Abstract
Biofilms provide bacteria with protection from environmental stresses and host immune defenses. The pathogenic marine bacterium Vibrio vulnificus forms biofilms and colonizes environmental niches such as oysters. The cabABC operon encodes an extracellular matrix protein CabA and the corresponding type I secretion system, which are essential for biofilm and rugose colony development of V. vulnificus. In this study, molecular biological analyses revealed the roles of three transcriptional regulators BrpR, BrpT, and BrpS in the regulatory pathway for the cabABC operon. BrpR induces brpT and BrpT in turn activates the cabABC operon in a sequential cascade, contributing to development of robust biofilm structures. BrpT also activates brpS, but BrpS represses brpT, constituting a negative feedback loop that stabilizes brpT expression. BrpT and BrpS directly bind to specific sequences upstream of cabA, and they constitute a feedforward loop in which BrpT induces brpS and together with BrpS activates cabABC, leading to precise regulation of cabABC expression. Accordingly, BrpS as well as BrpT plays a crucial role in complete development of rugose colonies. This elaborate network of three transcriptional regulators BrpR, BrpT, and BrpS thus tightly controls cabABC regulation, and contributes to successful development of robust biofilms and rugose colonies in V. vulnificus.
Keywords: Vibrio vulnificus, biofilm, extracellular matrix protein, colony morphology, transcriptional regulator
Introduction
Bacteria exist as free-living planktonic cells or surface-attached complex biofilm communities in the environment (Hall-Stoodley et al., 2004). Biofilms provide bacteria with protection from various stresses such as nutrient limitation and desiccation, as well as antimicrobial agents and host immune defenses during infection (Flemming et al., 2016). Thus, biofilm formation is important for niche colonization and development of persistent bacterial infections (Costerton et al., 1999; Yildiz and Visick, 2009). Biofilm formation involves sequential developmental stages comprised of initial surface attachment, microcolony formation, maturation into three-dimensional biofilms, and dispersal of bacterial cells from mature biofilms (Watnick and Kolter, 2000). Mature biofilms are specialized and highly differentiated communities of bacteria sheathed in an extracellular polymeric matrix (Flemming and Wingender, 2010). The biofilm matrix mainly consists of polysaccharides, proteins, nucleic acids, and lipids; the spatially organized matrix provides beneficial functions for biofilm communities such as resource capture, enzyme retention, and influx inhibition of toxic chemicals (Flemming et al., 2016). Variations in biofilm matrix production can alter the colony morphology, affecting the rugosity and opacity of the apparent colony morphotypes (Yildiz and Visick, 2009; Serra et al., 2013; Park et al., 2015a).
Bis-(3′–5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) is a universal bacterial second messenger and a key player in the molecular decision between the planktonic and biofilm lifestyles (Hengge, 2009). It is synthesized by diguanylate cyclases (DGCs) containing the GGDEF domain and degraded by c-di-GMP-specific phosphodiesterases (PDEs) harboring the EAL or HD-GYP domain (Krasteva et al., 2012). The DGCs and PDEs often carry sensory input domains, and thus control the intracellular c-di-GMP levels precisely in response to various extracellular signals such as light, oxygen, nitric oxide, and nutrient levels (Boyd and O’Toole, 2012; McDougald et al., 2012). Diverse effector components, including regulatory proteins and riboswitches, bind c-di-GMP molecules and regulate downstream target pathways at the transcriptional, post-transcriptional, and post-translational levels (Hengge, 2009; Conner et al., 2017). Generally, increased intracellular c-di-GMP levels lead to stimulation of biofilm formation and repression of motility and virulence (Hengge, 2009).
The pathogenic marine bacterium Vibrio vulnificus is a causative agent of food-borne diseases ranging from gastroenteritis to life-threatening septicemia (Oliver, 2015). Raw oysters serve as the primary infection route of V. vulnificus, and the pathogen colonizes and persists in oyster populations forming biofilms (Froelich and Oliver, 2013; Park et al., 2016; Pu and Rowe-Magnus, 2018; Pu et al., 2018). Biofilm formation of V. vulnificus is induced by elevated levels of intracellular c-di-GMP, which leads to significant changes in gene expression profiles (Nakhamchik et al., 2008; Park et al., 2015b; Chodur and Rowe-Magnus, 2018). Among the genes positively regulated by elevated c-di-GMP levels, the brp locus and the cabABC operon are essential for c-di-GMP-induced biofilm phenotypes (Guo and Rowe-Magnus, 2010; Park et al., 2015a). The brp locus consists of nine genes (brpABCDFHIJK), which are responsible for production and secretion of exopolysaccharide (Guo and Rowe-Magnus, 2010). The cabABC operon encodes a calcium-binding matrix protein CabA, which is essential for the development of biofilm structure and rugose colony morphology, along with CabB and CabC constituting a type I secretion system for CabA (Park et al., 2015a).
C-di-GMP-dependent biofilm formation by V. vulnificus also requires two transcriptional regulators BrpR and BrpT (Guo and Rowe-Magnus, 2010). BrpR and BrpT are homologs of Vibrio cholerae VpsR and VpsT, respectively, which are known to bind c-di-GMP directly and regulate transcription of biofilm-associated genes in V. cholerae (Krasteva et al., 2010; Zamorano-Sanchez et al., 2015; Hsieh et al., 2018). It was recently reported that the expression of the brp locus is positively regulated by BrpT and negatively regulated by BrpS in V. vulnificus. BrpS is another VpsT-type regulator whose gene is adjacent to brpT (Chodur and Rowe-Magnus, 2018). However, the roles of the three transcriptional regulators BrpR, BrpT, and BrpS in the regulation of cabABC, as well as their regulatory hierarchy in V. vulnificus have not been well defined. In the present study, we conducted transcript analyses and structural analyses of V. vulnificus biofilms and demonstrated that BrpR and BrpT activate the cabABC operon in a sequential manner, and thus play an essential role in development of robust biofilm structures. The role of BrpS in the regulatory pathway was also examined, and our results suggested that the expression of brpT is regulated by a negative feedback loop involving BrpS. Molecular biological analyses were performed and revealed that both BrpT and BrpS activate cabABC by directly binding to specific sequences in the regulatory region, constituting a coherent feedforward loop. Microscopic analyses indicated that BrpS, along with BrpR and BrpT, is required for the complete development of rugose colonies. Together, our results suggest that the three transcriptional regulators BrpR, BrpT, and BrpS collaboratively regulate the cabABC operon for robust biofilm and rugose colony development.
Results
BrpR and BrpT Positively Regulate cabABC as a Sequential Cascade
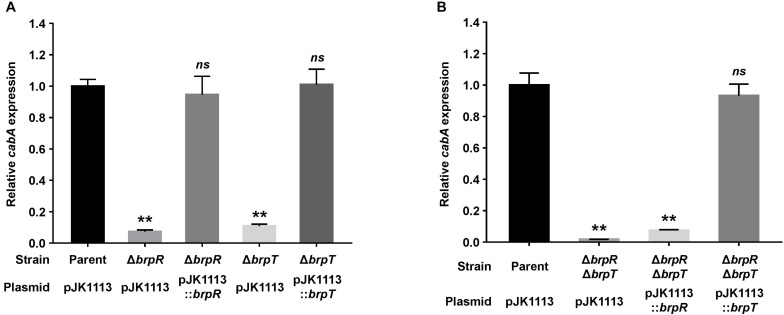
We previously observed that expression of the cabABC operon is induced by elevated intracellular c-di-GMP levels (Park et al., 2015b). Since BrpR and BrpT are involved in c-di-GMP-induced biofilm formation (Guo and Rowe-Magnus, 2010), we examined whether the two transcriptional regulators are also involved in the activation of cabABC. For this purpose, isogenic mutants lacking brpR or brpT were generated from V. vulnificus JN111, the parent strain previously constructed to manipulate the intracellular c-di-GMP levels (Park et al., 2015a), and the level of cabA transcript in biofilm cells of the parent and mutant strains was compared under elevated c-di-GMP levels. The cabA expression was almost 10-fold lower in the brpR and brpT mutants compared with that in the parent strain (Figure 1A). The level of cabA expression in the brpR and brpT mutants was restored to the level in the parent strain by introducing pJK1113::brpR and pJK1113::brpT, respectively (Figure 1A). The results indicated that both BrpR and BrpT are positive regulators that increase the expression of the cabABC operon in either an additive or a sequential manner.
FIGURE 1.
Deletion of brpR and brpT eliminates cabA expression. (A,B) Total RNA was isolated from biofilms of the V. vulnificus strains grown in microtiter plates. The level of cabA transcript was determined by qRT-PCR, and the parent strain was set to 1. Error bars represent the SD from three independent experiments. ∗∗, p < 0.005 relative to the parent strain; ns, not significant. Parent (pJK1113), parent strain; ΔbrpR (pJK1113), brpR mutant; ΔbrpT (pJK1113), brpT mutant; ΔbrpR (pJK1113::brpR) and ΔbrpT (pJK1113::brpT), complemented strains; ΔbrpRΔbrpT (pJK1113), brpR brpT double mutant; ΔbrpRΔbrpT (pJK1113::brpR) or ΔbrpRΔbrpT (pJK1113::brpT), brpR brpT double mutant expressing BrpR or BrpT, respectively.
To characterize the functional relationship of BrpR and BrpT in cabABC regulation, the brpR brpT double mutant was also constructed and the cabA expression was determined. In the absence of both brpR and brpT, there was a significant reduction of cabA expression (Figure 1B), but the level of residual cabA transcript was comparable with that in the brpR or brpT single mutant (Figure 1A). This result indicated that the positive regulation of BrpR and BrpT for cabA is not additive. When the brpR brpT double mutant was complemented with pJK1113::brpR, cabA expression was not activated in the absence of brpT (Figure 1B). However, complementation of the double mutant with pJK1113::brpT restored the level of cabA expression to the level in the parent strain in the absence of brpR (Figure 1B). These results suggested that BrpT activates cabA directly, and BrpR activates cabA through activation of brpT in a sequential manner. Consistent with the suggestion, the expression of brpT was greatly reduced in the brpR mutant compared with that in the parent strain (Supplementary Figure S1A), whereas brpR expression was not affected by the absence of brpT (Supplementary Figure S1B). Together, these results suggested that BrpR and BrpT positively regulate cabABC expression in a regulatory cascade, in which BrpR activates brpT, and BrpT in turn activates cabABC.
BrpR and BrpT Contribute to Robust Biofilm Structures Through Activation of cabABC
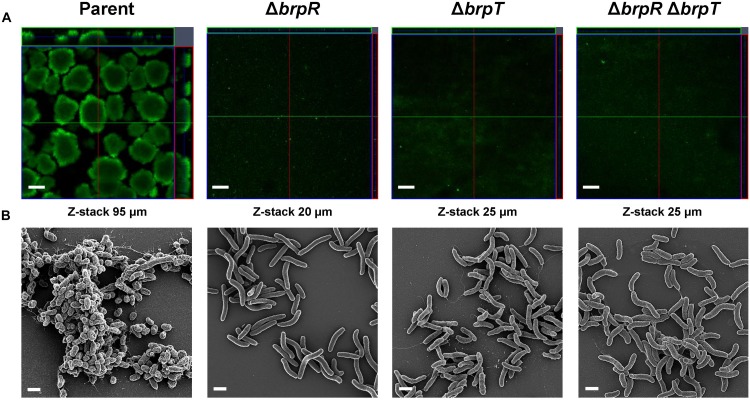
To assess the importance of BrpR and BrpT in biofilm development, biofilm structures of the V. vulnificus strains developed in response to elevated c-di-GMP levels were examined in flow cells. As shown in Figure 2A, confocal laser scanning microscopy (CLSM) images demonstrated that the parent strain produced biofilms with robust, large, uniform, and mushroom-like structures. However, the mutant strains lacking brpR, brpT, or both brpR and brpT produced weak, sparse, inconsistent, and markedly unstructured biofilms, which were similar to those previously observed when CabA was absent from the biofilm matrix (Park et al., 2015a). Z-stack measurements showed that the depth of the parent strain biofilms was 95 μm, whereas those of the mutant strain biofilms were between 20 and 25 μm. These results indicated that BrpR and BrpT are important for development of robust biofilms.
FIGURE 2.
Deletion of brpR and brpT disrupts biofilm structures. Biofilms of the V. vulnificus strains were grown in flow cell chambers under constant flow. (A) Biofilms were stained by LIVE/DEAD BacLight Viability Kit (Invitrogen), and CLSM (LSM710, Zeiss) images were acquired at 100 × magnification. The depth of the Z-stack is indicated below the images in μm. (B) Biofilms of the strains were fixed, dehydrated, coated with platinum, and visualized using SEM (Supra 55VP, Zeiss) at 10000 × magnification. Scale bars, 100 μm (A) and 1 μm (B); Parent, parent strain; ΔbrpR, brpR mutant; ΔbrpT, brpT mutant; ΔbrpRΔbrpT, brpR brpT double mutant.
To examine the effects of BrpR and BrpT on the biofilm structure in more detail, the biofilms developed in flow cells were further examined using scanning electron microscopy (SEM) (Figure 2B). The parent strain produced compact and thick biofilms in which bacterial cells were bound together and encased within an extracellular matrix. Filamentous materials that connected bacterial cells were visible in the biofilm matrix of the parent strain. In contrast, the mutant strains lacking brpR, brpT, or both brpR and brpT exhibited only a few bacterial cell clusters that were void of any extracellular matrix surrounding the cells. Since CabA is an essential component of the extracellular matrix (Park et al., 2015a), this observation indicated that cabABC expression activated by BrpR and BrpT is crucial for development of the structured matrix. Therefore, these results, combined with the results from transcript analyses (Figure 1), suggested that BrpR and BrpT are essential for activation of the cabABC operon, thereby contributing to the structural integrity of V. vulnificus biofilms.
BrpT and BrpS Constitute a Negative Feedback Loop
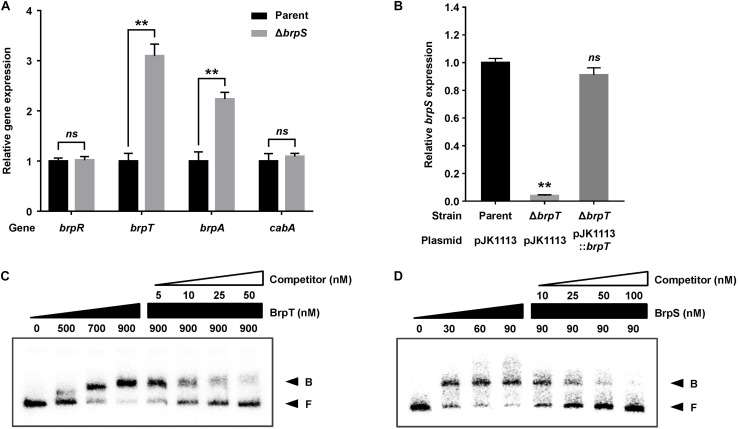
To examine whether BrpS is also involved in the pathway for cabABC regulation, the isogenic brpS mutant was constructed and expression of brpR, brpT, brpA, and cabA was compared to the parent strain. The brpS deletion did not significantly affect brpR expression but increased the level of brpT expression about 3-fold (Figure 3A), suggesting that BrpS represses brpT. In contrast, the expression of brpS was substantially reduced in the absence of brpT (Figure 3B), indicating that BrpT activates brpS. Electrophoretic mobility shift assays (EMSAs) also revealed that BrpT and BrpS could directly bind to the upstream region of brpS and brpT, respectively (Figures 3C,D). These results suggested that BrpT activates brpS, but BrpS represses brpT, forming a negative feedback loop. Since BrpT activates both the cabABC operon (Figure 1), as well as the brp locus (Chodur and Rowe-Magnus, 2018), both of these loci might be activated by the higher levels of BrpT in the brpS mutant. However, only brpA expression increased in the brpS mutant, and not cabA (Figure 3A). This result indicated that additional regulatory factor(s) other than BrpT, which counterbalances the effect of increased BrpT, likely participates in the precise regulation of the cabABC operon.
FIGURE 3.
BrpT and BrpS constitute a negative feedback loop. (A,B) Total RNA was isolated from biofilms of the V. vulnificus strains grown in microtiter plates. The levels of brpR, brpT, brpA, and cabA transcripts (A) or brpS transcript (B) were determined by qRT-PCR, and the parent strain was set to 1. Error bars represent the SD from three independent experiments. ∗∗, p < 0.005 relative to the parent strain; ns, not significant. Parent and Parent (pJK1113), parent strain; ΔbrpS, brpS mutant; ΔbrpT (pJK1113), brpT mutant; ΔbrpT (pJK1113::brpT), complemented strain. (C) A 358-bp DNA fragment of the brpS upstream region (2.5 nM) was radioactively labeled and incubated with increasing amounts of BrpT as indicated in the presence of c-di-GMP (50 μM). For competition analysis, the same but unlabeled 358-bp DNA fragment was used as self-competitor DNA. Various amounts of self-competitor DNA were added as indicated to a reaction mixture containing the probe DNA before the addition of BrpT. (D) A 388-bp DNA fragment of the brpT upstream region (2.5 nM) was radioactively labeled and incubated with increasing amounts of BrpS as indicated in the presence of c-di-GMP (50 μM). For competition analysis, the same but unlabeled 388-bp DNA fragment was used in the same manner as described above, except that BrpS was added. B, bound DNA; F, free DNA.
Determination and Deletion Analysis of the cabA Promoter
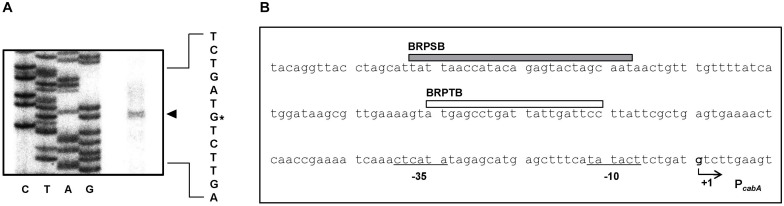
To map the promoter of the cabABC operon, the transcription start site was determined by primer extension analysis. A single reverse transcript was produced from primer extension of the RNA isolated from biofilms of the parent strain grown under elevated intracellular c-di-GMP levels (Figure 4A). The 5′-end of the cabABC transcript was located 79 bp upstream of the translation initiation codon of cabA and was subsequently designated +1 (Figure 4B). The putative promoter constituting the transcription start site was named PcabA to represent the cabA promoter, and the sequences for −10 and −35 regions of PcabA were assigned on the basis of similarity to the consensus sequences of Escherichia coli σ70 promoters (Figure 4B).
FIGURE 4.
Transcription start site and sequence of the PcabA regulatory region. (A) The transcription start site of PcabA was determined by primer extension of the RNA isolated from biofilms of the parent strain. Lanes G, A, T, and C represent the nucleotide sequencing ladders. The asterisk indicates the transcription start site of PcabA. (B) The transcription start site of PcabA is indicated by a bent arrow, and the position of the putative –10 and –35 regions are underlined. The sequences for binding of BrpS (BRPSB; gray box) and BrpT (BRPTB; white box) were determined later in this study (Figures 6C,D).
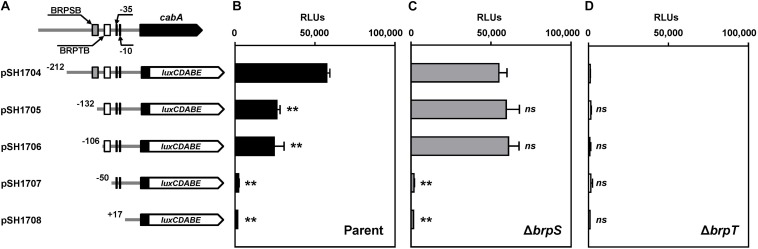
To delineate the cis-DNA sequences required for the PcabA activity, pSH reporters carrying varied PcabA regulatory regions, which were deleted up to different 5′-ends and fused transcriptionally to luxCDABE, were constructed (Figure 5A). The luminescence produced by pSH1704 carrying PcabA deleted up to −212 was comparable between the parent strain and brpS mutant (Figures 5B,C). This was consistent with the comparable levels of cabA expression in these two strains (Figure 3A). Compared with pSH1704, the reporters pSH1705 and pSH1706, carrying PcabA deleted up to −132 and −106, respectively, produced significantly reduced luminescence in the parent strain (Figure 5B). However, the relative light units (RLUs) of the brpS mutants containing pSH1704, pSH1705, or pSH1706 did not significantly differ (Figure 5C), indicating that the cis-DNA sequence required for BrpS-dependent activation of PcabA is deleted in pSH1705 and pSH1706. The reporters pSH1707 and pSH1708 produced the basal level of RLUs in the parent strain (Figure 5B), and the level was comparable with the level produced by any pSH reporter in the brpT mutant (Figure 5D). This observation indicated that BrpT-dependent activation is crucial for the PcabA activity and is probably impaired by deletion of PcabA regulatory region up to −50. The results implied that the cis-DNA sequences necessary for BrpS and BrpT to activate PcabA encompass the −132 and −50 regions, respectively.
FIGURE 5.
Deletion analysis of the PcabA regulatory region. (A) PCR fragments carrying the PcabA regulatory region with 5′-end deletions were subcloned into pBBR-lux to create each pSH reporter. The wild-type PcabA regulatory region is shown on top with the proposed –10 and –35 regions, and the binding sites for BrpS (BRPSB; gray box) and BrpT (BRPTB; white box) were determined later in this study (Figures 6C,D). Solid lines, the upstream region of cabA; black blocks, the cabA coding region; white blocks, luxCDABE. (B–D) Cellular luminescence was determined from the parent strain (B, black bars), brpS mutant (C, gray bars), and brpT mutant (D, white bars) containing each pSH reporter as indicated. Cultures grown to an A600 of 2.0 were used to measure the cellular luminescence. Error bars represent the SD from three independent experiments. ∗∗, p < 0.005 relative to the strain containing pSH1704; ns, not significant. RLUs, relative light units. Parent, parent strain; ΔbrpS, brpS mutant; ΔbrpT, brpT mutant.
BrpT and BrpS Directly Bind to PcabA
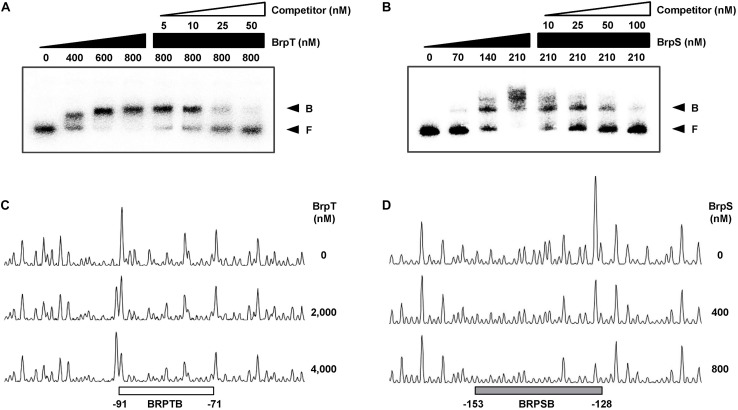
To examine whether BrpT and BrpS directly bind to the PcabA promoter, EMSAs were performed as shown in Figures 6A,B. For this purpose, the 337-bp labeled DNA probe encompassing the PcabA regulatory region (from −230 to +107) was incubated with increasing amounts of BrpT or BrpS and then subjected to electrophoresis. The addition of BrpT or BrpS to the DNA probe resulted in a retarded band in a concentration-dependent manner (Figures 6A,B). The binding of BrpT and BrpS was specific, because the assays were performed in the presence of poly(dI-dC) as a non-specific competitor. In addition, the same unlabeled 337-bp DNA fragment was used as a self-competitor to confirm the specific binding of BrpT and BrpS. The unlabeled DNA competed for binding of BrpT and BrpS in a dose-dependent manner (Figures 6A,B), confirming that BrpT and BrpS bind specifically to PcabA.
FIGURE 6.
Specific binding of BrpT and BrpS to PcabA. (A,B) A 337-bp DNA fragment of the PcabA regulatory region (2.5 nM) was radioactively labeled and incubated with increasing amounts of BrpT (A) or BrpS (B) as indicated in the presence of c-di-GMP (50 μM). For competition analysis, the same but unlabeled 337-bp DNA fragment was used as a self-competitor DNA. Various amounts of self-competitor DNA were added as indicated to a reaction mixture containing the probe DNA before the addition of BrpT (A) or BrpS (B). B, bound DNA; F, free DNA. (C,D) A 337-bp DNA fragment of same PcabA regulatory region as above was labeled with 6-FAM and then used as a probe DNA. The 6-FAM-labeled probe DNA (48 nM) was incubated with increasing amounts of BrpT (C) or BrpS (D) as indicated in the presence of c-di-GMP (50 μM), and then digested with DNase I. The regions protected from DNase I cleavage by BrpT or BrpS are indicated by a white (C, BRPTB) or gray box (D, BRPSB), respectively. Nucleotide numbers shown are relative to the transcription start site of PcabA.
DNase I protection assays were performed using the 337-bp DNA probe encompassing the same region as above to identify the binding sequences for BrpT and BrpS in the PcabA regulatory region. Upon addition of BrpT, the region extending from −91 to −71 (BRPTB, centered at −81) was protected from DNase I digestion (Figure 6C). The addition of BrpS resulted in protection of the region extending from −153 to −128 (BRPSB, centered at −140.5), as shown in Figure 6D. Taken together, these results suggested that BrpT and BrpS directly regulate cabABC by binding to the specific sequences of PcabA.
BrpS Is Important for Complete Development of Rugose Colonies
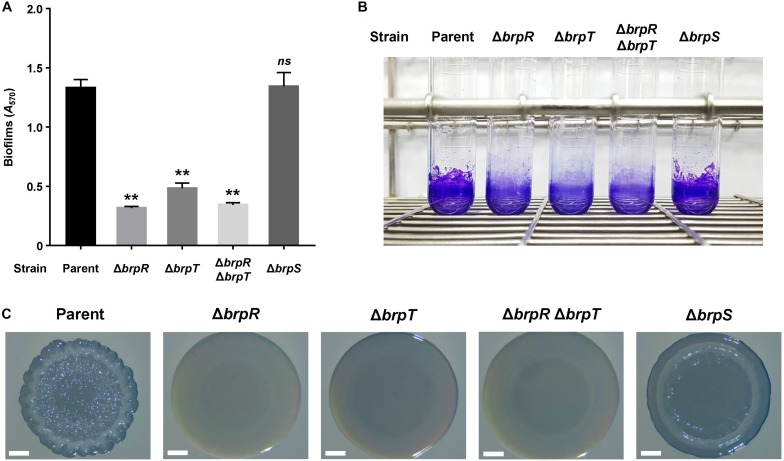
We previously demonstrated that CabA is essential for development of both biofilms and rugose colonies under elevated c-di-GMP levels (Park et al., 2015a). Thus, the role of BrpS as well as BrpR and BrpT on the biofilm- and rugose colony-forming abilities of the V. vulnificus strains was examined. Biofilms were grown in microtiter plates and quantified using crystal violet staining assays (Figure 7A). Compared to the parent strain, the level of biofilm formation was significantly reduced in the mutant strains lacking brpR, brpT, or both brpR and brpT. This was consistent with the severe reduction of cabA expression observed (Figure 1). Meanwhile, biofilm formation of the brpS mutant was comparable with that of the parent strain. This could be attributed to the comparable level of cabA expression in these two strains (Figure 3A). Biofilm formation of the strains, in the same condition used for Figure 7A but in a larger scale in test tubes, were also similar with those observed in microtiter plates (Figure 7B). This correlation of biofilm formation with the cabA expression supported our previous observation that expression of cabABC plays a decisive role in biofilm development of V. vulnificus.
FIGURE 7.
Effects of brpR, brpT, and brpS deletions on biofilm formation and colony morphology. (A) Biofilms of the V. vulnificus strains were grown in microtiter plates for 24 h, and then stained with 1% crystal violet. The crystal violet was eluted, and its absorbance at 570 nm (A570) was determined to quantify the biofilms. Error bars represent the SD from three independent experiments. ∗∗, p < 0.005 relative to the parent strain; ns, not significant. (B) Biofilms were grown and stained as described above, except that round-bottom test tubes were used. The stained biofilms were photographed using a digital camera (PowerShot G7X Mark II, Canon). (C) V. vulnificus strains were spotted onto VFMG agar plates and incubated for 36 h. Each colony that represented the mean rugosity from at least three independent experiments was visualized using a stereomicroscope (Stemi 305, Zeiss) at 8 × magnification. Scale bars, 1 mm. Parent, parent strain; ΔbrpR, brpR mutant; ΔbrpT, brpT mutant; ΔbrpRΔbrpT, brpR brpT double mutant; ΔbrpS, brpS mutant.
To further examine the effects of the regulatory proteins on colony rugosity, the parent and mutant strains were grown on agar plates and their colony morphologies were visualized (Figure 7C). The parent strain displayed the rugose colony morphology, but the mutant strains lacking brpR, brpT, or both brpR and brpT exhibited the smooth colony morphology. Again, this was expected, based on the reduced cabA expression in these mutants (Figure 1). Interestingly, the brpS mutant displayed a colony morphology that was distinct from the parent and other mutant strains. The colony of the brpS mutant was characterized by a concentric ridge around the rim, but it was less wrinkled than the rugose colony of the parent strain. It has also been previously reported that BrpS is required for the colony rugosity (Chodur and Rowe-Magnus, 2018). This could result from the imbalance between matrix components in the brpS mutant. V. vulnificus requires production of both the matrix protein CabA and brp-exopolysaccharide to develop rugose colony morphology (Park et al., 2015a). In contrast to the expression of the cabABC operon, that of the brp locus greatly increased in the brpS mutant (Figure 3), leading to overproduction of exopolysaccharides in the mutant strain (Chodur and Rowe-Magnus, 2018). Still, the brpS mutant did not show an increase in the level of biofilm formation (Figures 7A,B), indicating that the overproduced exopolysaccharides did not further increase the biofilm biomass. However, this imbalance could lead to significantly altered colony morphology, because proper balance between the matrix components is critical for development of a specific morphology (Serra et al., 2013). These results suggested that BrpS as well as BrpR and BrpT plays an important role in the complete development of rugose colonies as well as elaborate regulation of the cabABC operon.
Discussion
Vibrio vulnificus CabA is a structural protein of the extracellular matrix that plays a crucial role in development of robust biofilms and rugose colonies (Park et al., 2015a). Another component in the biofilm matrix of V. vulnificus is the exopolysaccharide encoded by the brp locus (Guo and Rowe-Magnus, 2010). CabA and brp-exopolysaccharide are functionally associated, and CabA requires brp-exopolysaccharides to develop a robust matrix structure (Park et al., 2015a). It was previously reported that BrpT activates the expression of the brp locus (Chodur and Rowe-Magnus, 2018). In the present study, we demonstrated that BrpR and BrpT activate the expression of the cabABC operon in a sequential cascade (Figure 1 and Supplementary Figure S1). These observations propose that expression of the brp locus and the cabABC operon are induced together through BrpR and BrpT under elevated c-di-GMP levels. Consistent with this proposition, the mutant strains deficient in brpR or brpT formed bare bacterial cells without any extracellular matrix in flow cells (Figure 2), indicating that production of both matrix protein CabA and brp-exopolysaccharide was suppressed. The combined results suggest that the cabABC operon and the brp locus are coordinately regulated for efficient and successful development of biofilms.
In the present study, we also revealed the regulatory relationship between BrpT and BrpS. A previous study reported that BrpT represses brpS in V. vulnificus strain ATCC 27562 (Chodur and Rowe-Magnus, 2018). However, in this study, we determined that BrpT and BrpS constitute a negative feedback loop, in which BrpT activates brpS, but brpT is repressed by BrpS (Figure 3). This discrepancy was not due to strain differences, as reexamination of the regulatory relationship between BrpT and BrpS in the ATCC 27562 strain also identified the same negative feedback loop (Supplementary Figure S2). Another possible explanation is that the transcriptional reporter system they used may not contain the whole cis-element sequences required for PbrpS activation, since the PbrpS reporter exhibited unusually low level of expression compared to other reporters used in the study (Chodur and Rowe-Magnus, 2018). They also reported that brpS expression is activated under elevated intracellular c-di-GMP levels, and it makes sense if BrpT, which is induced by elevated c-di-GMP levels (Chodur and Rowe-Magnus, 2018), activates brpS, as we proposed.
It is logical to expect that increasing brpT expression in the brpS mutant would increase expression of both the cabABC operon and the brp locus. However, although expression of the brp locus increased, that of cabABC did not in the brpS mutant (Figure 3). This result indicates that the effect of BrpT-mediated activation of cabABC is counterbalanced by another effect(s) resulting from the brpS deletion. One possible explanation is that BrpS also activates the cabABC operon. Indeed, BrpS as well as BrpT directly regulates the cabABC operon by binding to specific, but distinct sequences in the PcabA regulatory region (Figures 4–6). The positioning of BrpT- and BrpS-binding sequences suggested that both BrpT and BrpS may act as class I activators interacting with the C-terminal domain of RNA polymerase α subunits (Browning and Busby, 2016). These results indicate that BrpT and BrpS activate cabABC in a feedforward loop for additive and precise regulation (Alon, 2007). We have also tried to examine the simultaneous binding of BrpT and BrpS to the PcabA regulatory region in vitro. However, the specific requirements of buffer composition for the binding of BrpT or BrpS were so different from each other that we could not find any optimized composition for the simultaneous binding of both proteins (see Experimental Procedures). The possible interaction between BrpT and BrpS in activation of PcabA remains to be elucidated.
Although we proposed that BrpS activates the cabABC operon, the cabA expression (Figure 3A), as well as the PcabA activity (Figures 5B,C), was not reduced in the brpS mutant compared with that in the parent strain. This observation could result from the increased brpT expression in the absence of BrpS (Figure 3A), which may compensate for the loss of BrpS-dependent activation of cabA. In this case, BrpS might not be required for full activation of cabABC. However, BrpS was required for complete development of rugose colonies (Figure 7C), possibly by balancing matrix components. Moreover, the region encompassing the BrpS-binding sequence was required for full activation of cabA in the parent strain (Figure 5B). This effect was BrpS-dependent, because deletion of the region did not affect cabA activation in the absence of BrpS (Figure 5C). These results suggest that V. vulnificus requires BrpS for rugose colony development and, at the same time, BrpS contributes to proper cabABC expression by activating cabABC while repressing brpT.
Transcriptional regulators which are homologous to BrpR, BrpT, and BrpS have also been studied in V. cholerae and Vibrio parahaemolyticus. Best known are VpsR and VpsT, two major regulators of biofilm formation in V. cholerae. VpsR and VpsT regulate expression of each other, and both can directly regulate downstream exopolysaccharide and matrix protein genes (Beyhan et al., 2007; Zamorano-Sanchez et al., 2015). On the contrary, BrpR and BrpT act in a sequential manner, and BrpT, not BrpR, directly regulates the brp locus and cabABC operon. Three regulatory proteins CpsR, CpsQ, and CpsS, corresponding to BrpR, BrpT, and BrpS respectively, also constitute a regulatory pathway for biofilm formation in V. parahaemolyticus. These three act in a cascade; While CpsS represses cpsR, CpsR activates cpsQ, and CpsQ activates expression of downstream genes (Ferreira et al., 2012). In contrast to the role of CpsS, which acts as a master repressor of the pathway, BrpS plays two distinct roles. First, BrpS represses brpT in a negative feedback loop. It can stabilize the steady state expression of brpT (Dublanche et al., 2006; Chalancon et al., 2012). Second, BrpS, along with BrpT, activates cabABC. It makes a difference between the regulation of the cabABC operon and the brp locus, so the balance in expression of the two loci can be properly adjusted through BrpS. Thus, V. vulnificus has a distinct regulatory network, which may be reflected in the differences of biofilm life style and niche occupation between V. vulnificus and other Vibrio species.
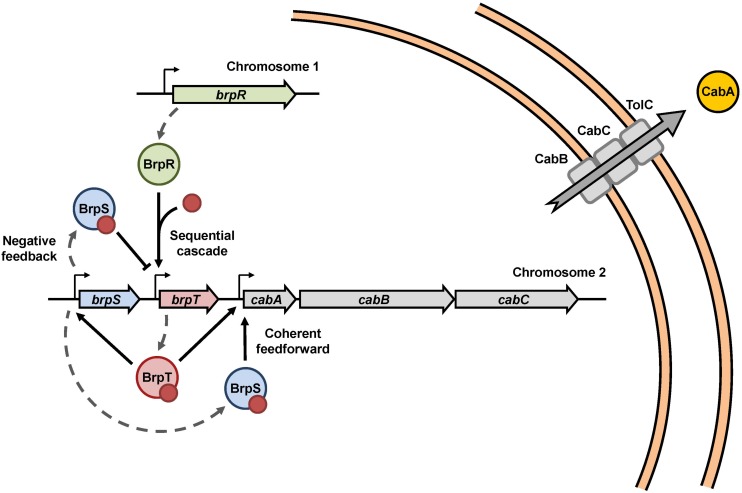
BrpR, BrpT, and BrpS combine to regulate the cabABC operon in V. vulnificus, and constitute a complex regulatory network as depicted in Figure 8. In summary, under elevated intracellular c-di-GMP levels, BrpR activates brpT, and BrpT in turn activates brpS and the cabABC operon. BrpT and BrpS together constitute a negative feedback loop and a coherent feedforward loop to regulate brpT and the cabABC operon, respectively. BrpT and BrpS activate cabABC expression by directly binding to the specific sequences in the PcabA regulatory region, leading to development of a structured biofilm matrix and rugose colony morphology. Consequently, the combination of three transcriptional regulators BrpR, BrpT, and BrpS elaborately controls cabABC expression, driving robust biofilm and rugose colony development by V. vulnificus.
FIGURE 8.
Proposed model for the cabABC regulatory network. A regulatory network comprised of the three transcriptional regulators BrpR, BrpT, and BrpS controls expression of the cabABC operon. Intracellular c-di-GMP levels can be altered in response to various environmental signals. Under elevated intracellular c-di-GMP levels, BrpR activates brpT and cabABC in a sequential cascade. BrpT and BrpS constitute a negative feedback loop for brpT regulation, where BrpT activates brpS, but BrpS represses brpT. BrpT and BrpS also form a feedforward loop for positive regulation of cabABC. CabA is secreted through a type I secretion system comprised of CabB, CabC, and the outer membrane protein TolC. CabA in the extracellular matrix contributes to development of robust biofilm structures and rugose colony morphology in cooperation with brp-exopolysaccharide. The red dots represent c-di-GMP molecules.
Materials and Methods
Strains, Plasmids, and Culture Conditions
The strains and plasmids used in this study are listed in Supplementary Table S1. Unless otherwise noted, the V. vulnificus strains were grown aerobically in LB medium supplemented with 2.0% (w/v) NaCl (LBS) at 30°C. The Vibrio fischeri minimal medium (Cao et al., 2012) containing glycerol (50 mM Tris–HCl, pH 7.2, 50 mM MgSO4, 300 mM NaCl, 10 mM KCl, 0.33 mM K2HPO4, 18.5 mM NH4Cl, 10 mM CaCl2, and 32.6 mM glycerol) (VFMG) was used for biofilm formation. When required, antibiotics were added to the media at the following concentrations: kanamycin, 100 μg/ml; chloramphenicol, 3 μg/ml. To manipulate intracellular c-di-GMP levels, V. vulnificus JN111, which carries dcpA encoding a diguanylate cyclase (Nakhamchik et al., 2008) on the chromosome under the control of arabinose-inducible promoter PBAD (Guzman et al., 1995), was constructed previously (Park et al., 2015a). JN111 was used as a parent strain in this study (Supplementary Table S1), and intracellular c-di-GMP levels of the V. vulnificus strains were manipulated by adding different concentrations of arabinose to the growth media.
Generation and Complementation of the Deletion Mutants
The brpR gene was inactivated in vitro by deletion of the ORF of brpR (967-bp of 1335-bp) using the PCR-mediated linker-scanning mutation method as described previously (Kim et al., 2014). Briefly, pairs of primers BRPR01-F and -R (for amplification of the 5′ amplicon) or BRPR02-F and -R (for amplification of the 3′ amplicon) were designed and used (Supplementary Table S2). The resulting ΔbrpR fragment was amplified by PCR using the mixture of both amplicons as the template and BRPR01-F and BRPR02-R as primers. Similar experimental procedures were adopted for amplification of the ΔbrpT and ΔbrpS fragments in vitro, except that primers BRPT01-F, BRPT01-R, BRPT02-F, and BRPT02-R (for 405-bp deleted brpT) and BRPS01-F, BRPS01-R, BRPS02-F, and BRPS02-R (for 627-bp deleted brpS) were used (Supplementary Table S2). The resulting ΔbrpR, ΔbrpT, and ΔbrpS fragments were ligated into SpeI-SphI-digested pDM4 (Milton et al., 1996) to generate pJN1302, pJN1607, and pSH1805, respectively (Supplementary Table S1). E. coli S17-1 λpir, tra strain (Simon et al., 1983) containing pJN1302, pJN1607, or pSH1805 was used as a conjugal donor to JN111 to generate the brpR mutant JN131D, the brpT mutant JN161D, or the brpS mutant SH181D, respectively (Supplementary Table S1). Similarly, E. coli S17-1 λpir, tra strain containing pJN1607 was used as a conjugal donor to JN131D to generate the brpR brpT double mutant JN162D (Supplementary Table S1). The conjugation and isolation of the transconjugants were conducted using the method described previously (Kim et al., 2011). In the same manner as above, the brpT mutant SH191 and the brpS mutant SH192 were generated from V. vulnificus strain ATCC 27562 using pJN1607 and pSH1805 (Supplementary Table S1).
To complement the brpR and brpT deletions, each ORF of brpR and brpT was amplified by PCR using a pair of specific primers, as listed in Supplementary Table S2. The amplified ORFs of brpR and brpT were cloned into pJK1113 (Lim et al., 2014) under an arabinose-inducible promoter PBAD to create pJN1601 and pJN1602, respectively (Supplementary Table S1). The plasmids were transferred into the appropriate mutants by conjugation as described above.
RNA Purification and Transcript Analysis
Total RNA was isolated by using an RNeasy® mini kit (Qiagen, Valencia, CA, United States) from biofilms of the V. vulnificus strains grown on 24-well polystyrene microtiter plates (SPL, Seoul, South Korea) containing VFMG supplemented with 0.01% (w/v) arabinose for 24 h. For quantitative reverse transcription-PCR (qRT-PCR), the concentrations of total RNAs were measured by using a NanoDropTM OneC spectrophotometer (Thermo Scientific, Waltham, MA, United States), and cDNA was synthesized from 1 μg of the total RNA by using an iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA, United States). Real-time PCR amplification of the cDNA was performed by using a CFX96 real-time PCR detection system (Bio-Rad) with pairs of specific primers (Supplementary Table S2), as described previously (Kim et al., 2012). Relative levels of the transcripts were calculated by using the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression level as the internal reference for normalization (Williams et al., 2014).
For primer extension analysis, a 24-base primer CABAUP-R (Supplementary Table S2) complementary to the coding region of cabA was end-labeled with [γ-32P]-ATP and added to the RNA. The primer was then extended with SuperScript II reverse transcriptase (Invitrogen). The cDNA products were purified and resolved on a sequencing gel alongside sequencing ladders generated from pBH1402 with the same primer. The plasmid pBH1402 was constructed by cloning the 337-bp cabA upstream region extending from −230 to +107, amplified by PCR using a pair of primers CABAUP-F and -R (Supplementary Table S2), into pGEM-T Easy (Promega, Madison, WI, United States). The primer extension product was visualized using a phosphor image analyzer (BAS1500, Fujifilm, Tokyo, Japan).
Construction of a Set of cabA-luxCDABE Transcriptional Fusions
The primer CABAR006 carrying a SpeI restriction site was used in conjunction with one of the primers carrying a SacI restriction site, CABAR001, CABAR002. CABAR003, CABAR004, and CABAR005 (Supplementary Table S2), to amplify the DNA of cabA extending up to −212, −132, −106, −50, and +17 bp, respectively. The amplified DNA fragments were inserted into the SpeI-SacI-digested pBBR-lux carrying promoterless luxCDABE genes (Lenz et al., 2004) to create five cabA-lux reporter constructs: pSH1704, pSH1705, pSH1706, pSH1707, and pSH1708 (Supplementary Table S1). The constructs were then transferred into the V. vulnificus strains by conjugation. The cellular luminescence of each culture grown to an A600 of 2.0 in LB medium supplemented with 0.1% arabinose was measured using a Tecan Infinite M200 microplate reader (Tecan, Männedorf, Switzerland) and expressed in arbitrary RLUs as described previously (Jeong et al., 2003).
Protein Purification, EMSA, and DNase I Protection Assay
The ORFs of brpT and brpS were amplified by PCR using pairs of primers BRPT04-F and -R or BRPS04-F and -R (Supplementary Table S2) and subcloned into pET-28a(+) (Novagen, Madison, WI, United States), resulting in pSH1819 and pSH1823 (Supplementary Table S1), respectively. The His6-tagged BrpT and BrpS were expressed in E. coli BL21 (DE3) and purified by affinity chromatography using Ni-NTA agarose (Qiagen).
For EMSA, The 337-bp cabA upstream region, extending from −230 to +107, was amplified by PCR using unlabeled CABAUP-F and [γ-32P]-ATP-labeled CABAUP-R as primers (Supplementary Table S2). Similarly, the 388-bp brpT upstream region or 358-bp brpS upstream region were amplified by PCR using unlabeled BRPTUP-F or BRPSUP-F in conjunction with [γ-32P]ATP-labeled BRPTUP-R or BRPSUP-R as primers (Supplementary Table S2), respectively. For binding of BrpT, the labeled DNA (2.5 nM) was incubated with purified BrpT for 25 min at 30°C in a 20-μl reaction mixture containing 1 × BrpT-binding buffer (20 mM HEPES (pH 7.6), 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% (v/v) Tween 20, 30 mM KCl, 75 mM NaCl, 50 μM c-di-GMP) and 0.1 μg of poly(dI-dC) (Sigma-Aldrich, St. Louis, MO, United States). The protein-DNA binding reactions with BrpS were performed in the same manner as those with BrpT, except that 1 × BrpS-binding buffer (10 mM Tris–HCl (pH 7.9), 50 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 50 μM c-di-GMP) was used. Electrophoretic analysis of the protein-DNA complexes was performed as described previously (Jang et al., 2017). When necessary, various concentrations of unlabeled DNA were added as competitors to the reaction mixture before incubation.
For DNase I protection assay, the same 337-bp cabA upstream region as above was amplified by PCR using unlabeled CABAUP-F and 6-carboxyfluorescein (6-FAM)-labeled CABAUP-R as primers (Supplementary Table S2). The binding of purified BrpT or BrpS to the labeled DNA (48 nM) and DNase I digestion of the protein-DNA complexes followed the procedures described previously (Lim and Choi, 2014), except that 1 × BrpT- or BrpS-binding buffer was used, respectively. The digested DNA products were precipitated with ethanol, eluted in H2O, and analyzed using an ABI 3730×l DNA analyzer (Applied Biosystems, Foster City, CA, United States) with Peak ScannerTM Software v1.0 (Applied Biosystems).
Quantitative and Structural Analyses of Biofilms
To quantify biofilms of the V. vulnificus strains, each well of the 96-well polystyrene microtiter plates (Nunc, Roskilde, Denmark) was inoculated with 200 μl of each culture diluted to an A600 of 0.05 in VFMG supplemented with 0.01% arabinose. After 24 h static incubation at 30°C, the biofilms were stained with 1% (w/v) crystal violet solution for 15 min and quantified by elution of crystal violet from the stained biofilms with ethanol and measurement of absorbance at 570 nm (A570) as described previously (Park et al., 2015a). To visualize the crystal violet-stained biofilms, biofilms of the strains were formed as described above but in a larger scale (1 ml) using 14 ml round-bottom test tubes (BD Biosciences, San Jose, CA, United States), and the stained biofilms were photographed using a digital camera (PowerShot G7X Mark II, Canon, Tokyo, Japan).
For structural analyses, biofilms of the strains were formed in flow cell chambers using the method described previously (Park et al., 2015a). Glass coverslips were attached on polycarbonate flow cells with individual channel dimensions of 1 × 4 × 40 mm. Each flow cell was inoculated with 100 μl of the culture diluted to A600 of 0.1, and inverted to allow bacteria to attach to the coverslip for 1 h without flow. Then VFMG supplemented with 0.01% arabinose was flowed at a constant rate of 8 ml/h using a Minipuls EvolutionTM peristaltic pump (Gilson, Villiers-le-Bel, France) to grow biofilms for 3 days.
For CLSM analysis, biofilms on the coverslips were stained with LIVE/DEAD BacLightTM bacterial viability kit containing SYTO 9 and propidium iodide (Invitrogen) for 15 min in the dark and visualized by CLSM (LSM710, Zeiss). The biofilm images were processed using Zeiss Zen software (Zeiss). For SEM analysis, biofilms on the coverslips were fixed and dehydrated as described previously (Park et al., 2015a). The resulting biofilms were mounted on an aluminum stub, coated with platinum using a putter coater (BAL-TEC SCD 005, BAL-TEC AG, Balzers, Liechtenstein), and visualized using SEM (Supra 55VP, Zeiss).
Colony Morphology Assay
For analysis of colony morphology, 2 μl of each culture grown to A600 of 0.8 was spotted onto VFMG agar plate supplemented with 0.02% arabinose. The resulting colonies grown at 30°C for 36 h were visualized using a Stemi 305 stereomicroscope (Zeiss, Oberkochen, Germany) equipped with an Axiocam 105 color camera (Zeiss).
Data Analysis
The mean and standard deviation (SD) values were calculated from at least three independent experiments. The experimental data were analyzed by Student’s t-test using GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, United States). Significance of differences between experimental groups was accepted at a p value of <0.05.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
All authors conceived and designed the research, analyzed and interpreted the data, reviewed the results, and approved the final version of the manuscript. S-HH, JP, and BL performed the experiments. S-HH and SC wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Research Foundation of Korea, funded by the Ministry of Science, ICT, and Future Planning (2017R1E1A1A01074639) to SC. This work was also supported by the BK21 Plus Program of the Department of Agricultural Biotechnology, Seoul National University, Seoul, South Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03063/full#supplementary-material
References
- Alon U. (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8 450–461. 10.1038/nrg2102 [DOI] [PubMed] [Google Scholar]
- Beyhan S., Bilecen K., Salama S. R., Casper-Lindley C., Yildiz F. H. (2007). Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189 388–402. 10.1128/JB.00981-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C. D., O’Toole G. A. (2012). Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28 439–462. 10.1146/annurev-cellbio-101011-155705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning D. F., Busby S. J. (2016). Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 14 638–650. 10.1038/nrmicro.2016.103 [DOI] [PubMed] [Google Scholar]
- Cao X., Studer S. V., Wassarman K., Zhang Y., Ruby E. G., Miyashiro T. (2012). The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. MBio 3:e00285-11. 10.1128/mBio.00285-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalancon G., Ravarani C. N., Balaji S., Martinez-Arias A., Aravind L., Jothi R., et al. (2012). Interplay between gene expression noise and regulatory network architecture. Trends Genet. 28 221–232. 10.1016/j.tig.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodur D. M., Rowe-Magnus D. A. (2018). Complex control of a genomic Island governing biofilm and rugose colony development in Vibrio vulnificus. J. Bacteriol. 200:e00190-18. 10.1128/JB.00190-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J. G., Zamorano-Sanchez D., Park J. H., Sondermann H., Yildiz F. H. (2017). The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr. Opin. Microbiol. 36 20–29. 10.1016/j.mib.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284 1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Dublanche Y., Michalodimitrakis K., Kummerer N., Foglierini M., Serrano L. (2006). Noise in transcription negative feedback loops: simulation and experimental analysis. Mol. Syst. Biol. 2:41. 10.1038/msb4100081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R. B., Chodur D. M., Antunes L. C., Trimble M. J., McCarter L. L. (2012). Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J. Bacteriol. 194 914–924. 10.1128/JB.05807-5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- Flemming H. C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14 563–575. 10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- Froelich B., Oliver J. D. (2013). The interactions of Vibrio vulnificus and the oyster Crassostrea virginica. Microb. Ecol. 65 807–816. 10.1007/s00248-012-0162-163 [DOI] [PubMed] [Google Scholar]
- Guo Y., Rowe-Magnus D. A. (2010). Identification of a c-di-GMP-regulated polysaccharide locus governing stress resistance and biofilm and rugose colony formation in Vibrio vulnificus. Infect. Immun. 78 1390–1402. 10.1128/IAI.01188-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177 4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Hsieh M. L., Hinton D. M., Waters C. M. (2018). VpsR and cyclic di-GMP together drive transcription initiation to activate biofilm formation in Vibrio cholerae. Nucleic Acids Res. 46 8876–8887. 10.1093/nar/gky606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K. K., Lee Z. W., Kim B., Jung Y. H., Han H. J., Kim M. H., et al. (2017). Identification and characterization of Vibrio vulnificus plpA encoding a phospholipase A2 essential for pathogenesis. J. Biol. Chem. 292 17129–17143. 10.1074/jbc.M117.791657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. S., Lee M. H., Lee K. H., Park S. J., Choi S. H. (2003). SmcR and cyclic AMP receptor protein coactivate Vibrio vulnificus vvpE encoding elastase through the RpoS-dependent promoter in a synergistic manner. J. Biol. Chem. 278 45072–45081. 10.1074/jbc.M308184200 [DOI] [PubMed] [Google Scholar]
- Kim B. S., Hwang J., Kim M. H., Choi S. H. (2011). Cooperative regulation of the Vibrio vulnificus nan gene cluster by NanR protein, cAMP receptor protein, and N-acetylmannosamine 6-phosphate. J. Biol. Chem. 286 40889–40899. 10.1074/jbc.M111.300988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Bang Y. J., Kim D., Lim J. G., Oh M. H., Choi S. H. (2014). Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Mol. Microbiol. 93 992–1009. 10.1111/mmi.12712 [DOI] [PubMed] [Google Scholar]
- Kim S. M., Lee D. H., Choi S. H. (2012). Evidence that the Vibrio vulnificus flagellar regulator FlhF is regulated by a quorum sensing master regulator SmcR. Microbiology 158 2017–2025. 10.1099/mic.0.059071-59070 [DOI] [PubMed] [Google Scholar]
- Krasteva P. V., Fong J. C., Shikuma N. J., Beyhan S., Navarro M. V., Yildiz F. H., et al. (2010). Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327 866–868. 10.1126/science.1181185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva P. V., Giglio K. M., Sondermann H. (2012). Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci. 21 929–948. 10.1002/pro.2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz D. H., Mok K. C., Lilley B. N., Kulkarni R. V., Wingreen N. S., Bassler B. L. (2004). The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118 69–82. 10.1016/j.cell.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Lim J. G., Bang Y. J., Choi S. H. (2014). Characterization of the Vibrio vulnificus 1-Cys peroxiredoxin Prx3 and regulation of its expression by the Fe-S cluster regulator IscR in response to oxidative stress and iron starvation. J. Biol. Chem. 289 36263–36274. 10.1074/jbc.M114.611020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. G., Choi S. H. (2014). IscR is a global regulator essential for pathogenesis of Vibrio vulnificus and induced by host cells. Infect. Immun. 82 569–578. 10.1128/IAI.01141-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald D., Rice S. A., Barraud N., Steinberg P. D., Kjelleberg S. (2012). Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10 39–50. 10.1038/nrmicro2695 [DOI] [PubMed] [Google Scholar]
- Milton D. L., O’Toole R., Horstedt P., Wolf-Watz H. (1996). Flagellin a is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178 1310–1319. 10.1128/jb.178.5.1310-1319.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhamchik A., Wilde C., Rowe-Magnus D. A. (2008). Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol. 74 4199–4209. 10.1128/AEM.00176-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J. D. (2015). The biology of Vibrio vulnificus. Microbiol. Spectr. 3 VE-0001-2014. 10.1128/microbiolspec.VE-0001-2014 [DOI] [PubMed] [Google Scholar]
- Park J. H., Jo Y., Jang S. Y., Kwon H., Irie Y., Parsek M. R., et al. (2015a). The cabABC operon essential for biofilm and rugose colony development in Vibrio vulnificus. PLoS Pathog. 11:e1005192. 10.1371/journal.ppat.1005192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Lim J. G., Choi S. H. (2015b). Effects of elevated intracellular cyclic di-GMP levels on biofilm formation and transcription profiles of Vibrio vulnificus. Food Sci. Biotechnol. 24 771–776. 10.1007/s10068-015-0100-105 [DOI] [Google Scholar]
- Park J. H., Lee B., Jo Y., Choi S. H. (2016). Role of extracellular matrix protein CabA in resistance of Vibrio vulnificus biofilms to decontamination strategies. Int. J. Food Microbiol. 236 123–129. 10.1016/j.ijfoodmicro.2016.07.032 [DOI] [PubMed] [Google Scholar]
- Pu M., Duriez P., Arazi M., Rowe-Magnus D. A. (2018). A conserved tad pilus promotes Vibrio vulnificus oyster colonization. Environ. Microbiol. 20 828–841. 10.1111/1462-2920.14025 [DOI] [PubMed] [Google Scholar]
- Pu M., Rowe-Magnus D. A. (2018). A Tad pilus promotes the establishment and resistance of Vibrio vulnificus biofilms to mechanical clearance. NPJ Biofilms Microbiomes 4:10. 10.1038/s41522-018-0052-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra D. O., Richter A. M., Hengge R. (2013). Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 195 5540–5554. 10.1128/Jb.00946-913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Puhler A. (1983). A broad host range mobilization system for invivo genetic-engineering - transposon mutagenesis in gram-negative bacteria. Bio-Technology 1 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Watnick P., Kolter R. (2000). Biofilm, city of microbes. J. Bacteriol. 182 2675–2679. 10.1128/Jb.182.10.2675-2679.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. C., Blackman E. R., Morrison S. S., Gibas C. J., Oliver J. D. (2014). Transcriptome sequencing reveals the virulence and environmental genetic programs of Vibrio vulnificus exposed to host and estuarine conditions. PLoS One 9:e114376. 10.1371/journal.pone.0114376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F. H., Visick K. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17 109–118. 10.1016/j.tim.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamorano-Sanchez D., Fong J. C., Kilic S., Erill I., Yildiz F. H. (2015). Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J. Bacteriol. 197 1221–1235. 10.1128/JB.02439-2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.