Abstract
DNA topoisomerase IIα protein (TOP2α) 170 kDa (TOP2α/170) is an important target for anticancer agents whose efficacy is often attenuated by chemoresistance. Our laboratory has characterized acquired resistance to etoposide in human leukemia K562 cells. The clonal resistant subline K/VP.5 contains reduced TOP2α/170 mRNA and protein levels compared with parental K562 cells. The aim of this study was to determine whether microRNA (miRNA)-mediated mechanisms play a role in drug resistance via decreased expression of TOP2α/170. miRNA-sequencing revealed that human miR-9-3p and miR-9-5p were among the top six of those overexpressed in K/VP.5 compared with K562 cells; validation by quantitative polymerase chain reaction demonstrated overexpression of both miRNAs. miRNA recognition elements (MREs) for both miRNAs are present in the 3ʹ-untranslated region (UTR) of TOP2α/170. Transfecting K562 cells with a reporter plasmid harboring the TOP2α/170 3ʹ-UTR together with either miR-9-3p or miR-9-5p mimics resulted in a statistically significant decrease in luciferase expression. Mutating the miR-9-3p or miR-9-5p MREs prevented this decrease, demonstrating direct interaction between these miRNAs and TOP2α/170 mRNA. Transfection of K562 cells with miR-9-3p or miR-9-5p mimics led to decreased TOP2α/170 protein levels without a change in TOP2α/170 mRNA and resulted in attenuated etoposide-induced DNA damage (gain-of-miRNA-inhibitory function). Conversely, transfection of miR-9-3p or miR-9-5p inhibitors in K/VP.5 cells (overexpressed miR-9 and low TOP2α/170) led to increased TOP2α/170 protein expression without a change in TOP2α/170 mRNA levels and resulted in enhancement of etoposide-induced DNA damage (loss-of-miRNA-inhibitory function). Taken together, these results strongly suggest that these miRNAs play a role in and are potential targets for circumvention of acquired resistance to etoposide.
SIGNIFICANCE STATEMENT
Results presented here indicate that miR-9-3p and miR-9-5p decrease DNA topoisomerase IIα protein 170 kDa expression levels in acquired resistance to etoposide. These findings contribute new information about and potential strategies for circumvention of drug resistance by modulation of microRNA levels. Furthermore, increased expression of miR-9-3p and miR-9-5p in chemoresistant cancer cells may support their validation as biomarkers of responsiveness to DNA topoisomerase II–targeted therapy.
Introduction
DNA topoisomerase IIα (TOP2α) 170 kDa (TOP2α/170) induces covalent complexes with DNA and produces transient double-strand DNA breaks crucial for processes such as replication and chromosomal dysjunction at mitosis (Deweese and Osheroff, 2009). The TOP2α/170 enzyme exists as a homodimer. Each monomer utilizes a tyrosine active site as a nucleophile to begin a reversible transesterification reaction with 5ʹ-nucleobase scissile sites on the phosphate backbones of the top or bottom strands of DNA (Deweese and Osheroff, 2009). This cleavage and subsequent rapid religation of DNA allows for the relaxation/separation of DNA entanglements (Vos et al., 2011; Chen et al., 2013; Pendleton et al., 2014; Ashour et al., 2015). TOP2α/170 has been used as an important target in cancer therapy (Larsen et al., 2003; Deweese and Osheroff, 2009; Nitiss, 2009; Pommier et al., 2010). Type IIA topoisomerase interfacial inhibitors, such as etoposide, stabilize the otherwise short-lived enzyme-DNA complexes by insertion within the four base pair scissile break sites generated by TOP2α/170 (Laponogov et al., 2010; Pommier et al., 2010), thereby inhibiting religation. Programmed cell death can ensue because of an aggregation of double-strand DNA breaks (Pommier et al., 2010).
Acquired resistance to TOP2α/170-targeted agents (e.g., etoposide, mitoxantrone, amsacrine, doxorubicin, and analogs) is important clinically in the treatment of a variety of malignancies (Chen and Liu, 1994; Vassetzky et al., 1995; Pilati et al., 2012; Ganapathi and Ganapathi, 2013). Though mutant forms of TOP2α have been reported in acquired resistance (Zwelling et al., 1989; Bugg et al., 1991; Hinds et al., 1991; Kubo et al., 1996), most evidence from selected cell lines (Ritke and Yalowich, 1993; Ritke et al., 1994; Mirski et al., 1993, 2000; Harker et al., 1995; Mirski and Cole, 1995; Wessel et al., 1997; Yu et al., 1997; Burgess et al., 2008) and patient samples (Ganapathi and Ganapathi, 2013) suggests that decreased levels of TOP2α are a major determinant of drug resistance. Our laboratory has previously demonstrated that a cloned K562 cell line (designated K/VP.5 cells) with acquired resistance (∼25-fold) to etoposide expresses less TOP2α/170 protein compared with the parental K562 cells with no change in drug accumulation and no overexpression of multidrug resistance protein 1/P-glycoprotein (ABCB1) (Ritke and Yalowich, 1993; Ritke et al., 1994). In addition, K/VP.5 cells overexpressed a novel TOP2α 90 kDa (TOP2α/90) isoform, the translation product of TOP2α mRNA that retains a processed intron 19 (Kanagasabai et al., 2017; Kanagasabai et al., 2018). Importantly, the TOP2α/90 isoform heterodimerizes with the TOP2α/170 isoform, resulting in a dominant-negative effect with respect to etoposide-induced covalent TOP2α-DNA complexes, DNA damage, and cytotoxicity (Kanagasabai et al., 2018). Together, literature indicates that acquired resistance to TOP2α-targeted agents can result from multiple mechanisms involving dysregulation of TOP2α.
MicroRNA (miRNA)-mediated post-transcriptional gene silencing is an established mechanism of genetic regulation (Bartel, 2009). Typically, mature noncoding miRNAs (21–25 nucleotides) are embedded in primary (pri)-miRNA transcripts as imperfectly based-paired stem-loops (Ha and Kim, 2014). In the nucleus, pri-miRNA is further processed to a hairpin-structured precursor miRNA (pre-miRNA) of ∼60–70 nucleotides. Subsequently, in the cytoplasm, the terminal loop is removed, the resulting miRNA duplexes of ∼22 nucleotides are loaded into the RNA-induced silencing complex (RISC), and then the duplex is matured (Treiber et al., 2019). Importantly, two distinct functional mature miRNAs can be processed from opposite sides of the same pre-miRNA (i.e., miRNA matured from the 5ʹ arm denoted -5p or from the 3ʹ arm denoted -3p) (Treiber et al., 2019). Finally, the miRNA-RISC complex (miRISC) is guided by a seed sequence (spanning position 2–8 at the 5ʹ end of the miRNA) that base pairs to a specific miRNA recognition element (MRE) typically harbored in the 3ʹ-untranslated region (UTR) of a target mRNA (Bushati and Cohen, 2007). Interactions of miRNAs with the 5ʹ-UTR and coding region of mRNAs have also been reported (Broughton et al., 2016). Upon mRNA binding, miRISC will reduce the expression of the target mRNA by blocking the ribosome from mRNA translation and/or by initiating mRNA degradation (Fabian and Sonenberg, 2012; Jonas and Izaurralde, 2015).
In this study, we posit that TOP2α/170 expression is attenuated in etoposide-resistant K/VP.5 cells, in part by miRNA-mediated mechanisms that are determinants of drug resistance. Results demonstrated that human miR-9-3p (previously denoted miR-9*) and miR-9-5p (previously denoted miR-9): 1) were overexpressed in K/VP.5 cells; 2) contained corresponding MREs in the 3ʹ-UTR of TOP2α/170; 3) functionally regulated the protein expression levels of TOP2α/170; and 4) modulated etoposide activity. Altogether results are consistent with a role for these miRNAs in acquired drug resistance.
Materials and Methods
Cell Culture.
Human K562 leukemia cells were maintained in Dulbecco’s modified Eagle’s medium (Corning, Manassas, VA) supplemented with 10% FBS. Cloned etoposide-resistant K562 cells (K/VP.5) were generated by continuous incubation in 0.5 µM etoposide followed by isolation via limiting dilution, as previously described (Ritke and Yalowich, 1993). K/VP.5 cells were maintained in Dulbecco’s modified Eagle’s medium/10% FBS with 0.5 µM etoposide added every other week. All experiments described below were performed utilizing K562 and K/VP.5 cells growing in log phase.
miRNA-Sequencing.
A mirVana miRNA Isolation Kit with phenol was used to isolate total RNA from K562 and K/VP.5 cells following the manufacturer’s recommended instructions (cat. no. AM1560; ThermoFisher, Waltham). The total RNA was subsequently DNase I–treated and concentrated on a spin-column following the manufacturer’s recommended instructions (cat. no. R1013; Zymo Research, Irvine, CA). miRNA-sequencing (Seq) analysis was performed by the Biomedical Genomics Core of The Research Institute at Nationwide Children’s Hospital, Columbus, OH. Briefly, NEB Next Small RNA libraries were prepared from the purified K562 and K/VP.5 total RNA samples and the libraries were size selected using Sage Pippin (130–200 bp range, adapters are ∼128 bp) with an average final library size of 180 bp and sequenced on MiniSeq as 2 × 75 bp reads.
Computational Processing of smRNA-Seq Data.
First, 3′ adaptor sequences and low-quality bases were trimmed from 75 base pair paired-end sequence reads using Cutadapt v2.4 (Martin, 2011). Trimmed reads with a sequence length between 15 and 50 base pairs were aligned to potential contaminant sequences of human genome (ribosomal RNAs and transfer RNAs) using Bowtie v1.1.2 (Langmead et al., 2009) with a seed length of 18 and allowing zero mismatches in the seed. Then, remaining reads were aligned to miRBase (Griffiths-Jones, 2004) v21 mature sequences using Bowtie v1.1.2 with a seed length of 18, allowing one mismatch in seed and reporting maximum five alignments per read. Finally, remaining reads were aligned to human reference genome, Genome Reference Consortium human genome build 38, using Bowtie v1.1.2 with a seed length of 18, allowing one mismatch in seed and reporting only unique alignments for each read. Using Samtools (Li et al., 2009) and in-house R scripts, total number of reads for each mature miRNA was counted. Fractional counts were used when a sequence read aligned to more than one miRNA. Genomic alignment was quantified using featureCounts function of SubReads (Liao et al., 2014) with Gencode v22 annotation. Using limma R/Biconductor package (Ritchie et al., 2015), raw read counts were normalized as log2 counts per million, and differential expression analysis was performed between three K562 and four K/VP.5 RNA samples.
TOP2α/170 mRNA/miRNA Bioinformatic Analyses.
To predict putative MREs harbored in the 3′-UTR of TOP2α/170 mRNAs, multiple computational algorithms were used (TargetScan 7.2, http://www.targetscan.org; Agarwal et al., 2015 and DIANA-microT-CDS, http://www.microrna.gr/microT-CDS, fifth version of the microT algorithm, Paraskevopoulou et al., 2013). GEO accession number for small (sm)RNA-seq data: GSE141687.
Luciferase Reporter Constructs.
The psiCHECK-2 dual luciferase vector was purchased from Promega Corporation (cat. no. C8021; Madison, WI) and linearized by XhoI digestion (cat. no. R0146S; New England Biolabs, Ipswich, MA) according to the manufacturer’s protocol. A 998-bp fragment encompassing the entire TOP2α/170 3′-UTR was polymerase chain reaction (PCR)-amplified by utilizing sense (5′-AGGCGATCGCTCGAGAATGTGAGGCGATTATTTTAAGTAA-3′) and antisense (5′-ATTCCCGGGCTCGAGTGGGTTGCAATGTTTAGAACATT-3′) primers using standard PCR procedures with a proofreading polymerase, CloneAmp HiFi (cat. no. 639298; Takara Bio Inc., Kusatsu, Shiga, Japan). Genomic DNA isolated from K562 cells was used as template. The primer sequences homologous to the regions immediately downstream of the TOP2α/170 stop codon and immediately upstream of the TOP2α/170 poly(A) tail are bolded above. The sense and antisense primers also harbor 15 nucleotide extensions (not bolded) that are homologous to ends of the XhoI linearized dual luciferase psiCHECK-2 reporter plasmid and are necessary for “In-Fusion” cloning methodology (In-Fusion HD Cloning Kit, cat. no. 638916; Takara Bio Inc). The TOP2α/170 3′-UTR PCR product was gel purified and subcloned into the XhoI site, which is located downstream of the Renilla luciferase (luc) reporter open reading frame in the psiCHECK-2 reporter plasmid, utilizing the manufacturer’s protocol. The authenticity and orientation of the inserts relative to the Renilla luciferase gene were confirmed by dideoxy sequencing. The resulting recombinant plasmid was designated psiTOP2α/170/UTR.
The mutant reporter construct psiTOP2α/170/UTR-mutant (mut)-5p was generated utilizing the psiTOP2α/170/UTR vector as template and mutating the miR-9-5p MRE (located at 703–709 bp) harbored in the TOP2α/170 3′-UTR using a Q5 Site-Directed Mutagenesis kit from New England Biolabs (cat. no. E0554S) following the manufacturer’s instructions. The primers used for mutagenesis were sense (5′-AAGCGTGGAGAAACCAATTTCTAAG-3′) and antisense (5′-GAGATTCAGACTCAGAGGCAGC-3′). Importantly, these primers were designed with their 5′ ends annealing back-to-back (inverse PCR). The nucleotides that were mutated are shown in bold print. The mutation of the miR-9-5p seed sequence was confirmed by dideoxy sequencing. The mutant reporter construct psiTOP2α/170/UTR-mut-3p was generated in the same manner as psiTOP2α/170/UTR-mut-5p but the predicted miR-9-3p site was modified (466–471 bp of the 3′-UTR) with the following sense (5′-GAAACTGGTTCTAGTACAGATAC-3′) and antisense (5′-TTCTATCTGATGGTAAA-TTATG-3′) primers. Finally, transformed bacterial cultures were grown, and each reporter construct was isolated with a Qiagen Plasmid Midi Kit (cat. no. 12143; Hilden, Germany).
Transfection Experiments.
K562 and K/VP.5 cells (5 × 105 in 5 ml per condition) were transfected with the psiTOP2α/170/UTR dual luciferase construct utilizing 1 µg of the plasmid, 5.5 µl Lipofectamine 3000, 10 µl P3000 (cat. no. L3000008; ThermoFisher), and 250 µl Opti-Mem Medium (cat. no. 31985062; ThermoFisher). Cotransfection experiments utilizing K562 cells were performed as described above with the respective dual luciferase construct (psiTOP2α/170/UTR, psiTOP2α/170/UTR-3p-mut, or psiTOP2α/170/UTR-5p-mut) and either a miRNA mimic negative control (cat. no. 4464058; ThermoFisher), miR-9-3p mimic (cat. no. MC13072), or miR-9-5p mimic (cat. no. MC10022). In addition, K562 cells were transfected with only the mimics described above for subsequent TOP2α/170 immuno-assays and for DNA damage studies. Finally, K/VP.5 cells were transfected with either a miRNA inhibitor negative control (cat. no. MH4464076; ThermoFisher), miR-9-3p inhibitor (cat. no. MH13072), or miR-9-5p inhibitor (cat. no. MH10022). In all transfection experiments employing miRNA mimics or miRNA inhibitors, the final concentration was 25 nM. Forty-eight hours after transfection, total RNA and/or cellular extracts were prepared for quantitative PCR (qPCR), luciferase, immunoblotting, and DNA damage assays as outlined below.
Quantitative PCR Assays.
Total RNA was isolated from nontransfected and transfected K562 and K/VP.5 cells using PureZOL RNA Isolation Reagent from Bio-Rad Laboratories (cat. no. 7326880; Hercules, CA). RNA samples were subsequently treated with DNase I (TURBO DNA-free Kit, cat. no. AM2238; ThermoFisher Scientific). RNA (1 µg) used for TOP2α/170 mRNA quantification experiments was reverse transcribed using random hexamers and MultiScribe Reverse Transcriptase (High Capacity cDNA Reverse Transcription Kit, cat. no. 4368814; ThermoFisher Scientific) as previously described by our laboratory (Kanagasabai et al., 2017, 2018). qPCR evaluations (20 µl/reaction) were performed using 5 µl of a 1:10 dilution of cDNA with primer/probe sets specific for TOP2α/170 (Assay ID Hs01032135_m1; ThermoFisher Scientific) and TATA-binding protein (TBP) (Assay ID Hs99999910_ml; ThermoFisher Scientific). The relative gene expression level of TOP2α/170 in each experimental condition was normalized to TBP expression using the 2−ΔΔCt method (Schmittgen and Livak, 2008). In addition, RNA samples (50 ng) were reverse transcribed using miR-9-3p, miR-9-5p, and RNU48 antisense primers and MultiScribe Reverse Transcriptase in a 15 µl reaction according to manufacturer instructions. qPCR for miRNA quantification (20 µl/reaction) was performed using 1.33 µl of cDNA with primer/probe sets specific for miR-9-3p (Assay ID 00231), miR-9-5p (Assay ID 000583), and RNU48 (Assay ID 001006). The relative gene expression level of miR-9-3p and/or miR-9-5p in each experimental condition was normalized to RNU48 expression using the 2−ΔΔCt method (Schmittgen and Livak, 2008). Finally, cDNA synthesized from RNA isolated from nontransfected K562 and K/VP.5 cells was used in qPCR experiments with primer/probe sets specific for pri hsa-miRNA-9s (mir-9-1, Assay ID Hs03303201_pri; mir-9-2, Assay ID Hs03303202_pri; and mir-9-3, Assay ID Hs03293595_pri; ThermoFisher Scientific). The relative gene expression level of mir-9-1, mir-9-2, and mir-9-3 in K562 and K/VP.5 cells was normalized to TBP expression using the 2−ΔΔCt method (Schmittgen and Livak, 2008).
Luciferase Assay.
Forty-eight hours after transfection, K562 and/or K/VP.5 cells were washed and lysed with passive lysis buffer (cat. no. E1941; Promega), and firefly and Renilla luciferase activities were determined by using the Dual-Luciferase Reporter Assay System (cat. no. E1910; Promega) with a Synergy H1 Hybrid Multi-Mode Reader (BioTek, Winooski, VT). Renilla luciferase expression in the psiCHECK-2 vector is generated via an SV40 promoter, whereas firefly luciferase in this vector is generated via a herpes simplex virus-1 thymidine kinase promoter and has been specifically designed to be an intraplasmid transfection normalization reporter. The Renilla luciferase signal was therefore normalized to the firefly luciferase signal.
Western Blot Analysis.
Forty-eight hours after transfection, K562 and/or K/VP.5 cells were pelleted and washed in PBS. After washing, 1.5 million cells were lysed in 2× Laemmli sample buffer (cat. no. 1610737; Bio-Rad Laboratories) containing 5% β-mercaptoethanol and boiled at 100°C for 5 minutes. DNA was sheared using a sonicator (Tekmar, Mason, OH) at 35 W three times in 10-second intervals, and the samples were subsequently electrophoresed on a NuPAGE 4%–12% [bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol gel (cat. no. NP0322BOX; ThermoFisher). Duplicate samples were run with an equal amount of protein (16.6 µg) loaded into each well, and an equal volume of Precision Plus Protein Dual Color Standards was also run for molecular weight reference (cat. no. 1610374; Bio-Rad Laboratories). Protein was transferred to a polyvinylidene difluoride (PVDF) membrane (cat. no. 1620177; Bio-Rad Laboratories) using a semidry electroblotter (ThermoFisher). The PVDF membrane was then sectioned. The 75–250 kDa portion was labeled by overnight incubation at 4°C with rabbit polyclonal antibodies raised against the N-terminal sequence of human TOP2α/170 (cat. no. ab74715; Abcam, Cambridge, MA; used at 1:1000 dilution). The 25–75 kDa portion was labeled by overnight incubation at 4°C with a [glyceraldehyde 3-phosphate dehydrogenase (GAPDH)] mouse monoclonal antibody (1:5000; cat. no. sc-47724; Santa Cruz Biotechnology, Santa Cruz, CA). PVDF membranes were subsequently incubated at room temperature for 1.5 hours with a donkey anti-rabbit or donkey anti-mouse secondary antibody, respectively (1:8000 dilution; Jackson ImmunoResearch, West Grove, PA). Antibody-labeled TOP2α/170 and GAPDH were detected using the Immun-Star horseradish peroxidase Chemiluminescence Kit (cat. no. 170-5040; Bio-Rad Laboratories). All immunoassay images were taken with the ChemiDoc XRS+ imaging system and analyzed with ImageLab software (Bio-Rad Laboratories).
Comet DNA Damage Assays.
Single-cell gel electrophoresis (Comet) assays were performed according to the manufacturer’s protocol (cat. no. 4250-050-K; Trevigen, Gaithersburg, MD) and as previously described by our laboratory (Vlasova et al., 2011; Kanagasabai et al., 2017). Briefly, K562 or K/VP.5 cells were transfected with miR-9 mimics or inhibitors, respectively, as described above. Forty-eight hours after transfection, cells were washed and resuspended in buffer (25 mM HEPES, 10 mM glucose, 1 mM MgCl2, 5 mM KCl, 130 mM NaCl, 5 mM monosodium phosphate, pH 7.4). Transfected K562 cells were subsequently incubated with 2 or 5 µM etoposide or DMSO (solvent control) for 1 hour at 37°C. Transfected K/VP.5 cells were incubated with 10 or 25 µM etoposide or DMSO for 1 hour at 37°C. Cells were subsequently washed with ice-cold buffer and resuspended to 0.28 × 106 cells/ml and then diluted so that ∼2000 cells in low melt agarose underwent alkaline electrophoresis. After staining with a fluorescent DNA intercalating dye, SYBR Gold, the migrating fragments (comet tail) from the nucleoid (comet head) were visualized and the images captured by fluorescence microscopy. The Olive tail moment, which is defined as the product of the comet tail length and the fraction of total DNA in the comet tail (Olive, 2002), was quantified by the ImageJ processing program with the open-source software tool Open Comet (Gyori et al., 2014). Olive tail moments from greater than 100 cells per sample condition were determined.
Data Analysis.
Statistical analysis was performed using SigmaPlot 14. All data are expressed as the mean ± S.D. Group-wise differences were analyzed using a two-tailed paired Student’s t test with no correction for multiple comparisons. qPCR gene expression data (2−ΔCt values) were subjected to log transformation to assure distribution normality prior to paired Student’s t test analysis (Ganger et al., 2017). A P value of <0.05 was considered statistically significant.
Results
TOP2α/170 Expression Is Attenuated in Etoposide-Resistant K562 Cells.
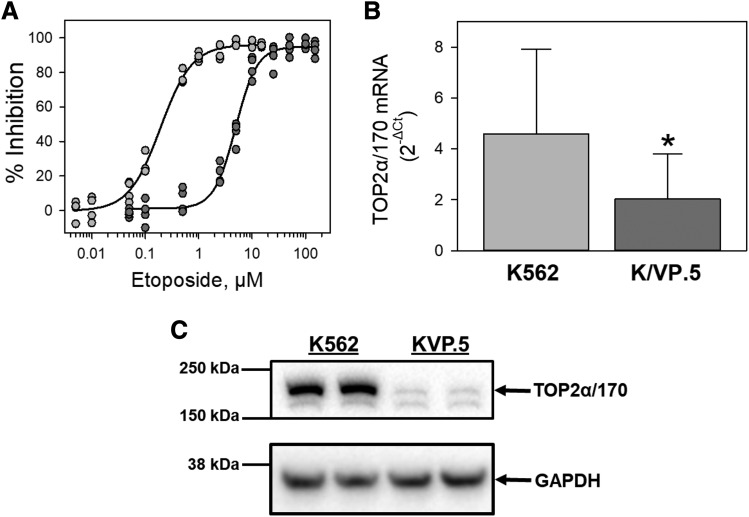
Our laboratory previously established and characterized an etoposide-resistant K562 cell line, designated K/VP.5 cells (Ritke and Yalowich, 1993), which are cross-resistant also to other TOP2α/170-targeted agents (Ritke et al., 1994). To demonstrate resistance to etoposide, cell growth inhibition of K562 and K/VP.5 cells was evaluated by 48-hour incubation with increasing concentrations of etoposide. Results show that K/VP.5 cells are 26-fold resistant to etoposide (Fig. 1A). To investigate the relative TOP2α/170 mRNA expression levels, K562 and K/VP.5 cDNAs were subjected to qPCR utilizing TaqMan hydrolysis primers/probes. The results of these experiments demonstrated that TOP2α/170 mRNA expression levels were reduced in K/VP.5 cells to 41.2% the level expressed in parental K562 cells (Fig. 1B), recapitulating the 2.5-fold reduction of TOP2α/170 mRNA in K/VP.5 cells found previously by Northern blot analysis (Ritke and Yalowich, 1993). TOP2α/170 protein expression was also decreased in K/VP.5 cells to a level 10.6% that of parental K562 cells (Fig. 1C).
Fig. 1.
K/VP.5 cell resistance to etoposide is associated with attenuated TOP2α/170 mRNA and protein levels. (A) Parental K562 and K/VP.5 cells with acquired resistance to etoposide were incubated with increasing concentrations of etoposide for 48 hours, and the cells were then counted. The extent of growth (beyond initial concentration of 1 × 105 cell/ml) in drug-treated vs. control was ultimately expressed as percent inhibition of control growth. Results are shown as a scattergram from four independent experiments performed on separate days. (B) qPCR experiments were performed utilizing K562 and K/VP.5 cDNAs and a TaqMan hydrolysis assay specific for TOP2α/170. Results shown are the mean ± S.D. from five RNA/cDNA isolations/determinations performed on separate days, K/VP.5 vs. K562 for expression of TOP2α/170 mRNA (*P < 0.001). Calculated 2−ΔCt values from qPCR assays were log transformed to assure distribution normality prior to data analysis using a two-tailed paired Student’s t test. (C) Representative immunoassay using K562 and K/VP.5 cellular lysates. Blots were probed with antibodies specific for the N-terminal portion of TOP2α/170 (i.e., amino acids 14–27) or for GAPDH. Averaging results from five separate paired collections of K562 and K/VP.5 cells on different days, there was a reduction of TOP2α/170 in K/VP.5 cells to 10.6% the level found in parental K562 cells, taking into account the GAPDH loading control (P = 0.014).
miR-9-3p and miR-9-5p Are Overexpressed in K/VP.5 Cells.
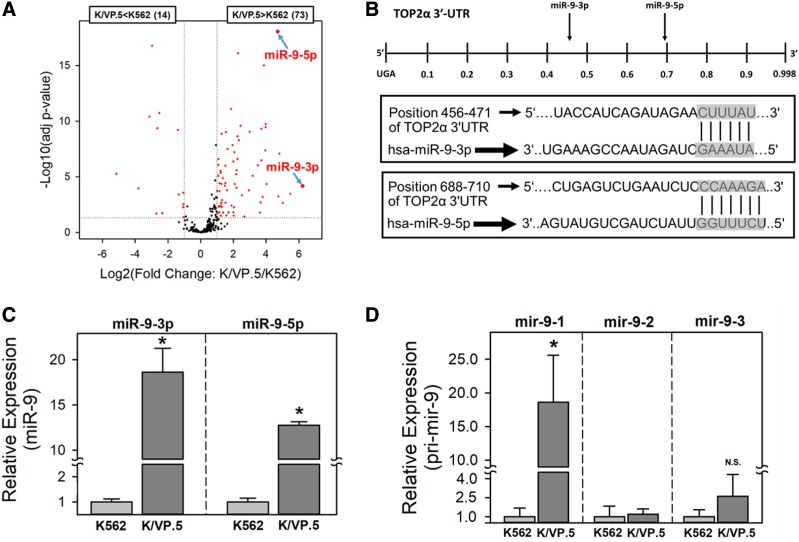
Because miRNAs play an integral role in almost all known biologic processes (reviewed in Bushati and Cohen, 2007; Bartel, 2009; Fabian and Sonenberg, 2012), we hypothesized that TOP2α/170 expression is attenuated in K/VP.5 cells, in part by a miRNA-mediated mechanism. As an initial test of this hypothesis, total mRNA was isolated from K562 and K/VP.5 cells, and small RNA libraries were generated, size selected, and sequenced to examine the global miRNA expression levels in both cell lines. The volcano plot (Fig. 2A) illustrates that, of the ∼300 mature miRNAs that were detected in these cells, 73 miRNAs were overexpressed (fold change >2; adjusted P value <0.05) and 14 were underexpressed (fold change >2; adjusted P value <0.05) in K/VP.5 compared with K562 cells.
Fig. 2.
miR-9-3p and miR-9-5p are overexpressed in K/VP.5 cells. (A) Volcano plot analysis of miRNAs overexpressed or underexpressed in K/VP.5 compared with K562 cells. Differentially expressed miRNAs are defined as those with at least a ± 2-fold change [red circles outside vertical dotted lines and above the horizontal dotted line (P < 0.05)]. (B) Schematic representation of the location of putative MREs harbored in the 3ʹ-UTR of TOP2α/170 mRNA. Additionally, the “seed sequence” complementarity is shown between miR-9-3p and miR-9-5p and the putative TOP2α/170 3ʹ-UTR MREs. (C) qPCR utilizing K562 and K/VP.5 cDNAs and TaqMan hydrolysis assays specific for miR-9-3p and miR-9-5p. Results shown are the mean ± S.D. for three determinations made from three separate RNA/cDNA preparations made on separate days; *P < 0.001, comparing K/VP.5 to K562 cell levels of miR-9-3p and miR-9-5p. (D) qPCR utilizing K562 and K/VP.5 cDNAs and TaqMan hydrolysis assays specific for pri-miRNAs (i.e., mir-9-1, mir-9-2, and mir-9–3) transcribed from three independent genes: MIR9-1, MIR9-2, and MIR9-3. Results shown are the mean ± S.D. from four to seven determinations made from separate RNA/cDNA preparations; *P < 0.001 (N = 7), comparing K/VP.5 to K562 cell levels of mir-9-1; *P = 0.078 (N = 4), comparing K/VP.5 to K562 cell levels of mir-9-3. For Fig. 2, C and D, calculated 2−ΔCt values from qPCR assays were log transformed to assure distribution normality prior to data analysis using a two-tailed paired Student’s t test. N.S., not significant.
The miRNA-Seq data demonstrated that miR-9-3p (75.1-fold) and miR-9-5p (26.2-fold) were among the top six overexpressed pre-miRNAs in K/VP.5 cells (Fig. 2A). TargetScan (Agarwal et al., 2015) and DIANA-microT-CDS (Paraskevopoulou et al., 2013) algorithms identified a putative mature miR-9-5p MRE located at position 703–709 nt in the TOP2α/170 3ʹ-UTR (Fig. 2B). In addition, a putative seed sequence for miR-9-3p binding was also predicted - by DIANA-microT-CDS at position 466–471 nt of the TOP2α/170 3ʹ-UTR (Fig. 2B). Based on these evaluations and literature establishing that miR-9-3p and miR-9-5p can be simultaneously matured from pre-miR (mir-9) (Packer et al., 2008; Schraivogel et al., 2011, Nowek et al., 2016, 2018), the potential effects of both these miRNAs were studied further.
To independently validate the miR-9 miRNA-Seq data (Fig. 2A), qPCR experiments were performed utilizing TaqMan primer/probe sets specific for mature miR-9-3p and miR-9-5p. Both miR-9-3p and miR-9-5p were statistically significantly overexpressed in K/VP.5 cells compared with K562 cells: 18.6-fold and 12.7-fold, respectively; P < 0.001 (Fig. 2C). Because miR-9-3p and miR-9-5p can be matured from pri-miRNAs (mir-9s) transcribed from three independent genes, with MIR9-1, MIR9-2, and MIR9-3 located on chromosomes 1, 5, and 15, respectively (Yuva-Aydemir et al., 2011), qPCR experiments were performed utilizing TaqMan primer/probe sets specific for each MIR9 gene. These experiments demonstrated that mir-9-1 was the only miRNA precursor statistically significantly overexpressed in K/VP.5 cells compared with K562 cells (18.6-fold; P < 0.001) (Fig. 2D). Together, these observations suggest that it is the activation of the MIR9-1 gene that results in the overexpression of miR-9-3p and miR-9-5p in K/VP.5 cells.
miR-9-3p and miR-9-5p Directly Interact with the TOP2α/170 3′-UTR.
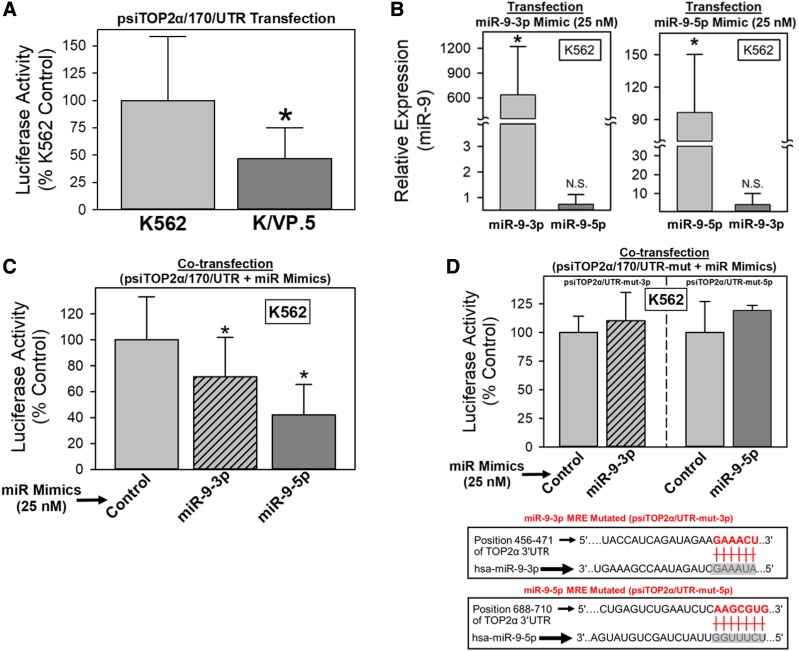
Initial evaluation of putative miRNA regulation of TOP2α/70 expression used a dual luciferase reporter assay system in which binding of a given miRNA to its distinct MRE may repress reporter protein production, thereby reducing Renilla-luc activity (Kuhn et al., 2008; Sansom et al., 2010; Elton and Yalowich 2015). Therefore, the TOP2α/170 3ʹ-UTR (998-bp), which harbored MREs for miR-9-3p and miR-9-5p (Fig. 2B), was subcloned immediately downstream of the Renilla-luc open reading frame, and the plasmid construct was designated psiTOP2α/170/UTR. After transfection of cells with psiTOP2α/170/UTR, there was a statistically significant reduction in luciferase activity in K/VP.5 compared with K562 cells (P < 0.001), suggesting a miRNA-mediated mechanism regulating TOP2α/170 protein expression levels (Fig. 3A).
Fig. 3.
miR-9-3p and miR-9-5p directly interact with the TOP2α/170 3′-UTR. (A) K562 and K/VP.5 cells were transfected with the psiTOP2α/170/UTR luciferase reporter construct (1 µg). Forty-eight hours later, luciferase activities were measured. Renilla luciferase activity was normalized to firefly luciferase activity. Results shown are the mean ± S.D. from 17 experiments performed on separate days; *P < 0.001, K/VP.5 luciferase vs. K562 luciferase levels. (B) K562 cells were cotransfected with the psiTOP2α/170/UTR luciferase reporter construct (1 µg) and either control, miR-9-3p, or miR-9-5p mimics (25 nM). Forty-eight hours later, total RNA was isolated and reverse transcribed. qPCR experiments were performed utilizing K562 cDNAs and TaqMan hydrolysis assays specific for miR-9-3p and miR-9-5p. Results shown are the mean ± S.D. from five experiments performed on separate days; *P < 0.001, miR-9-3p levels in miR-9-3p mimic–transfected K562 vs. nontransfected K562 cells, and miR-9-5p levels in miR-9-5p mimic–transfected K562 vs. nontransfected K562 cells, respectively. mir-9-5p was not significantly increased in mir-9-3p mimic–transfected cells (P = 0.340) nor was mir-9-3p significantly increased in mir-9-5p mimic–transfected cells (P = 0.223). Calculated 2−ΔCt values from qPCR assays were log transformed to assure distribution normality prior to data analysis using a two-tailed paired Student’s t test. (C) K562 cells were cotransfected with the psiTOP2α/170/UTR luciferase reporter construct (1 µg) and either control, miR-9-3p, or miR-9-5p mimics (25 nM). Forty-eight hours later, luciferase activities were measured. Results shown are the mean ± S.D. from nine experiments performed on separate days; P < 0.05 for miR-9-3p mimic and miR-9-5p mimic vs. control mimic–transfected K562 cells. (D) K562 cells were cotransfected with luciferase constructs, wherein the miR-9-3p and miR-9-5p putative binding sites were mutated (shown schematically) and either control, miR-9-3p, or miR-9-5p mimics (25 nM). Forty-eight hours later, luciferase activities were measured. Results shown are the mean ± S.D. from three to five experiments performed on separate days. N.S., not significant.
To evaluate the putative role of miR-9-3p and/or miR-9-5p in this regulation, K562 cells, which express low levels of miR-9-3p and miR-9-5p, were cotransfected with the psiTOP2α/170/UTR construct (1 μg) plus 25 nM miR-9-3p, miR-9-5p, or control nontargeting miRNA mimics (chemically modified double-stranded RNAs that mimic endogenous miRNAs) for gain-of-inhibitory-function experiments. Forty-eight hours after transfection, qPCR experiments utilizing TaqMan probes specific for miR-9-3p or miR-9-5p demonstrated that transfection with the corresponding miRNA mimic significantly increased the detectable levels of miR-9-3p (600-fold) and miR-9-5p (94-fold), with P < 0.001 for both (Fig. 3B). No significant increase was observed for miR-9-5p after miR-9-3p mimic transfection nor was there a statistically significant increase in miR-9-3p after mir-9-5p mimic transfection, with P > 0.220 for both (Fig. 3B), demonstrating the specificity of miR-9 mimics and the qPCR assays. K562 cells were cotransfected with psiTOP2α/170/UTR and miR-9 mimics, and this was followed by measurement of luciferase activity 48 hours later (Fig. 3C). There was a statistically significant reduction in luciferase activity to 71.4% (P = 0.033) and 42.1% (P = 0.001) of the nontargeted miR-9 mimic control in cells treated with miR-9-3p mimic and miR-9-5p mimic, respectively (Fig. 3C).
To validate that miR-9-3p and miR-9-5p interact with the predicted MREs harbored in the 3ʹ-UTR of TOP2α/170 mRNA, additional luciferase reporter constructs were generated wherein the MRE “seed” sequences (CUUUAU for miR-9-3p and CCAAAGA for miR-9-5p), which are complementary to the 5ʹ ends of miR-9-3p and miR-9-5p, were mutated (GAAACU for miR-9-3p and AAGCGUG for miR-9-5p) (Fig. 3D bottom). The mutant constructs were designated psiTOP2α/170/UTR-mut-3p and psiTOP2α/170/UTR-mut-5p. Cotransfection of K562 cells (48 hours) with psiTOP2α/170/UTR-mut-3p plus miR-9-3p mimic or with psiTOP2α/170/UTR-mut-5p plus miR-9-5p mimic revealed that, compared with wild-type psiTOP2α/170/UTR transfection results (Fig. 3C), these miRNAs no longer had any effect on luciferase activity (Fig. 3D). Together, results from Fig. 3, C and D indicate that miR-9-3p and miR-9-5p can interfere with luciferase expression by directly interacting with the predicted miR-9-3p and miR-9-5p MREs harbored within the 3′-UTR TOP2α/170 mRNA.
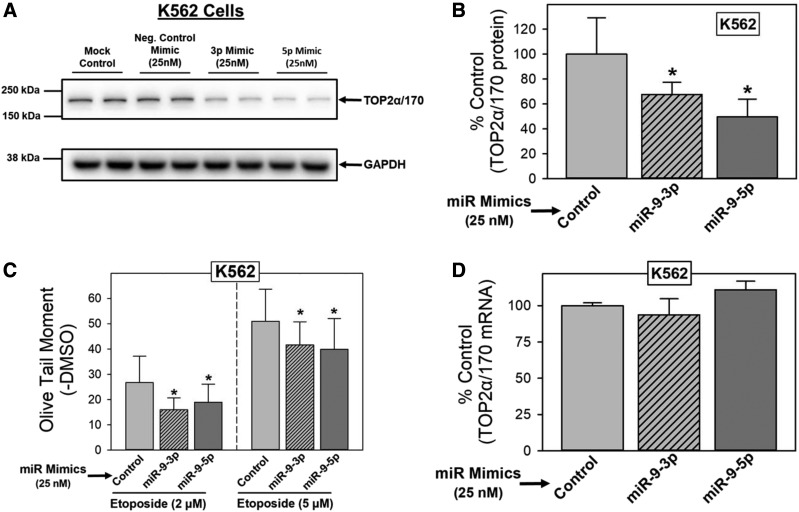
miR-9-3p and miR-9-5p Overexpression Results in Decreased TOP2α/170 Protein and Reduced Etoposide-Induced DNA Damage in K562 Cells.
If TOP2α/170 mRNA is a biologic target of miR-9-3p and miR-9-5p, alteration of the endogenous expression levels of these miRNAs should correspond to predictable changes in TOP2α/170 protein. Therefore, additional gain-of-inhibitory-function experiments were performed by transfecting K562 cells (48 hours) with 25 nM miR-9-3p, miR-9-5p, or control (nontargeting) miRNA mimics, which was followed by immunoblotting of cell lysates to evaluate TOP2α/170 protein levels. Figure 4A shows that TOP2α/170 levels were similar in mock transfected (lipofectamine only) and miRNA mimic control conditions (Fig. 4A). Consistent with our hypothesis, TOP2α/170 protein levels were decreased when miR-9-3p or miR-9-5p were overexpressed (Fig. 4A). Averaging results from five separate experiments run on separate days, there was a reduction of TOP2α/170 levels to 67.6% (P < 0.001) and 49.6% (P = 0.004) of miRNA mimic control in cells treated with miR-9-3p and miR-9-5p, respectively (Fig. 4B).
Fig. 4.
miR-9-3p and miR-9-5p overexpression results in decreased TOP2α/170 protein and reduced etoposide-induced DNA damage in K562 cells. (A) Representative immunoassay using cellular lysates from K562 cells that were transfected (48 hours) with control, miR-9-3p, miR-9-5p mimics (25 nM), or transfectant reagent alone (Lipofectamine: mock control). Blots were probed with antibodies specific for the N-terminal portion of TOP2α/170 (i.e., amino acids 14–27) or for GAPDH. (B) Expression of TOP2α/170 protein levels in K562 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p mimics (25 nM). Results shown are the mean ± S.D. from five experiments performed on separate days; *P < 0.05 comparing miR-9-3p and miR-9-5p mimic–transfected K562 lysates vs. control mimic–transfected K562 lysates. (C) Etoposide (2 and 5 µM)-induced DNA damage in control, miR-9-3p, or miR-9-5p mimic (25 nM)-transfected (48 hours) K562 cells was determined by Comet assays after a 1-hour incubation and subtraction of DMSO vehicle controls, as described in Materials and Methods. Results shown are the mean ± S.D. from seven experiments performed on separate days; *P < 0.05 comparing miR-9-3p and miR-9-5p mimic–transfected vs. control mimic–transfected K562 cells. (D) TOP2α/170 mRNA levels in K562 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p mimics (25 nM). Results shown are the mean ± S.D. from three experiments performed on separate days.
Etoposide-induced DNA damage correlates most often with levels of the target type II Topoisomerases (Mirski et al., 1993, 2000; Ritke and Yalowich, 1993; Ritke et al., 1994; Harker et al., 1995; Mirski and Cole, 1995; Wessel et al., 1997; Yu et al., 1997; Burgess et al., 2008; Ganapathi and Ganapathi, 2013). Hence, we next investigated whether forced expression of miR-9-3p and miR-9-5p would diminish etoposide activity in parallel with a reduction in TOP2α/170 levels (as demonstrated in Fig. 4, A and B). K562 cells were transfected with 25 nM miR-9-3p, mir-9-5p, or nontargeting control miRNA mimics. Forty-eight hours after transfection, cells were incubated for 1 hour with etoposide (2 or 5 μM) or DMSO vehicle and subsequently evaluated for DNA damage by alkaline single-cell gel electrophoresis (Comet) assays (Kanagasabai et al., 2017, 2018). Etoposide-induced DNA strand breaks were attenuated in miR-9-3p and miR-9-5p mimic–transfected K562 cells compared with control miR-mimic (Fig. 4C). Averaging results from seven experiments run on separate days, there was a reduction in: 1) etoposide (2 µM)-induced DNA damage to 59.7% (P = 0.011) and 70.7% (P = 0.025) of control and 2) etoposide (5 µM)-induced DNA damage to 80.7% (P = 0.006) and 77.4% (P = 0.002) of control in K562 cells transfected with miR-9-3p or mir-9-5p mimics, respectively (Fig. 4C). Together these results indicate that forced overexpression of miR-9-3p or miR-9-5p reduces TOP2α/170 protein expression and, as a consequence, decreases etoposide-mediated DNA damage.
The potential mechanism by which miR-9-3p and miR-9-5p mimics reduce TOP2α/170 protein levels was assessed by qPCR. RNA isolated from miR-9-3p and mir-9-5p mimic–transfected K562 cells was used to quantify TOP2α/170 mRNA levels. Transfection with miR-9-3p or miR-9-5p mimics did not alter TOP2α/170 steady-state mRNA levels (Fig. 4D). Therefore, decreased TOP2α/170 protein expression observed in K562 cells by forced expression of miR-9-3p and miR-9-5p is not due to the degradation of TOP2α/170 mRNA, suggesting that these miRNAs/miRISC function by blocking the ribosome from translating TOP2α/170 mRNA.
miR-9-3p and miR-9-5p Inhibitors Increase TOP2α/170 Protein and DNA Damage in K/VP.5 Cells.
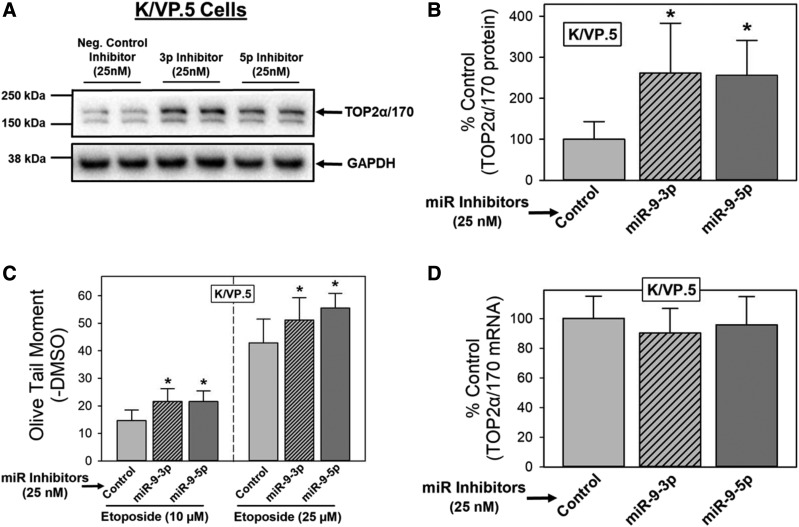
Because the miR-9-3p and miR-9-5p forced expression (gain-of-inhibitory-function) experiments described (Fig. 4) may lead to off-target effects due to supra-physiologic miRNA levels (Fig. 2C) (Bracken et al., 2008; Khan et al., 2009), reciprocal loss-of-miR-9-inhibitory-function experiments were also performed in K/VP.5 cells. Utilizing miRNA inhibitors (chemically modified single stranded antisense nucleic acids designed to bind to and inhibit miRNA function), effects on TOP2α/170 protein expression and subsequent etoposide-induced DNA damage were evaluated. K/VP.5 cells, which express high levels of miR-9-3p and miR-9-5p (Fig. 2B), were transfected with 25 nM miR-9-3p, miR-9-5p, or control miRNA inhibitors (48 hours), which was followed by immunoblotting of cell lysates to evaluate TOP2α/170 protein levels. Figure 5A shows that TOP2α/170 protein in K/VP.5 cells was increased after transfection of miR-9-3p and miR-9-5p inhibitors. Averaging results from five separate experiments run on separate days, there was an increase in TOP2α/170 levels to 261% (P = 0.035) and 256% (P = 0.027) of miRNA inhibitor control in K/VP.5 cells treated with miR-9-3p inhibitor and miR-9-5p, respectively (Fig. 5B).
Fig. 5.
miR-9-3p and miR-9-5p inhibitors increase TOP2α/170 protein and enhance etoposide-induced DNA damage in K/VP.5 cells. (A) Representative immunoassay using cellular lysates from K/VP.5 cells that were transfected (48 hours) with either control, miR-9-3p, or miR-9-5p inhibitors (25 nM). Blots were probed with antibodies specific for the N-terminal portion of TOP2α/170 (i.e., amino acids 14–27) or for GAPDH. (B) Expression of TOP2α/170 protein levels in K/VP.5 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p inhibitors (25 nM). Results shown are the mean ± S.D. from five experiments performed on separate days; *P < 0.05 comparing miR-9-3p and miR-9-5p inhibitor–transfected K/VP.5 lysates vs. control inhibitor–transfected K/VP.5 lysates. (C) Etoposide (10 and 25 µM)-induced DNA damage in control, miR-9-3p, or miR-9-5p inhibitor (25 nM)-transfected K/VP.5 cells (48 hours) was determined by Comet assays after a 1-hour incubation and subtraction of DMSO vehicle controls, as described in Materials and Methods. Results shown are the mean ± S.D. from six experiments performed on separate days; *P < 0.05 comparing miR-9-3p and miR-9-5p inhibitor–transfected vs. control inhibitor–transfected K/VP.5 cells. (D) Expression of TOP2α/170 mRNA levels in K/VP.5 cells transfected (48 hours) with either control, miR-9-3p, or miR-9-5p inhibitors (25 nM). Results shown are the mean ± S.D. from five experiments performed on separate days.
We next investigated whether transfection of K/VP.5 cells with miR-9-3p and miR-9-5p inhibitors would increase etoposide activity in parallel with the increase in TOP2α/170 protein levels (as demonstrated in Fig. 5, A and B). K/VP.5 cells were transfected with 25 nM miR-9-3p, miR-9-5p, or control miRNA inhibitors. Forty-eight hours after transfection, cells were incubated for 1 hour with etoposide (10 or 25 μM) or DMSO vehicle and subsequently evaluated for DNA damage by alkaline single-cell gel electrophoresis (Comet) assays (Kanagasabai et al., 2017, 2018) as above in Fig. 4C. Averaging results from six separate experiments performed on separate days, there was an increase in: 1) etoposide (10 µM)-induced DNA damage to 147% (P = 0.002) and 147% (P = 0.001) of control and 2) etoposide (25 µM)-induced DNA damage to 119% (P = 0.040) and 130% (P = 0.008) of control in K/VP.5 cells transfected with miR-9-3p or miR-9-5p inhibitors, respectively (Fig. 5C). Together, results indicate that miR-9-3p and miR-9-5p inhibitors allow for increased TOP2α/170 protein expression in drug-resistant K/VP.5 cells and, as a consequence, enhance etoposide-induced DNA damage.
Similar to results in miR-9 mimic–transfected K562 cells (Fig. 4D), neither miR-9-3p nor miR-9-5p inhibitor elicited changes in TOP2α/170 mRNA when transfected in K/VP.5 cells (Fig. 5D), again suggesting that miR-9 effects are not related to alteration in mRNA stability but may impact on translation of TOP2α/170 mRNA, in this case by loss of miR-9-3p and miR-9-5p inhibitory activity.
Discussion
Acquired drug resistance is a major therapeutic challenge in the treatment of cancers. Chemoresistance can result from a number of molecular mechanisms, including aberrant drug transport/trafficking processes, altered drug metabolism, increased or modified drug targets, abnormal cell cycling, cell death evasion, and DNA damage/repair dysregulation (Zahreddine and Borden, 2013; Cree and Charlton, 2017). In recent years, numerous studies have shown that miRNAs can be involved in acquired drug resistance by directly targeting mRNAs that encode proteins regulating the biologic pathways described above (Gabra and Salmena, 2017; Anfossi et al., 2018; Corrà et al., 2018). Our present study focused on whether decreased TOP2α/170 levels (Fig. 1C) and attenuated etoposide activity in K/VP.5 cells (Fig. 1A) result, in part, from miRNA mechanisms.
miRNA-Seq data revealed that 73 mature miRNAs were overexpressed and 14 miRNAs were repressed in K/VP.5 cells compared with K562 cells (Fig. 2A). Algorithm searches (Agarwal et al., 2015; Paraskevopoulou et al., 2013) suggested that of the 73 overexpressed miRNAs in K/VP.5 cells, nine miRNAs were predicted to interact with MREs harbored within the TOP2α/170 mRNA (unpublished data). miR-9-3p and miR-9-5p were highly overexpressed in K/VP.5 cells (Fig. 2A), and MREs were identified in the TOP2α/170 3ʹ-UTR (Fig. 2B) for both these miRNAs, leading to further investigation of their function(s).
During RISC loading, the miRNA duplex is bound by a member of the Argonaute protein family, which is followed by strand selection (Treiber et al., 2019). Typically, the strand with the least thermodynamically stable base pair at its 5′ end in the miRNA duplex is selected as the guide strand. The “passenger” strand with the stronger 5′ end interaction is released and degraded (Schwarz et al., 2003; Khvorova et al., 2003). However, the passenger strand can also be loaded into the miRISC and exhibit inhibitory activity like any other miRNA (Packer et al., 2008). The ratio of Argonaute-loaded 5p or 3p mature strand varies greatly, with coexpression of 5p/3p pairs demonstrated in about half of the miRNA populations analyzed (Ro et al., 2007; Kuchenbauer et al., 2011). Such seems to be the case with miR-9-3p and miR-9-5p in K562 and K/VP.5 cells.
Using qPCR, we confirmed miRNA-Seq data (Fig. 2A) to demonstrate overexpression of both miR-9-3p (18.6-fold) and miR-9-5p (12.7-fold) in K/VP.5 compared with K562 cells (Fig. 2C). Additionally, transfection of luciferase constructs containing TOP2α/170 3ʹ-UTR established that TOP2α/170 mRNA is a direct target of both miRNAs (Fig. 3, C and D). Transfection of miR-9 inhibitors increased TOP2α/170 expression and enhanced etoposide-induced DNA damage in resistant K/VP.5 cells, whereas forced overexpression of both miRNAs in parental K562 cells decreased TOP2α/170 levels and reduced etoposide activity (Figs. 4 and 5). Because decreased TOP2α/170 expression is most often associated with resistance (Ganapathi and Ganapathi, 2013), our results strongly suggest that increased expression of miR-9-3p and miR-9-5p in K/VP.5 cells are determinants of acquired resistance.
Consistent with our observations of dual expression/function of miR-9-3p and miR-9-5p, both miRNAs cooperate in regulating the balance between self-renewal and differentiation of human neural stem cells by simultaneously targeting members of the notch pathway (Roese-Koerner et al., 2016). Additionally, miR-9-3p and miR-9-5p can be coexpressed in primary cases of human acute myeloid leukemia (AML) and cotarget erythroblast transformation-specific–related gene mRNA (Nowek et al., 2016). Finally, miR-9-3p and miR-9-5p concurrently target calmodulin-binding transcription activator one mRNA in glioblastoma stem cells (Schraivogel et al., 2011).
Importantly, dysregulation of miR-9-3p and/or miR-9-5p expression has been implicated in a wide range of diseases including cancers (Nass et al., 2009; Barbano et al., 2017; Nowek et al., 2018), diabetes (Chakraborty et al., 2014), neurodegenerative diseases (Packer et al., 2008; Juźwik et al., 2019), osteoarthritis (Nugent, 2016), and sporadic premature aging syndromes (Frankel et al., 2018).
Increased levels of miR-9-3p and/or miR-9-5p have been shown to play an important role in mediating chemoresistance (Munoz et al., 2013, 2015b,a). In temozolomide-resistant glioblastoma cells, miR-9-3p and miR-9-5p were overexpressed (Munoz et al., 2015b), miR-9-5p led to increased expression of ABCB1 through activation of the Sonic Hedgehog pathway (Munoz et al., 2015a), and delivery of anti–miR-9-5p decreased ABCB1 and sensitized resistant glioblastoma cells to temozolomide (Munoz et al., 2013).
Conversely, Li et al. (2017) demonstrated that miR-9-3p expression was significantly reduced in Adriamycin (ADR)-resistant K562/ADR compared with parental K562 cells. These investigators also established that miR-9-3p targeted ABCB1 and that transfection of miR-9-3p mimics increased the chemosensitivity of K562/ADR cells. Li et al., 2015 demonstrated that miR-9-5p was down-regulated in both paclitaxel-resistant epithelial ovarian carcinoma (EOC) patient samples and EOC cell lines. Similarly, Sun et al. (2013) established that cisplatin-resistant ovarian cancer patients manifest lower miR-9-5p expression levels. In AML cell lines resistant to daunorubicin, transfection of miR-9-5p mimics enhanced chemosensitivity to daunorubicin by targeting eukaryotic translation initiation factor 5A-2 (Liu et al., 2019). Collectively, this literature indicates that overexpression or underexpression of miR-9-3p or miR-9-5p can play a critical role in chemoresistance depending on cell type. Additionally, these observations support the conclusion that miR-9-3p and/or miR-9-5p act on many mRNA targets important in chemoresistance.
miRISC mediates post-transcriptional silencing of specific mRNAs that contain MREs that are partially or fully complementary to the miRNA (Bartel, 2009). Silencing occurs by target mRNA translational repression and/or mRNA degradation (Fabian and Sonenberg, 2012; Jonas and Izaurralde, 2015). Ribosome profiling studies that compared changes in translational efficiency to changes in mRNA levels demonstrated that the majority of the negative post-transcriptional gene regulatory effects of miRNAs/miRISCs resulted from mRNA degradation (Guo et al., 2010; Eichhorn et al., 2014); true translational repression accounts for only 6%–26% of the decrease of each endogenous target in mammalian cells (Eichhorn et al., 2014).
Our studies demonstrated that there was no reduction in TOP2α/170 steady-state mRNA levels after transfection of K562 cells with miR-9-3p or miR-9-5p mimics (Fig. 4D), which attenuated TOP2α/170 protein levels (Fig. 4, A and B), or after transfection of K/VP.5 cells with miR-9-3p or miR-9-5p inhibitors (Fig. 5D), which increased TOP2α/170 protein levels (Fig. 5, A and B). These results strongly suggest that TOP2α/170 mRNA is translationally repressed by these miRNAs. In contrast, Hua et al. (2015) demonstrated that TOP2α/170 mRNA (and protein) was reduced by miR-139 in luminal type breast cancer cells, again pointing to the pleiotropic effects of miRNAs.
Though we conclude that miR-9-3p and miR-9-5p impact translation, steady-state mRNA levels of TOP2α/170 are reduced in K/VP.5 compared with K562 cells (Fig. 1B) attributable to alternative RNA processing of TOP2α/170 precursor mRNA, as we have reported previously (Kanagasabai et al., 2017, 2018). Hence, reduced TOP2α/170 protein levels (and acquired drug resistance) can derive from both translational repression (via miRNA mechanisms) and aberrant precursor mRNA processing.
Given that three distinct MIR9 genes transcribe primary transcripts processed into identical mature miR-9-3p and miR-9-5p miRNAs (Yuva-Aydemir et al., 2011), we performed qPCR experiments to investigate whether MIR9-1, MIR9-2, MIR9-3, or a combination of these genes was significantly up-regulated in etoposide-resistant K/VP.5 cells. Interestingly, only pri-miR-9-1 was significantly up-regulated in K/VP.5 cells (Fig. 2D); likely leading to the observed increase in mature miR-9-3p and miR-9-5p expression levels.
It is not clear how MIR9-1 gene expression is activated in K/VP.5 cells. However, because the promoters of all three MIR9 genes are embedded within CpG islands, they are frequently aberrantly hypomethylated or hypermethylated in cancer (Kunej et al., 2011; Suzuki et al., 2013). Li et al., 2015 established that the MIR9-1 and MIR9-3 genes were hypermethylated in paclitaxel-resistant EOC cell lines, suggesting that decreased miR-9-5p expression resulted from aberrant DNA methylation. Recently, Fu et al. (2017) demonstrated that the MIR9-1 gene underwent heterochromatic silencing by the binding of the runt-related family transcription factor 1 (RUNX1) to the MIR9-1 promoter region and recruitment of chromatin-remodeling enzymes that resulted in the hypermethylation of MIR9-1 in t(8;21) AML. Given that RUNX1 is a direct target of both miR-9-3p and miR-9-5p (Tian et al., 2015; Raghuwanshi et al., 2018; TargetScan), we speculate that that the observed increase in MIR9-1 gene expression in etoposide-resistant K/VP.5 cells may result from the hypomethylation of the MIR9-1 promoter due to decreased RUNX1 expression and the subsequent lack of recruitment of chromatin-remodeling enzymes. Future experiments will test this hypothesis.
In conclusion, TOP2α/170 is an established therapeutic target, the expression levels of which are important for the efficacy of topoisomerase II–targeted drugs. Hence, it is critical to understand the mechanisms by which TOP2α/170 expression is regulated, especially in acquired drug resistance. Results demonstrated that miR-9-3p and miR-9-5p were overexpressed in etoposide-resistant K/VP.5 cells and regulated the expression of TOP2α/170, likely through inhibition of translation. Additionally, miR-9-3p and miR-9-5p impact etoposide-induced DNA damage secondary to effects on TOP2α/170 levels. This functional aspect of both miR-9-3p and miR-9-5p with regards to chemoresistance demonstrates their potential as drug targets for and/or biomarkers of resistance to TOP2α interfacial inhibitors.
Future studies aim to validate miR-9-3p and miR-9-5p as biomarkers of chemoresistance by analyzing expression in cells from newly diagnosedand relapsed leukemia patients who received TOP2α/170-targeted agents as well as establish the in vivo effects of these miRNAs on etoposide efficacy in xenograft models of leukemia.
Acknowledgments
The authors thank Dr. Junan Li, The Ohio State University College of Pharmacy, for excellent biostatistical support.
Abbreviations
- ABCB1
multidrug resistance protein 1/P-glycoprotein
- ADR
Adriamycin
- AML
acute myeloid leukemia
- EOC
epithelial ovarian carcinoma
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- luc
luciferase
- miRNA
microRNA
- miRISC
miRNA-RISC complex
- MRE
miRNA recognition element
- mut
mutant
- PCR
polymerase chain reaction
- pre-miRNA
precursor miRNA
- pri
primary
- PVDF
polyvinylidene difluoride
- qPCR
quantitative PCR
- RISC
RNA-induced silencing complex
- RUNX1
runt-related family transcription factor 1
- Seq
sequencing
- TBP
TATA-binding protein
- TOP2α
DNA topoisomerase IIα protein
- TOP2α/170
TOP2α 170 kDa
- TOP2α/90
TOP2α 90 kDa
- UTR
untranslated region
Authorship Contributions
Participated in research design: Kania, Carvajal-Moreno, Hernandez, English, Papa, Ozer, Yalowich, Elton.
Conducted experiments: Kania, Carvajal-Moreno, Hernandez, English, Papa, Shkolnikov.
Contributed new reagents or analytical tools: Kania, Ozer.
Performed data analysis: Kania, Carvajal-Moreno, Hernandez, English, Papa, Shkolnikov, Ozer, Yilmaz, Yalowich, Elton.
Wrote or contributed to the writing of the manuscript: Kania, Hernandez, Ozer, Yalowich, Elton.
Footnotes
This work was supported by the National Institutes of Health National Cancer Institute [Grant R01 CA226906-01A1].
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi S, Fu X, Nagvekar R, Calin GA. (2018) MicroRNAs, regulatory messengers inside and outside cancer cells. Adv Exp Med Biol 1056:87–108. [DOI] [PubMed] [Google Scholar]
- Ashour ME, Atteya R, El-Khamisy SF. (2015) Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat Rev Cancer 15:137–151. [DOI] [PubMed] [Google Scholar]
- Barbano R, Pasculli B, Rendina M, Fontana A, Fusilli C, Copetti M, Castellana S, Valori VM, Morritti M, Graziano P, et al. (2017) Stepwise analysis of MIR9 loci identifies miR-9-5p to be involved in Oestrogen regulated pathways in breast cancer patients. Sci Rep 7:45283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68:7846–7854. [DOI] [PubMed] [Google Scholar]
- Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. (2016) Pairing beyond the seed supports MicroRNA targeting specificity. Mol Cell 64:320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg BY, Danks MK, Beck WT, Suttle DP. (1991) Expression of a mutant DNA topoisomerase II in CCRF-CEM human leukemic cells selected for resistance to teniposide. Proc Natl Acad Sci USA 88:7654–7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, Hannon GJ, Lowe SW, Hemann MT. (2008) Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci USA 105:9053–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. (2007) microRNA functions. Annu Rev Cell Dev Biol 23:175–205. [DOI] [PubMed] [Google Scholar]
- Chakraborty C, Doss CG, Bandyopadhyay S, Agoramoorthy G. (2014) Influence of miRNA in insulin signaling pathway and insulin resistance: micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip Rev RNA 5:697–712. [DOI] [PubMed] [Google Scholar]
- Chen AY, Liu LF. (1994) Mechanisms of resistance to topoisomerase inhibitors. Cancer Treat Res 73:263–281. [DOI] [PubMed] [Google Scholar]
- Chen SH, Chan NL, Hsieh TS. (2013) New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170. [DOI] [PubMed] [Google Scholar]
- Corrà F, Agnoletto C, Minotti L, Baldassari F, Volinia S. (2018) The network of non-coding RNAs in cancer drug resistance. Front Oncol 8:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree IA, Charlton P. (2017) Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer 17:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese JE, Osheroff N. (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res 37:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, Bartel DP. (2014) mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 56:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton TS, Yalowich JC. (2015) Experimental procedures to identify and validate specific mRNA targets of miRNAs. EXCLI J 14:758–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19:586–593. [DOI] [PubMed] [Google Scholar]
- Frankel D, Delecourt V, Harhouri K, De Sandre-Giovannoli A, Lévy N, Kaspi E, Roll P. (2018) MicroRNAs in hereditary and sporadic premature aging syndromes and other laminopathies. Aging Cell 17:e12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Shi J, Liu A, Zhou L, Jiang M, Fu H, Xu K, Li D, Deng A, Zhang Q, et al. (2017) A minicircuitry of microRNA-9-1 and RUNX1-RUNX1T1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Int J Cancer 140:653–661. [DOI] [PubMed] [Google Scholar]
- Gabra MM, Salmena L. (2017) microRNAs and acute myeloid leukemia chemoresistance: a mechanistic overview. Front Oncol 7:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi RN, Ganapathi MK. (2013) Mechanisms regulating resistance to inhibitors of topoisomerase II. Front Pharmacol 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganger MT, Dietz GD, Ewing SJ. (2017) A common base method for analysis of qPCR data and the application of simple blocking in qPCR experiments. BMC Bioinformatics 18:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. (2004) The microRNA Registry. Nucleic Acids Res 32:D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement MV. (2014) OpenComet: an automated tool for comet assay image analysis. Redox Biol 2:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. [DOI] [PubMed] [Google Scholar]
- Harker WG, Slade DL, Parr RL, Holguin MH. (1995) Selective use of an alternative stop codon and polyadenylation signal within intron sequences leads to a truncated topoisomerase II alpha messenger RNA and protein in human HL-60 leukemia cells selected for resistance to mitoxantrone. Cancer Res 55:4962–4971. [PubMed] [Google Scholar]
- Hinds M, Deisseroth K, Mayes J, Altschuler E, Jansen R, Ledley FD, Zwelling LA. (1991) Identification of a point mutation in the topoisomerase II gene from a human leukemia cell line containing an amsacrine-resistant form of topoisomerase II. Cancer Res 51:4729–4731. [PubMed] [Google Scholar]
- Hua W, Sa KD, Zhang X, Jia LT, Zhao J, Yang AG, Zhang R, Fan J, Bian K. (2015) MicroRNA-139 suppresses proliferation in luminal type breast cancer cells by targeting Topoisomerase II alpha. Biochem Biophys Res Commun 463:1077–1083. [DOI] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16:421–433. [DOI] [PubMed] [Google Scholar]
- Juźwik CA, Drake SS, Zhang Y, Paradis-Isler N, Sylvester A, Amar-Zifkin A, Douglas C, Morquette B, Moore CS, Fournier A. (2019) microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog Neurobiol 182:101664. [DOI] [PubMed] [Google Scholar]
- Kanagasabai R, Karmahapatra S, Kientz CA, Yu Y, Hernandez VA, Kania EE, Yalowich JC, Elton TS. (2018) The novel C-terminal truncated 90-kDa isoform of topoisomerase IIα (TOP2α/90) is a determinant of etoposide resistance in K562 leukemia cells via heterodimerization with the TOP2α/170 isoform. Mol Pharmacol 93:515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagasabai R, Serdar L, Karmahapatra S, Kientz CA, Ellis J, Ritke MK, Elton TS, Yalowich JC. (2017) Alternative RNA processing of topoisomerase IIα in etoposide-resistant human leukemia K562 cells: intron retention results in a novel C-terminal truncated 90-kDa isoform. J Pharmacol Exp Ther 360:152–163. [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216. [DOI] [PubMed] [Google Scholar]
- Kubo A, Yoshikawa A, Hirashima T, Masuda N, Takada M, Takahara J, Fukuoka M, Nakagawa K. (1996) Point mutations of the topoisomerase IIalpha gene in patients with small cell lung cancer treated with etoposide. Cancer Res 56:1232–1236. [PubMed] [Google Scholar]
- Kuchenbauer F, Mah SM, Heuser M, McPherson A, Rüschmann J, Rouhi A, Berg T, Bullinger L, Argiropoulos B, Morin RD, et al. (2011) Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood 118:3350–3358. [DOI] [PubMed] [Google Scholar]
- Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. (2008) Experimental validation of miRNA targets. Methods 44:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. (2011) Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res 717:77–84. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. (2010) Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One 5:e11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen AK, Escargueil AE, Skladanowski A. (2003) Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol Ther 99:167–181. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pan Q, Wan X, Mao Y, Lu W, Xie X, Cheng X. (2015) Methylation-associated Hsa-miR-9 deregulation in paclitaxel- resistant epithelial ovarian carcinoma. BMC Cancer 15:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao L, Li N, Miao Y, Zhou H, Jia L. (2017) miR-9 regulates the multidrug resistance of chronic myelogenous leukemia by targeting ABCB1. Oncol Rep 37:2193–2200. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lei P, Qiao H, Sun K, Lu X, Bao F, Yu R, Lian C, Li Y, Chen W, et al. (2019) miR-9 enhances the chemosensitivity of AML cells to daunorubicin by targeting the EIF5A2/MCL-1 axis. Int J Biol Sci 15:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. [Google Scholar]
- Mirski SE, Cole SP. (1995) Cytoplasmic localization of a mutant M(r) 160,000 topoisomerase II alpha is associated with the loss of putative bipartite nuclear localization signals in a drug-resistant human lung cancer cell line. Cancer Res 55:2129–2134. [PubMed] [Google Scholar]
- Mirski SE, Evans CD, Almquist KC, Slovak ML, Cole SP. (1993) Altered topoisomerase II alpha in a drug-resistant small cell lung cancer cell line selected in VP-16. Cancer Res 53:4866–4873. [PubMed] [Google Scholar]
- Mirski SE, Sparks KE, Yu Q, Lang AJ, Jain N, Campling BG, Cole SP. (2000) A truncated cytoplasmic topoisomerase IIalpha in a drug-resistant lung cancer cell line is encoded by a TOP2A allele with a partial deletion of exon 34. Int J Cancer 85:534–539. [DOI] [PubMed] [Google Scholar]
- Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. (2013) Delivery of functional anti-miR-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol Ther Nucleic Acids 2:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz JL, Rodriguez-Cruz V, Rameshwar P. (2015b) High expression of miR-9 in CD133+ glioblastoma cells in chemoresistance to temozolomide. J Cancer Stem Cell Res 3:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz JL, Rodriguez-Cruz V, Ramkissoon SH, Ligon KL, Greco SJ, Rameshwar P. (2015a) Temozolomide resistance in glioblastoma occurs by miRNA-9-targeted PTCH1, independent of sonic hedgehog level. Oncotarget 6:1190–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O, et al. (2009) MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol 19:375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL. (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowek K, Sun SM, Bullinger L, Bindels EM, Exalto C, Dijkstra MK, van Lom K, Döhner H, Erkeland SJ, Löwenberg B, et al. (2016) Aberrant expression of miR-9/9* in myeloid progenitors inhibits neutrophil differentiation by post-transcriptional regulation of ERG. Leukemia 30:229–237. [DOI] [PubMed] [Google Scholar]
- Nowek K, Wiemer EAC, Jongen-Lavrencic M. (2018) The versatile nature of miR-9/9* in human cancer. Oncotarget 9:20838–20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent M. (2016) MicroRNAs: exploring new horizons in osteoarthritis. Osteoarthritis Cartilage 24:573–580. [DOI] [PubMed] [Google Scholar]
- Olive PL. (2002) The comet assay. An overview of techniques. Methods Mol Biol 203:179–194. [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci 28:14341–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. (2013) DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41:W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton M, Lindsey RH, Jr, Felix CA, Grimwade D, Osheroff N. (2014) Topoisomerase II and leukemia. Ann N Y Acad Sci 1310:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati P, Nitti D, Mocellin S. (2012) Cancer resistance to type II topoisomerase inhibitors. Curr Med Chem 19:3900–3906. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Leo E, Zhang H, Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 17:421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuwanshi S, Gutti U, Kandi R, Gutti RK. (2018) MicroRNA-9 promotes cell proliferation by regulating RUNX1 expression in human megakaryocyte development. Cell Prolif 51:e12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritke MK, Yalowich JC. (1993) Altered gene expression in human leukemia K562 cells selected for resistance to etoposide. Biochem Pharmacol 46:2007–2020. [DOI] [PubMed] [Google Scholar]
- Ritke MK, Roberts D, Allan WP, Raymond J, Bergoltz VV, Yalowich JC. (1994) Altered stability of etoposide-induced topoisomerase II-DNA complexes in resistant human leukaemia K562 cells. Br J Cancer 69:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, Park C, Young D, Sanders KM, Yan W. (2007) Tissue-dependent paired expression of miRNAs. Nucleic Acids Res 35:5944–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roese-Koerner B, Stappert L, Berger T, Braun NC, Veltel M, Jungverdorben J, Evert BO, Peitz M, Borghese L, Brüstle O. (2016) Reciprocal regulation between bifunctional miR-9/9(∗) and its transcriptional modulator notch in human neural stem cell self-renewal and differentiation. Stem Cell Reports 7:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SE, Nuovo GJ, Martin MM, Kotha SR, Parinandi NL, Elton TS. (2010) miR-802 regulates human angiotensin II type 1 receptor expression in intestinal epithelial C2BBe1 cells. Am J Physiol Gastrointest Liver Physiol 299:G632–G642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- Schraivogel D, Weinmann L, Beier D, Tabatabai G, Eichner A, Zhu JY, Anton M, Sixt M, Weller M, Beier CP, et al. (2011) CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J 30:4309–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208. [DOI] [PubMed] [Google Scholar]
- Sun C, Li N, Yang Z, Zhou B, He Y, Weng D, Fang Y, Wu P, Chen P, Yang X, et al. (2013) miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J Natl Cancer Inst 105:1750–1758. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Maruyama R, Yamamoto E, Kai M. (2013) Epigenetic alteration and microRNA dysregulation in cancer. Front Genet 4:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Rui K, Tang X, Ma J, Wang Y, Tian X, Zhang Y, Xu H, Lu L, Wang S. (2015) MicroRNA-9 regulates the differentiation and function of myeloid-derived suppressor cells via targeting Runx1. J Immunol 195:1301–1311. [DOI] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Meister G. (2019) Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 20:5–20. [DOI] [PubMed] [Google Scholar]
- Vassetzky YS, Alghisi GC, Gasser SM. (1995) DNA topoisomerase II mutations and resistance to anti-tumor drugs. BioEssays 17:767–774. [DOI] [PubMed] [Google Scholar]
- Vlasova II, Feng WH, Goff JP, Giorgianni A, Do D, Gollin SM, Lewis DW, Kagan VE, Yalowich JC. (2011) Myeloperoxidase-dependent oxidation of etoposide in human myeloid progenitor CD34+ cells. Mol Pharmacol 79:479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Tretter EM, Schmidt BH, Berger JM. (2011) All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol 12:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel I, Jensen PB, Falck J, Mirski SE, Cole SP, Sehested M. (1997) Loss of amino acids 1490Lys-Ser-Lys1492 in the COOH-terminal region of topoisomerase IIalpha in human small cell lung cancer cells selected for resistance to etoposide results in an extranuclear enzyme localization. Cancer Res 57:4451–4454. [PubMed] [Google Scholar]
- Yu Q, Mirski SE, Sparks KE, Cole SP. (1997) Two COOH-terminal truncated cytoplasmic forms of topoisomerase II alpha in a VP-16-selected lung cancer cell line result from partial gene deletion and alternative splicing. Biochemistry 36:5868–5877. [DOI] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. (2011) MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol 8:557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahreddine H, Borden KL. (2013) Mechanisms and insights into drug resistance in cancer. Front Pharmacol 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwelling LA, Hinds M, Chan D, Mayes J, Sie KL, Parker E, Silberman L, Radcliffe A, Beran M, Blick M. (1989) Characterization of an amsacrine-resistant line of human leukemia cells. Evidence for a drug-resistant form of topoisomerase II. J Biol Chem 264:16411–16420. [PubMed] [Google Scholar]