium, the bioactive compounds were extracted from mycelium with methanol and then analyzed by HPLC on reverse phase C18 column. Two active purplish blue fractions were purified. The chemical structures of these molecules were determined on the basis of spectroscopic and spectrometric analyses (1H and 13C NMR, and mass spectra). They were identified to be novel angucycline derivative antibiotics. The pure molecules showed activity against some pathogenic Gram-positive bacteria which have multiple antibiotic resistance, such as Staphylococcus aureus MRSA 639c and Listeria monocytogenes ATCC 13932.
Keywords: Antimicrobial compounds, Angucycline antibiotics, L-Tryptophan, Synthetic medium, Streptomyces
1. Introduction
Actinobacteria are Gram-positive bacteria with a genomic guanine-cytosine content higher than 55%, and most of them are mycelial. These bacteria are very interesting due to their large capacity to produce secondary metabolites with diversified chemical structures (Kemung et al., 2018, Takahashi and Nakashima, 2018). They are well-known for the production of antibacterial and antifungal antibiotics and are the source of nearly 45% of the known molecules of microbial origin (Solecka et al., 2012) and 70% of actively marketed molecules (Solanki et al., 2008). However, they are also known for the production of diverse bioactive molecules such as antivirals, antiparasitics, immunostimulants and immunosuppressants (Solecka et al., 2012, Flatt et al., 2013, Nakae et al., 2013, Takahashi and Nakashima, 2018).
The genus Streptomyces is known as the producer of the largest number of antibiotics. It produces about 80% of the antibiotics secreted by actinobacteria (Demain, 2006, Demain and Sanchez, 2009). Many of these molecules have found an important therapeutic application (Jose and Jebakumar, 2014), and some of them may have cytostatic and antitumor properties, such as urdamycins and langkocyclines (Drautz et al., 1986, Kalyon et al., 2013).
Considering the increasing resistance of pathogenic microorganisms to antibiotics (Messai et al., 2008, Fair and Tor, 2014, Li and Webster, 2018), and the toxicity of several antibiotic compounds (Berdy, 2005), it is essential to perpetuate research on antibiotics in the hope of finding new effective and less toxic molecules in order to control pathogenic microorganisms.
Our previous works have already demonstrated the richness and biodiversity of actinobacteria in the Saharan soils of Algeria. These studies have led to the discovery of several novel interesting antibiotics (Zitouni et al., 2004a, Yekkour et al., 2015, Khebizi et al., 2018, Lahoum et al., 2019) and several new species of actinobacteria (Aouiche et al., 2015a, Bouras et al., 2015, Chaabane Chaouch et al., 2017). The actinobacterium strain PAL114 was isolated from Saharan soil collected from Ghardaïa province, Mzab region, south Algeria (Aouiche et al., 2014). This strain exhibited a strong antagonistic potential against several microorganisms and was found to be a producer of four bioactive molecules, saquayamycins A and C (Aouiche et al., 2014), and chaetoglobosin A and vineomycin A1 (Aouiche et al., 2015b), which were yellow and extracellular, and were produced in complex ISP2 broth medium (Shirling and Gottlieb, 1966).
In this work, we used a synthetic medium, containing starch and L-tryptophan, in order to control the culture conditions and allow the synthesis of new molecules that we could have missed on complex ISP2 (International Streptomyces Project) medium. We highlight the production of novel purplish blue intracellular antibiotics. These compounds were extracted and purified, and their structure and activity were determined.
2. Materials and methods
2.1. Actinobacterium strain and target-microorganisms
The actinobacterium strain PAL114 was isolated from a Saharan soil in Béni Isguen, Ghardaïa province, Mzab region, southern Algeria (Aouiche et al., 2014). Based on a polyphasic study, this strain was linked to the species Streptomyces griseoflavus (Aouiche et al., 2015b). The strain was cultivated on ISP2 medium (Shirling and Gottlieb, 1966) composed of malt extract (10 g/l), yeast extract (4 g/l), glucose (4 g/l) and agar (20 g/l). The pH of the medium was adjusted to 7.2. The aerial and substrate mycelia were grey and brownish-yellow, respectively. In ISP2 broth, PAL114 strain grows by forming pellets that are pale brownish-yellow in color.
The target-microorganisms included Gram-positive and Gram-negative bacteria, a yeast and filamentous fungi. They are mostly pathogenic or toxigenic for humans, and many of them have multiple antibiotic resistance (Table 1). Indeed, the strains of Staphylococcus aureus MRSA 639c, S. aureus S1, Pseudomonas aeruginosa IPA1 and Candida albicans M3 were isolated from sick patients in Algerian hospitals.
Table 1.
Resistance patterns of target-microorganisms.
| Microorganisms | Resistance to |
|---|---|
| Bacillus subtilis ATCC 6633 | NEO |
| Micrococcus luteus ATCC 9314 | NEO |
| Escherichia coli E52 | ATM, CAZ, CTX, FEP, GEN, PIP, TIC, TOB |
| Pseudomonas aeruginosa IPA1 | AMX, CAR, ERY, GEN, NEO, SPI, SSS, VAN |
| Staphylococcus aureus S1 | CLD, GEN, K, PEN, VAN |
| Staphylococcus aureus MRSA 639c | FA, K, OXA, PEN, TE |
| Listeria monocytogenes ATCC 13932 | OXA, FOS, CAZ, CTX, CXC, FEP, FOX, LIN, CLD, PRL, CIP |
| Candida albicans M3 | CHX, ITR, NYS, TER, TIZ |
| Umbelopsis ramanniana NRRL 1829 | CHX, ITR, TER, TIZ |
| Aspergillus carbonarius M333 | CHX, NYS |
AMX: amoxicillin; ATM: aztreonam; CAR: carbenicillin; CAZ: ceftazidim; CHX: cycloheximide; CIP: ciprofloxacin; CLD: clindamycin; CTX: cefotaxime; CXC: cefotaxime + clavulanic acid; ERY: erythromycin; FEP: cefepime; FA: fusidic acid; FOS: fosfomycin; FOX: cefoxitin; ITR: itraconazole; GEN: gentamicin; K: kanamycin; NEO: neomycin; NYS: nystatine; LIN: lincomycin; OXA: oxacillin; PEN: penicillin G; PRL: pirlimycin ; PIP: piperacillin; SPI: spiramycin; SSS: sulfamide; TE: tetracycline; TER: terbinafine; TIC: ticarcillin; TIZ: thioconazole; TOB: tobramycin; VAN: vancomycin.
2.2. Production, extraction and purification of antibiotics
Strain PAL114 was grown in two synthetic media, M1 and M2. Both of these media contain 10 g starch, 2 g NaCl, 0.5 g KH2PO4, 1 g K2HPO4, 0.5 g MgSO4, 7 H2O and 2 g CaCO3 in 1 l distilled water. However, M1 medium contains 0.25% (w/v) of (NH4)2SO4 and M2 medium contains 0.05% (w/v) of L-tryptophan as nitrogen sources. The final pH of the media was adjusted to 7.2. The production of bioactive compounds was conducted in these two media. A seed culture was prepared with the same medium and used to inoculate (for each medium) sixteen 500 ml Erlenmeyer flasks, each containing 100 ml of culture media. The cultures were incubated on a rotary shaker (250 rpm) for 10 days at 30 °C. The extraction of the active compounds was carried out after centrifugation (5000g, 20 min) of the culture broth to eliminate cells. Half of the cell-free supernatant was extracted with the same volume of dichloromethane and the other half with n-butanol. These two solvents were chosen because they extract the antibiotics (saquayamycins A and C, chaetoglobosin A and vineomycin A1) secreted by strain PAL114 (Aouiche et al., 2014, Aouiche et al., 2015b). Extraction of antibiotics from the mycelium was carried out according to the method of Mechlinski (1978). After centrifugation of the cultures, the mycelium was collected and washed several times with distilled water. Ten grams of wet mycelium were extracted with 500 ml of methanol, stirring for 2 h at room temperature. The organic layers (dichloromethane, n-butanol and methanol extracts) were dehydrated with Na2SO4 and concentrated to dryness by a rotary evaporator under a vacuum at a temperature lower than 40 °C. The residues of each extract were dissolved in 1 ml of methanol and subjected to biological assay (paper disk of 6 mm in diameter, Institute Pasteur) against the ten target-microorganisms listed in Table 1.
The purification of bioactive compounds was performed by Agilent reverse phase HPLC (Agilent 1260) using a C18 column (250 mm × 10 mm; 5 µm). The elution was at a flow rate of 1 ml/min with a continuous linear gradient solvent system from 20 to 100% methanol in water. The detection of products was carried out by UV at 220 nm. In order to detect the active fractions, all peak fractions were collected and tested by the paper disk diffusion method against the ten target-microorganisms (Table 1). Final purification of the active fractions was achieved after the second re-injection in the HPLC under the same conditions.
2.3. Structure determination of the antibiotics
The structure determination of the antibiotics was made with the pure bioactive compounds. The UV spectra were determined with a Shimadzu UV 1605 spectrophotometer. The mass spectra were recorded on a LCQ ion-trap mass spectrometer (Finnigan MAT, San Jose, CA, USA) with a nanospray ion electro-spray ionization (ESI) source (positive and negative ion modes).
1H and 13C NMR spectroscopy were used for the characterization of compounds X3 and X4. NMR samples were prepared by dissolving 5 mg of X3 and X4 compounds in 600 µl of CD3CN. All spectra were recorded on a Bruker Avance 500 spectrometer equipped with a 5 mm triple resonance inverse Z-gradient probe (TBI 1H, 31P, BB). All chemical shifts for 1H and 13C are relative to TMS using 1H (residual) or 13C chemical shifts of the solvent as a secondary standard. The temperature was set at 298 K. All the 1H and 13C signals were assigned on the basis of chemical shifts, spin-spin coupling constants, splitting patterns and signal intensities, and by using 1H–1H COSY45, 1H–13C HSQC and 1H–13C HMBC experiments. Gradient-enhanced 1H COSY45 was realized included 36 scans for per increment. 1H–13C correlation spectra using a gradient-enhanced HSQC sequence (delay was optimised for 1JCH of 145 Hz) was obtained with 200 scans per increment. Gradient-enhanced HMBC was performed allowing 62.5 ms for long-range coupling evolution (340 scans were accumulated). Typically, 2048 t2 data points were collected for 256 t1 increments.
2.4. Determination of minimum inhibitory concentrations
Minimum inhibitory concentrations (MICs) of pure bioactive compounds were investigated using the conventional agar dilution method of Oki et al. (1990) against the ten target-microorganisms (Table 1). The target bacterial strains were inoculated onto Mueller Hinton medium and the fungal strains on Sabouraud medium. The media contained different concentrations of each active compound (1, 2, 3, 5, 10, 15, 20, 30, 40, 50, 60, 80 and 100 µg/ml). After a growth period of 24–48 h at 37 °C for bacteria and 48–72 h at 28 °C for fungi, the plates were examined for growth and the lowest antibiotic concentration that inhibited the growth of each organism (MIC) was determined. Mueller Hinton and Sabouraud media, without active compound and inoculated with target organisms, were used as control treatments. All the experiments were performed in duplicate.
3. Results and discussion
3.1. Production and purification of the antibiotics
After four days of fermentation in M1 and M2 media, the culture filtrates were separated from the mycelial biomass by centrifugation, and then extracted with dichloromethane and n-butanol. The extracts of culture filtrates from both media were brownish yellow and inactive against all target-microorganisms. The mycelial biomass and the corresponding methanolic extracts were pale brownish yellow from M1 medium, which contains NH4SO4, whereas they were dark purplish blue from M2 medium containing L-tryptophan. Furthermore, this dark purplish blue methanolic extract was active against Gram-positive bacteria (B. subtilis ATCC 6633, M. luteus ATCC 9314, L. monocytogenes ATCC 13932, S. aureus MRSA 639c and S. aureus S1) but not against Gram-negative bacteria (E. coli E52, P. aeruginosa IPA1), yeast (C. albicans M3) and filamentous fungi (A. carbonarius M333 and U. ramanniana NRRL 1829), whereas, the extract from M1 medium was found to be inactive against all target-microorganisms. The active dark purplish blue extract, obtained from M2 medium, was analyzed by HPLC. Two active fractions against Gram-positive bacteria (cited above) were detected and named X3 (retention time, 64.7 min) and X4 (retention time, 65.19 min), with the latter being predominant (Supplementary data – Fig. 1S).
These intracellular and antimicrobial fractions are produced only in the presence of L-tryptophan (in M2 but not in M1 medium). L-Tryptophan seems to play an essential role in the biosynthesis of the two bioactive molecules and could therefore be a precursor of the two compounds. However, we have not detected, in any extract (from supernatant and mycelium), the saquayamycins A and C, or vineomycin A1 (angucycline antibiotics), or chaetoglobosin A. These molecules, which are yellow and active against B. subtilis ATCC 6633 and S. aureus MRSA 639c, were detected only in the ISP2 (complex medium) culture filtrate of strain PAL114 (Aouiche et al., 2014, Aouiche et al., 2015b).
Through these results, it appears that strain PAL114 produces bioactive compounds with different chemical structures depending on the culture conditions. Several previous works showed the ability of strains to produce many secondary metabolites with related chemical structures depending on the available precursors. Indeed, the results of Rohr et al. (1989) on the biosynthesis of urdamycins (angucycline antibiotics) by Streptomyces fradiae showed that this species used different labeled precursors to produce different urdamycin molecules. Thus, this species uses the 2-methyl-tryptophan as precursor to produce urdamycin D, the tyrosine to produce urdamycin C and the acetate to produce urdamycin A. All these molecules have the same central chromophore. Similar results were obtained with Saccharothrix algeriensis NRRL B-24137, which produces five dithiolopyrrolone antibiotics in ISP2 medium (Lamari et al., 2002) and several other dithiolopyrrolone molecules, in semi-synthetic medium, induced by the addition of organic acids and amino acids as precursors (Bouras et al., 2008, Merrouche et al., 2010, Merrouche et al., 2011). Lam et al. (2001) reported a similar approach of using precursor-directed biosynthesis to produce novel fluoroindolocarbazoles A and B by adding DL-6-fluorotryptophan, and fluoroindolocarbazole C by adding DL-5-fluorotryptophan in cultures of Saccharothrix aerocolonigenes ATCC 39243. This is particularly interesting research strategy for producing new antibiotic molecules.
3.2. Elucidation of the structure of the antibiotics
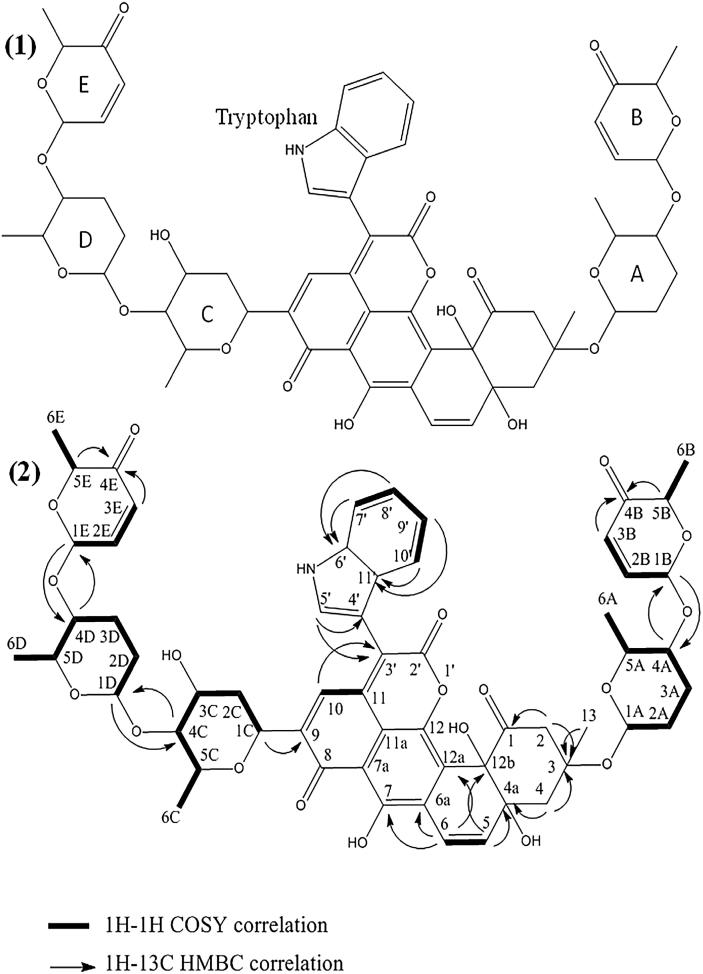
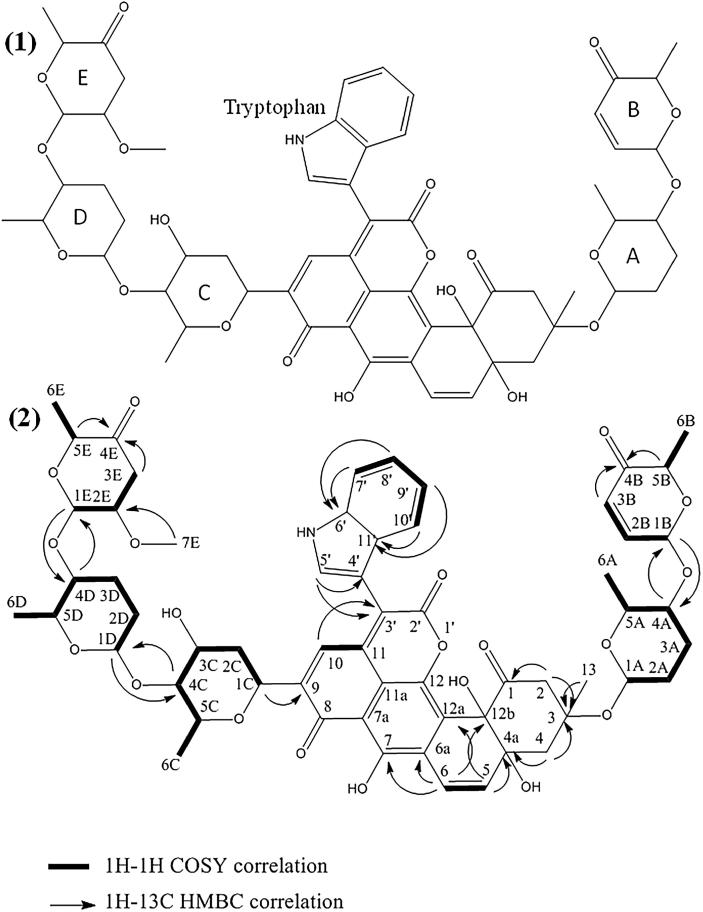
The structure of the compounds X3 and X4 was determined by NMR and mass spectrometry. The results showed that these two compounds are novel antibiotics belonging to the angucycline family. They were named mzabimycin A (for the major compound X4) and mzabimycin B (for the minor compound X3), with reference to Mzab region, southern Algeria, the source of the soil from which Streptomyces strain PAL114 was isolated. The structure of mzabimycins A and B is shown in Fig. 1, Fig. 2 respectively.
Fig. 1.
Structure of mzabimycin A (X4 compound) (1) and HMBC and COSY correlations (2). A and D, rhodinose; B and E, aculose; C, olivose.
Fig. 2.
Structure of mzabimycin B (X3 compound) (1) and HMBC and COSY correlations (2). A and D, rhodinose; B, aculose; C, olivose; E, reduced and methoxylated aculose.
Mzabimycin A (X4) was obtained as a purplish blue powder. The UV–visible spectrum (Supplementary data – Fig. 2S) showed the maximal absorbance at 218, 325 and 575 nm. The ESIMS spectrum (Supplementary data – Fig. 3S) contained an ion peak at m/z 1088.31 [M - H]−. Thus, the molecular weight of this compounds was M = 1089. The 1H and 13C chemical shifts are given in Table 2. The 13C, HSQC and HMBC spectra, showed 59 carbon signals. It was possible to discern 4 ketone group (δc 186.72 to 207.23), 3 hydroxyl group (δc 78.90 το 155.90), 11 ether function (δc 67.51 to 99.05), 24 sp2–hybridized carbons (δc from 107.00 to 144.00) and 11 sp3-hybridized carbons (δc 14.04 to 42.61). The 2D 1H–1H and 1H–13C experiments and especially the long range 1H–13C couplings observed in the HMBC spectrum (see Fig. 1) permitted to established the connectivity between all the groups of the molecule.
Table 2.
1H and 13C NMR data assignments of X3 (mzabimycin B) and X4 (mzabimycin A) in CD3CN at 298 K. See Fig. 1, Fig. 2 for numbering of hydrogen and carbon atoms.
|
1H and 13C number |
1H chemical shift, ppm |
13C chemical shift, ppm |
||||
|---|---|---|---|---|---|---|
| X3 | X4 | X3 | X4 | |||
| 1 | – | – | – | – | 206.90 | 206.00 |
| 2 | 3.03 | (m,2H | 3.03 | (m,2H) | 49.87 | 49.82 |
| 3 | – | – | – | – | 81.00 | 81.46 |
| 4 | 1.97–2.31 | (d,15.6,2H) | 1.97–2.30 | (d,15.6,2H) | 43.43 | 43.50 |
| 4a | – | – | – | – | 79.70 | 79.80 |
| 5 | 6.09 | (d,10.0,1H | 6.09 | (d,10.0,1H) | 137.30 | 138.00 |
| 6 | 7.03 | (d,10.0,1H | 7.03 | (d,10.0,1H) | 118.00 | 118.00 |
| 6a | – | – | – | – | 123.30 | 123.50 |
| 7 | – | – | – | – | 155.90 | 155.90 |
| 7a | – | – | – | – | 114.9 | 114.8 |
| 8 | – | – | – | – | 186.27 | 186.84 |
| 9 | – | – | – | – | 142.80 | 148.89 |
| 10 | 8.05 | (s,1H) | 8.05 | (s,1H) | 134.93 | 134.93 |
| 11 | – | – | – | – | 118.60 | 116.60 |
| 11a | – | – | – | – | 127.25 | 127.00 |
| 12 | – | – | – | – | 142.8 | 142.6 |
| 12a | – | – | – | – | 128.10 | 127.00 |
| 12b | – | – | – | – | 78.90 | 79.80 |
| 13 | 1.33 | (m,3H | 1.32 | (m,3H) | 24.83 | 24.80 |
| 1′ | – | – | – | – | – | – |
| 2′ | – | – | – | – | 159.20 | 159.20 |
| 3′ | – | – | – | – | 131.00 | 131.00 |
| 4′ | – | – | – | – | 107.00 | 107.00 |
| 5′ | 7.83 | (s,1H) | 7.83 | (s,1H) | 131.24 | 131.24 |
| 6′ | – | – | – | – | 136.00 | 136.00 |
| 7′ | 7.59 | (d,7.8,1H) | 7.59 | (d,7.8,1H) | 112.00 | 112.00 |
| 8′ | 7.29 | (t,7.8,1H) | 7.29 | (t,7.8,1H) | 123.00 | 123.00 |
| 9′ | 7.19 | (t,7.8,1H) | 7.19 | (t,7.8,1H) | 121.00 | 121.00 |
| 10′ | 7.53 | (d,7.8,1H) | 7.53 | (d,7.8,1H) | 120.20 | 120.20 |
| 11′ | – | – | – | – | 127.00 | 127.00 |
| 1A | 5.26 | (m,1H) | 5.25 | (m,1H) | 91.88 | 92.00 |
| 2A | 1.98 | (m,2H) | 1.98 | (m,2H) | 24.04 | 24.04 |
| 3A | 1.94–1.98 | (m,2H) | 1.94–1.98 | (m,2H) | 24.25 | 24.25 |
| 4A | 3.72 | (m,1H) | 3.71 | (m,1H) | 76.18 | 76.00 |
| 5A | 4.14 | (m,1H) | 4.14 | (m,1H) | 66.56 | 66.72 |
| 6A | 1.24 | (d,6.5,3H) | 1.24 | (d,6.5,3H) | 16.52 | 16.47 |
| 1B | 5.30 | (dd,9.0–3.4,1H) | 5.30 | (dd,9.0–3.4,1H) | 95.00 | 95.00 |
| 2B | 6.07 | (d,9.0,1H) | 6.07 | (d,9.0,1H) | 126.48 | 128.48 |
| 3B | 7.00 | (d,9.0,1H) | 7.00 | (d,9.0,1H) | 144.00 | 144.00 |
| 4B | – | – | – | – | 197.00 | 197.00 |
| 5B | 4.60 | (d,4.0, 1H) | 4.60 | (d,4.0, 1H) | 70.14 | 70.14 |
| 6B | 1.31 | (d,4.0, 3H) | 1.31 | (d,4.0, 3H) | 14.43 | 14.43 |
| 1C | 4.73 | (d,11.0,1H) | 4.73 | (d,11.0,1H) | 70.65 | 70.65 |
| 2C | 1.26–2.41 | (m,2H) | 1.72–2.41 | (m,2H) | 39.18 | 39.18 |
| 3C | 3.74 | (m,1H) | 3.74 | (m,1H) | 70.96 | 70.78 |
| 4C | 2.85 | (m,1H) | 2.86 | (m,1H) | 87.81 | 87.78 |
| 5C | 3.43 | (m,1H) | 3.43 | (m,1H) | 74.50 | 74.55 |
| 6C | 1.07 | (dd,6.0–1.8,3H) | 1.06 | (dd,6.0–1.8,3H) | 17.86 | 17.59 |
| 1D | 4.89 | (m,1H) | 4.88 | (m,1H) | 99.05 | 99.18 |
| 2D | 1.60–1.94 | (m,2H) | 1.60–1.99 | (m,2H) | 24.50 | 24.50 |
| 3D | 1.90–1.93 | (m,2H) | 1.90–1.93 | (m,2H) | 24.25 | 24.25 |
| 4D | 3.75 | (m,1H) | 3.72 | (m,1H) | 75.42 | 76.00 |
| 5D | 4.23 | (m,1H) | 4.22 | (m,1H) | 67.51 | 67.44 |
| 6D | 1.93 | (dd,8.7–2.0,3H) | 1.71 | (dd,7.9–3.5,1H) | 16.16 | 16.18 |
| 1E | 4.96 | (m,1H) | 5.30 | (m,1H) | 99.40 | 95.00 |
| 2E | 3.81 | (m,1H) | 6.07 | (d,9.0,1H) | 79.00 | 126.48 |
| 3E | 2.54–2.83 | (dt,15.4–4.8,2H) | 7.00 | (d,9.0,1H) | 40.13 | 144.00 |
| 4E | – | – | – | – | 207.23 | 197.00 |
| 5E | 4.29 | (m,1H) | 4.60 | (m,1H) | 71.53 | 70.14 |
| 6E | 1.19 | (d,3.6,3H) | 1.30 | (m,3H) | 13.94 | 14.43 |
| 7E | 1.45 | (d,5.5,3H) | – | – | 15.50 | – |
The numbering of the atoms was made like that of antibiotics urdamycin D (Rohr et al., 1989) and langkocyclines B1 and B2 (Kalyon et al., 2013), which are closely related to the structures of mzabimycins A and B.
The NMR data (Supplementary data – Fig. 4S) showed that mzabimycin A represents a new antibiotic belonging to the angucycline family. This compound has a central chromophore with L-tryptophan linked to carbon number 3′, and five sugars, two rhodinoses, two aculoses and one olivose.
Mzabimycin B was obtained as a purplish blue powder. The UV–visible spectrum (Supplementary data – Fig. 2S) showed the maximal absorbance at 218, 280, 330 and 575 nm. The ESIMS spectrum (Supplementary data – Fig. 3S) contained an ion peak at m/z 1120.36 [M - H]−. Thus, the molecular weight of this compound was M = 1121. The 1H and 13C chemical shifts are given in Table 2. The HSQC and HMBC spectra, showed 59 carbon signals. It was possible to discern 4 ketone group (δc 186.72 to 207.23), 3 hydroxyl group (δc 78.90 το 155.90), 11 ether function (δc 67.51 to 99.05), 22 sp2–hybridized carbons (δc from 107.00 to 142.89) and 13 sp3-hybridized carbons (δc 14.04 to 42.61). The 2D 1H–1H and 1H–13C experiments and especially the long range 1H–13C couplings observed in the HMBC spectrum (see Fig. 2) permitted to establish the connectivity between all the groups of the molecule.
The NMR data (Supplementary data – Fig. 5S) showed that mzabimycin B represent a novel antibiotic belonging to the angucycline family. This compound has a central chromophore typical of angucycline compounds, but with L-tryptophan linked to carbon number 3′, five sugars, two rhodinoses, one olivose, one aculose and one reduced and methoxylated aculose. These sugars were linked to the central chromophore at carbon number 3 (rhodinose and aculose) and carbon number 9 (olivose, rhodinose and reduced and methoxylated aculose). It differs from mzabimycin A only by the second aculose molecule that is reduced and methoxylated.
The structure of mzabimycins A and B do not correspond to any structure reported in the literature, notably in the www.sciencefinder.com and www.chemspider.com databases, or antibiotics described in The Dictionary of Natural Products (Buckingham, 1997), or in Berdy’s review of bioactive microbial metabolites (Berdy, 2005). The mzabimycins A and B are, therefore, two new angucyclines that possess a chromophore containing L-tryptophan and osidic derivatives. These two compounds have the same central chromophore, which is similar to that of the urdamycin D (Rohr et al., 1989) and the langkocyclines B1 and B2 (Kalyon et al., 2013). Furthermore, the urdamycin D (Drautz et al., 1986) and the langkocyclines B1 and B2 (Kalyon et al., 2013) are purple blue and have L-tryptophan linked to carbon number 3′ of the central chromophore, like mzabimycins A and B. However, langkocycline B1 has four sugars (two rhodinoses and two olivoses) and langkocycline B2 has five sugars (three rhodinoses and two olivoses); these sugars are linked to carbon number 12b of the central chromophore. Urdamycin D contained one olivose linked to carbon number 12b of the central chromophore and two olivoses and one rhodinose linked to carbon number 9 of the chromophore. Mzabimycins A and B differ from these antibiotics in the number and the composition of sugars (presence of aculose and reduced and methoxylated aculose) and in the linkage to the central chromophore. Mzabimycins A and B contain in their structure identical sugars to those of vineomycin A1, a yellow extracellular angucycline also produced by strain PAL114, not in synthetic media M1 and M2, but in complex ISP2 medium (Aouiche et al., 2015b). However, there are some differences in the central chromophore structure and the absence of L-tryptophan. These results showed some similarities between the biosynthesis processes of vineomycin A1 and mzabimycins A and B.
The production of antibiotics belonging to the families of angucyclines and anthracyclines (close to angucyclines) has already been demonstrated in some strains of actinobacteria isolated from Saharan soils as strain PAL114. This is the case of the antibiotic R2 secreted by Streptosporangium sp. Sg3 (Boudjella et al., 2010) and mutactimycins C and PR secreted by Saccharothrix sp. SA103 (Zitouni et al., 2004b).
3.3. Minimum inhibitory concentrations
Minimum inhibitory concentrations (MICs) of mzabimycin A (X4) and mzabimycin B (X3), purified by HPLC, are summarized in Table 3. The results showed that these compounds have very similar activity directed only against Gram-positive bacteria. The strains of Micrococcus luteus (MIC, 15 μg/ml for mzabimycin A and B) and Listeria monocytogenes ATCC 13932 (MICs, 20 μg/ml for mzabimycin B and 40 μg/ml for mzabimycin A) were the most sensitive. The other Gram-positive bacteria, including the two strains of Staphylococcus aureus (60–80 μg/ml) and Bacillus subtilis ATCC 6633 (50 μg/ml), were found to be less sensitive. All tested Gram-negative bacteria, yeasts and filamentous fungi were resistant (>100 μg/ml).
Table 3.
Minimum inhibitory concentrations (MICs) of X3 (mzabimycin B) and X4 (mzabimycin A) produced by Streptomyces sp. PAL114 against several target microorganisms.
| Target microorganisms | MICs (µg/ml)* | |
|---|---|---|
| X3 | X4 | |
| Bacillus subtilis ATCC 6633 | 50 | 50 |
| Micrococcus luteus ATCC 9314 | 15 | 15 |
| Staphylococcus aureus MRSA 639c | 60 | 60 |
| Staphylococcus aureus S1 | 80 | 80 |
| Listeria monocytogenes ATCC 13932 | 20 | 40 |
| Escherichia coli E52 | >100 | >100 |
| Pseudomonas aeruginosa IPA1 | >100 | >100 |
| Candida albicans M3 | >100 | >100 |
| Umbelopsis ramanniana NRRL 1829 | >100 | >100 |
| Aspergillus carbonarius M333 | >100 | >100 |
MIC values represent the mean of two replicates;
It should be noted that the antimicrobial activity of urdamycin D (Drautz et al., 1986) and langkocyclines B1 and B2 (Kalyon et al., 2013), which are angucyclines close in structure to mzabimycins A and B, is also directed only against Gram-positive bacteria.
Angucycline antibiotics are a group of biologically active compounds with interesting activities including antibacterial, antifungal and antiviral (Kharel et al., 2012), enzyme inhibitory (Eguchi et al., 2017), and platelet aggregation inhibitory properties (Kawashima et al., 1989). Therefore, they are cytotoxic, and some molecules were used as anticancer agents in chemotherapy (Abdelfattah et al., 2008). Yu and O'Doherty (2008) showed the role of vineomycin B2 trisaccharide, consisting of aculose, rhodinose and olivose, in anticancer activity (against a panel of cancer cell lines). The same authors also showed the role of antibiotic PI-080 trisaccharide, consisting of aculose and two olivoses, in anticoagulant activity.
4. Conclusion
Strain PAL114, related to Streptomyces griseoflavus, produced two novel intracellular antibiotics, mzabimycins A and B, in synthetic medium containing L-tryptophan as a precursor. These antibiotics, which belonged to the angucycline family, showed activity against some pathogenic Gram-positive bacteria with multiple antibiotic resistance. Considering the important biological activities of angucyclines, it would be interesting to explore other properties of these new angucyclines molecules, as for example anticancer activity, and in vivo evaluation studies which could prove promising.
Declaration of Competing Interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2019.06.004.
Contributor Information
Florence Mathieu, Email: Florence.Mathieu@ensat.fr.
Nasserdine Sabaou, Email: sabaou@yahoo.fr.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abdelfattah M.S., Kharel M.K., Hitron J.A., Baig I., Rohr J. Moromycins A and B isolation and structure elucidation of C-glycosylangucycline-type antibiotics from Streptomyces sp. KY002. J. Nat Prod. 2008;71(9):1569–1573. doi: 10.1021/np800281f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouiche A., Bijani C., Zitouni A., Mathieu F., Sabaou N. Antimicrobial activity of saquayamycins produced by Streptomyces sp. PAL114 isolated from a Saharan soil. J. Mycol. 2014;24(2):e17–e23. doi: 10.1016/j.mycmed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Aouiche A., Bouras N., Mokrane S., Zitouni A., Schumann P., Spröer C., Sabaou N., Klenk H.P. Actinokineospora mzabensis sp. nov., a novel actinomycete isolated from Saharan soil. Antonie van Leeuwenhoek. 2015;107(1):291–296. doi: 10.1007/s10482-014-0328-8. [DOI] [PubMed] [Google Scholar]
- Aouiche A., Meklat A., Bijani C., Zitouni A., Sabaou N., Mathieu F. Production of vineomycin A1 and chaetoglobosin A by Streptomyces sp. PAL114. Ann. Microbiol. 2015;65(3):1351–1359. [Google Scholar]
- Berdy J. Bioactive microbial metabolites. J. Antibiot. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Boudjella H., Zitouni A., Coppel Y., Mathieu F., Monje M., Sabaou N. Antibiotic R2, a new angucyclinone compound from Streptosporangium sp. Sg3. J. Antibiot. 2010;63(12):709–711. doi: 10.1038/ja.2010.111. [DOI] [PubMed] [Google Scholar]
- Bouras N., Merrouche R., Lamari L., Mathieu F., Sabaou N., Lebrihi A. Precursor directed biosynthesis of new dithiolopyrrolone analogs by Saccharothrix algeriensis NRRL B-24137. Process Biochem. 2008;43(11):1244–1252. [Google Scholar]
- Bouras N., Meklat A., Zitouni A., Mathieu F., Schumann P., Spröer C., Sabaou N., Klenk H.P. Nocardiopsis algeriensis sp. nov., an alkalitolerant actinomycete isolated from Saharan soil. Antonie van Leeuwenhoek. 2015;107(2):313–320. doi: 10.1007/s10482-014-0329-7. [DOI] [PubMed] [Google Scholar]
- Buckingham J. Chapman and Hall/CRC; UK: 1997. Dictionary of natural products. [Google Scholar]
- Chaabane Chaouch F., Bouras N., Mokrane S., Bouznada K., Zitouni A., Schumann P., Pӧtter G., Spröer C., Klenk H.P., Sabaou N. Planomonospora algeriensis sp. nov., an actinobacterium isolated from a Saharan soil of Algeria. Antonie van Leeuwenhoek. 2017;110(2):245–252. doi: 10.1007/s10482-016-0795-1. [DOI] [PubMed] [Google Scholar]
- Demain A.L. From natural products discovery to commercialization: a success story. J. Ind. Microbiol. Biotechnol. 2006;33(7):486–495. doi: 10.1007/s10295-005-0076-x. [DOI] [PubMed] [Google Scholar]
- Demain A.L., Sanchez S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009;62(1):5–16. doi: 10.1038/ja.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drautz H., Zähner H., Rohr J., Zeeck A. Metabolic products of microorganisms. 234. Urdamycins, new angucycline antibiotics from Streptomyces fradiae. I. Isolation, characterization and biological properties. J. Antibiot. 1986;39(12):1657–1669. doi: 10.7164/antibiotics.39.1657. [DOI] [PubMed] [Google Scholar]
- Eguchi Y., Okajima T., Tochio N., Inukai Y., Shimizu R., Ueda S., Shinya S., Kigawa T., Fukamizo T., Igarashi M., Utsumi R. Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases. J. Antibiot. 2017;70(3):251–258. doi: 10.1038/ja.2016.151. [DOI] [PubMed] [Google Scholar]
- Fair R.J., Tor Y. Antibiotics and bacterial resistance in the 21st Century. Perspect. Medicin. Chem. 2014;6(6):25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt P., Wu X., Perry S., Mahmud T. Genetic insights into pyralomicin biosynthesis in Nonomuraea spiralis IMC A-0156. J. Nat Prod. 2013;76(5):939–946. doi: 10.1021/np400159a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose P.A., Jebakumar S.R.D. Unexplored hypersaline habitats are sources of novel actinomycetes. Front. Microbiol. 2014;5:242. doi: 10.3389/fmicb.2014.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima A., Kishimura Y., Tamai M., Hanada K. New platelet aggregation inhibitors. Chem. Pharm. Bull. 1989;37(12):3429–3431. doi: 10.1248/cpb.37.3429. [DOI] [PubMed] [Google Scholar]
- Kalyon B., Tan G.Y.A., Pinto J.M., Foo C.Y., Wiese J., Imhoff J.F., Süssmuth R.D., Sabaratnam V., Fiedler H.P. Langkocyclines: novel angucycline antibiotics from Streptomyces sp. Acta 3034. J. Antibiot. 2013;66(10):609–616. doi: 10.1038/ja.2013.53. [DOI] [PubMed] [Google Scholar]
- Kemung H.M., Tan L.T.H., Khan T.M., Chan K.G., Pusparajah P., Goh B.H., Lee L.H. Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 2018;9:2221. doi: 10.3389/fmicb.2018.02221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharel M.K., Pahari P., Shepherd M.D., Tibrewal N., Nybo S.E., Shaaban K.A., Rohr J. Angucyclines: biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012;29:264–325. doi: 10.1039/c1np00068c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khebizi N., Boudjella H., Bijani C., Bouras N., Klenk H.P., Pont F., Mathieu F., Sabaou N. Oligomycins A and E, major bioactive secondary metabolites produced by Streptomyces sp. strain HG29 isolated from a Saharan soil. J. Mycol. 2018;28(1):150–160. doi: 10.1016/j.mycmed.2017.10.007. [DOI] [PubMed] [Google Scholar]
- Lahoum A., Sabaou N., Bijani C., Bouras N., Pont F., Snini S.P., Mathieu F. Antimicrobial activities of novel bipyridine compounds produced by a new strain of Saccharothrix isolated from Saharan soil. Saudi Pharm. J. 2019;27(1):56–65. doi: 10.1016/j.jsps.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K.S., Schroeder D.R., Veitch J.M., Colson K.L., Matson J.A., Rose W.C., Doyle T.W., Forenza S. Production, isolation and structure determination of novel fluoroindolocarbazoles from Saccharothrix aerocolonigenes ATCC39243. J. Antibiot. 2001;54(1):1–9. doi: 10.7164/antibiotics.54.1. [DOI] [PubMed] [Google Scholar]
- Lamari L., Zitouni A., Boudjella H., Badji B., Sabaou N., Lebrihi A. New dithiolopyrrolone antibiotics from Saccharothrix sp. SA 233 I. Taxonomy, production, isolation and biological properties. J. Antibiot. 2002;55(8):696–701. doi: 10.7164/antibiotics.55.696. [DOI] [PubMed] [Google Scholar]
- Li B., Webster T.J. Bacteria antibiotic resistance: new challenges and opportunities for implant-associated orthopaedic infections. J. Orthop Res. 2018;36(1):22–32. doi: 10.1002/jor.23656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrouche R., Bouras N., Coppel Y., Mathieu F., Monje M.C., Sabaou N., Lebrihi A. Dithiolopyrrolone antibiotic formation induced by adding valeric acid to the culture broth of Saccharothrix algeriensis. J. Nat. Prod. 2010;73(6):1164–1166. doi: 10.1021/np900808u. [DOI] [PubMed] [Google Scholar]
- Merrouche R., Bouras N., Coppel Y., Mathieu F., Sabaou N., Lebrihi A. New dithiolopyrrolone antibiotics induced by adding sorbic acid to the culture medium of Saccharothrix algeriensis NRRL B-24137. FEMS Microbiol. Lett. 2011;318(1):41–46. doi: 10.1111/j.1574-6968.2011.02246.x. [DOI] [PubMed] [Google Scholar]
- Messai Y., Iabadence H., Benhassine T., Alouache S., Tazir M., Guatier V., Arlet G., Bakour R. Prevalence and characterization of extended-spectrum β-lactamases in Klebsiella pneumoniae in Algiers hospitals (Algeria) Pathol. Biol. 2008;56(5):319–325. doi: 10.1016/j.patbio.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Mechlinski W. The polyene antifungal antibiotics. In: Laskin A.I., Lechevalier H.A., editors. Vol. III. CRC Press; 1978. pp. 93–107. (Handbook of microbiology). [Google Scholar]
- Nakae K., Kurata I., Kojima F., Igarashi M., Hatano M., Sawa R. Sacchathridine, A, a prostaglandin release inhibitor from Saccharothrix sp. J. Nat. Prod. 2013;76(4):720–722. doi: 10.1021/np3006327. [DOI] [PubMed] [Google Scholar]
- Oki T., Tenmyo O., Tomatsu K., Kamei H. Pradimicins A, B and C: new antifungal antibiotics. II. In vitro and in vivo biological activities. J. Antibiot. 1990;43(7):763–770. doi: 10.7164/antibiotics.43.763. [DOI] [PubMed] [Google Scholar]
- Rohr J., Beale J.M., Floss H.G. Urdamycins new angucycline antibiotics from Streptomyces fradiae. IV. Biosynthetic studies of urdamycins A ∼ D. J. Antibiot. 1989;41(7):1151–1157. doi: 10.7164/antibiotics.42.1151. [DOI] [PubMed] [Google Scholar]
- Shirling B., Gottlieb D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966;16(3):3313–3340. [Google Scholar]
- Solanki R., Kahanna M. Bioactive compounds from marine actinomycetes. Indian J. Microbiol. 2008;48(4):410–431. doi: 10.1007/s12088-008-0052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecka J., Zajko J., Postek M., Rajnisz A. Biologically active secondary metabolites from actinomycetes. Cent Eur. J. Biol. 2012;7(3):373–390. [Google Scholar]
- Takahashi Y., Nakashima T. Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics. 2018;7(3):74. doi: 10.3390/antibiotics7020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekkour A., Meklat A., Bijani A., Toumatia O., Errakhi R., Lebrihi A., Mathieu F., Zitouni A., Sabaou N. A novel hydroxamic acid-containing antibiotic produced by a Saharan soil-living Streptomyces strain. Lett. Appl. Microbiol. 2015;60(6):589–596. doi: 10.1111/lam.12412. [DOI] [PubMed] [Google Scholar]
- Yu X., O’Doherty A. De novo asymmetric synthesis and biological evaluation of the trisaccharide portion of PI-080 and vineomycin B2. Org. Lett. 2008;10(20):4529–4532. doi: 10.1021/ol801817f. [DOI] [PubMed] [Google Scholar]
- Zitouni A., Boudjella H., Mathieu F., Sabaou N., Lebrihi A. Mutactimycin PR, a new anthracycline antibiotic from Saccharothrix sp. SA 103. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 2004;57(6):367–372. doi: 10.7164/antibiotics.57.367. [DOI] [PubMed] [Google Scholar]
- Zitouni A., Lamari L., Boudjella H., Badji B., Sabaou N., Gaouar A., Mathieu F., Lebrihi A., Labeda D.P. Saccharothrix algeriensis sp. nov., isolated from Saharan soil. Int. J. Syst. Evol. Microbiol. 2004;54(4):1377–1381. doi: 10.1099/ijs.0.02679-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.