Abstract
Bacterial infection influences genomic stability and integrity by causing DNA damage, which increases the possibility of tumor initiation and development. We aimed to investigate whether Fusobacterium nucleatum, one of the periodontal pathogens, promoted oral squamous cell carcinoma (OSCC) by causing DNA double-strand break (DSB). Tca8113 tongue squamous cell carcinoma cells were infected with F. nucleatum. The expression of γH2AX was detected by western blots and immunofluorescence. The proliferation and cell cycle alterations were tested by CCK8 and flow cytometry, respectively. The expression levels of Ku70, p53, and p27 were evaluated by quantitative real-time polymerase chain reaction and western blots. A plasmid was used for the overexpression of Ku70 to verify the possible relationship between Ku70 and p53. We confirmed the presence of DSBs in the response to F. nucleatum by detecting the expression of γH2AX. The cell proliferation ability was increased with an accelerated cell cycle while the expression of p27 was decreased. Meanwhile, the expression of Ku70 and wild p53 was downregulated. When Ku70 was overexpressed, the expression of wild p53 in response to F. nucleatum infection was upregulated and cell proliferation was accordingly inhibited. We concluded that F. nucleatum infection promoted the proliferation ability of Tca8113 by causing DNA damage via the Ku70/p53 pathway.
Keywords: OSCC, Fusobacterium nucleatum, DNA damage, γH2AX, Ku70
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of head and neck malignancy with a high prevalence and a poor survival rate. It is widely accepted that inflammatory microenvironment is regarded as an important risk factor of all stages of tumor. Approximately 15–20% of malignancies are followed by inflammation that is sustained long before tumor formation (Parkin, 2006; Grivennikov et al., 2010; Greten and Grivennikov, 2019).
Bacterial infections, one of the important inflammatory factors, are being increasingly accepted as a risk factor for various types of cancer including OSCC (Moergel et al., 2013). Based on a recent report, the oral microbiome in OSCC lesions shifts significantly compared to healthy epithelium. Specifically, Fusobacterium species are regarded as one of the major components of the oral microbiome in OSCC (Perera et al., 2018). The role of Fusobacterium nucleatum in OSCC is being gradually recognized (Binder Gallimidi et al., 2015; Chang et al., 2018; Hsiao et al., 2018).
DNA damage is known to contribute to tumor initiation and development. In addition to traditional environmental factors such as carcinogens and radiations, inflammation is also involved in the regulation of DNA damage response (DDR) genes (Gronke et al., 2019). Specifically, DNA damage and repair are reported to be affected by bacteria, such as Helicobacter pylori or Pneumococci (Obst et al., 2000; Rai et al., 2016; Shi et al., 2019).
Histone H2A (H2AX) is a meiosis-specific isoform of histone H2A. γH2AX, the phosphorylated form of H2AX, is rapidly and accurately recruited when there is a DNA double-strand break (DSB), the most serious type of DNA damage, and it is applied as a DSB molecular marker (Rogakou et al., 1998; Bach et al., 2017). Once the DNA damage occurs, DSBs are repaired by homologous recombination or nonhomologous end joining (NHEJ). NHEJ, initiated by Ku, can bind to any two DNA break ends by sensing and binding DSBs, which makes it the most common DSB repair pathway in mammalian cells (Mari et al., 2006). Of note, a deficiency in NHEJ results in malignant transformation. Ku is a stable heterodimer structure formed by Ku70 and Ku80 (Puebla-Osorio et al., 2014; Lamaa et al., 2016). Current research has shown that Ku70 participates in DDR signaling through activating cell cycle detection points and initiating apoptosis programs.
Cell cycle disregulation is one of the common features of human cancers (Malumbres and Barbacid, 2009). Cell cycle progression is determined by cyclin-dependent kinases (CDKs) that are activated by binding cyclins and are inhibited by CDK inhibitors (CDKIs). Accumulation of DNA alterations might induce cell cycle progression by abnormal regulations of CDKIs, which results in genomic instability and ultimate oncogenesis (Kastan and Bartek, 2004; Malumbres and Barbacid, 2009). p27, a tumor suppressor, is one of the major members of the CDKI protein family and functions to negatively regulate cell cycle progression (Keles et al., 2003; Breitenstein et al., 2013). Studies have shown that p27 can cause abnormal cell proliferation, and downregulation of p27 is associated with a poor tumor prognosis (Pereira et al., 2013). As recently reported, F. nucleatum infection was associated with the activation of cyclin D1, which facilitated intestinal tumorigenesis (Wu et al., 2018; Rubinstein et al., 2019). However, the role of F. nucleatum in regulating the expression of CDKIs such as p27 remained investigations.
The Ku complex can bind to DSBs ends with the activation of protein sensors such as p53. To date, the interactions of Ku and p53 remain controversial. As DNA damage occurs, Ku is acetylated in the DNA binding region and its bond with p53 is thereby released, which leads to upregulation of p53 expression and initiation of DNA repair (Lamaa et al., 2016). However, on the contrary, some studies have shown that the NHEJ repair of DSBs is significantly different in colorectal cancer tissues and adjacent normal tissues, revealing a positive correlation between Ku and p53 changes (Komuro et al., 2002; Lu et al., 2015). To date, the role of F. nucleatum in DNA damage and the possible association with OSCC has not been clearly elucidated.
In the present study, we established a DNA damage model of OSCC cells infected with F. nucleatum at an MOI of 500 and a DSB molecular marker was evaluated. With further investigation, we found that the resulting increased proliferation ability and accelerated cell cycle of OSCC cells in response to DNA damage was dependent on the Ku70/p53 pathway.
Materials and Methods
Bacteria and eukaryotic cell culture
Frozen stock of F. nucleatum ATCC 25586 (provided by the Department of Oral Biology, Stomatology of China Medical University) was recovered on tryptic soy broth (TSB) agar plates and anaerobically incubated at 37°C for 3–5 days. Appropriate colonies from the plate were resuspended in TSB liquid medium and anaerobically cultured for 16 h before use. F. nucleatum in the mid-log phase was adjusted to 1 × 109 CFU/mL (OD = 1) in RPMI 1640 cell culture medium with a spectrophotometer at a wavelength of 600 nm.
Tca8113 tongue squamous cell carcinoma cells were purchased from the Shanghai Institute of the Chinese Academy of Sciences. Cells were routinely cultured in RPMI 1640 medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin and incubated at 37°C, 5% CO2.
Establishment of the DNA damage model
Cells (2 × 105 cells/well) were cultured at 37°C for 24 h. Then, the cells were incubated with fresh medium without penicillin and streptomycin. Actively growing F. nucleatum at an MOI of 500 was added to the cell culture plate and cultured for 36 h. The expression of γH2AX was detected at 0, 4, 12, 24, and 36 h, respectively.
Cell immunofluorescence assay
Actively growing cells were subcultured and inoculated on sterilized glass slides. After the cells were infected by F. nucleatum for 24 h, cells on the glass slides were treated with precooled 4% paraformaldehyde for 30 min and 0.2% Triton X-100 at room temperature for 10 min. The cell slides were blocked with 1% BSA for 30 min at room temperature and incubated with a primary antibody against γH2AX (1:1000) overnight at 4°C. The cell slides were then incubated with fluorescent secondary antibody (1:500) for 1 h at room temperature. After being stained with DAPI at room temperature for 5 min, the slides were mounted on glass slides with antifluorescence quenching tablets and observed under a fluorescence microscope. The culture medium and cells in medium without bacterial infection were used as negative controls.
Cell proliferation assay by CCK-8
Cells were inoculated into 96-well plates (3000/well) and the DNA damage model was constructed 24 h later. At 0, 4, 12, 24, and 36 h, CCK-8 assay solution (10%) was added to each well and incubated at 37°C in the dark for 2 h. The absorbance was then measured at 450 nm using a microplate reader. The culture medium and cells in medium without bacterial infection were used as negative controls.
Cell cycle analysis by flow cytometry
Cells were starved for 24 h in serum-free medium before infection with F. nucleatum. At 0, 4, 12, 24, and 36 h, cells were digested and fixed in 75% ethanol (precooled) overnight at 4°C. Then, the fixed cells were washed with ice-cold PBS and resuspended in mixed media containing propidium iodide (50 μg/mL) and RNase (100 μg/mL) for incubation at 37°C in the dark for 30 min. The cell cycle was determined by a flow cytometer (FACS, Becton-Dickinson, Islandia, NY). The culture medium and cells in medium without bacterial infection were used as negative controls.
Quantitative real-time polymerase chain reaction
Total RNAs of each group were extracted and quantitative real-time polymerase chain reaction (qPCR) was performed according to the manufacturer's instructions as described previously (Liu et al., 2015). The fold induction of the selected genes was calculated compared using the 2−ΔΔCT method (Schmittgen and Livak, 2008). The primer sequences are shown in the Supplementary (Table S1).
Western blot assay
Total protein was extracted with RIPA lysates and the protein concentration was determined with a bicinchoninic acid protein assay on ice. An equal amount of sample proteins were separated with 12% SDS-polyacrylamide gel electrophoresis, followed by being transferred to a nitrocellulose filter membrane. After being blocked in BSA medium for 1 h, the blots were probed with the following primary antibodies: anti-γH2AX (1:1000) (Abcam); anti-Ku70 (1:1000) (Santa Cruz Biotechnology); anti-p27 (1:500) (Affinity); anti-wild p53 (1:1000) (Wanleibio Co., Ltd., China); or anti-GAPDH (Proteintech Group) overnight. The blots were washed with TBST three times and exposed to the secondary antibody (1:500) (LI-COR) at room temperature for 1 h. The protein bands were detected with Odyssey CLX (LI-COR) and analyzed with ImageJ software (1.42q).
Overexpression of Ku70 by plasmid transfection
The overexpression plasmid for Ku70 was constructed by the GenePharma cooperation (Shanghai, China) and the plasmid was extracted according to the instructions of the Rapid Mini Plasmid Kit (Biomed, Beijing, China). Actively growing cells at 60% confluence were used for transfection with Lipo2000 for 24 h. The normally cultured cells, cells transfected with an empty vector and cells treated with Lipo2000 by itself were used as controls. The transfection efficiency was measured by qPCR. After transfection, the proliferation ability induced by F. nucleatum and the expression of wild p53 were measured.
Statistical analysis
The one-way ANOVA-LSD multiple comparison method or a rank sum test was used for statistical analysis. The level of detection was double-sided α = 0.05. All of the experiments were repeated at least three times. All statistical results were performed using the statistical analysis software IBM SPSS Statistics version 23. p < 0.05 was considered statistically significant.
Results
Successful construction of the cellular DNA damage model
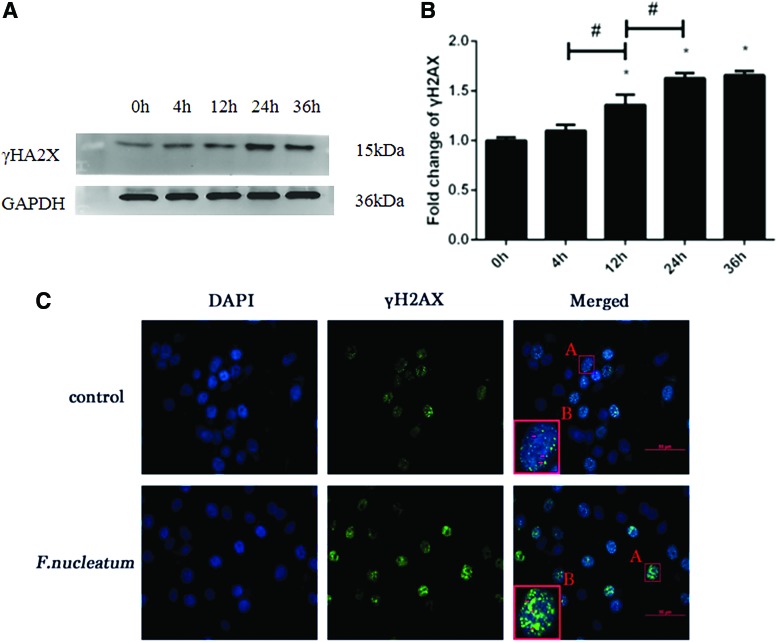
In the present cellular model, an MOI of 500 was applied for F. nucleatum infection based on the results of a preliminary study (shown in Supplementary Fig. S1). After the cells were infected with F. nucleatum at an MOI of 500, western blotting was used to detect the expression of γH2AX (shown in Fig. 1A, B). The expression of γH2AX protein was increased continuously in a time-dependent manner within 36 h, indicating that the DNA damage model of Tca8113 lingual squamous carcinoma cell was successfully constructed. The expression of γH2AX was significantly increased at 24 h, suggesting that DNA damage was significantly aggravated at 24 h (p < 0.05). Moreover, based on the immunofluorescence results, the fluorescence intensity of γH2AX at 24 h was increased compared to the blank control group and the number of positive cells was also increased (Fig. 1C). The time point of 24 h was therefore selected for use in the exploration of the molecular mechanisms.
FIG. 1.
At an MOI of 500, the protein expression of γH2AX was increasingly upregulated from 0 to 36 h (A). The expression of γH2AX was normalized to GAPDH and the relative fold change was compared with the control group (B). The data are presented as the mean ± standard deviation of three independent assays. *Significant difference (p < 0.05) compared with the control group (0 h). #Significant difference (p < 0.05) compared between the tested groups. After the infection with Fusobacterium nucleatum at an MOI of 500 for 24 h, the expression of γH2AX was further analyzed with immunofluorescence (C). The nuclei labeled with DAPI are shown in blue and the expression of γH2AX is presented in green fluorescence (red arrow). As we can see, the number of cells stained with green fluorescence and the fluorescence intensity of γH2AX were significantly increased in response to F. nucleatum infection. Color images are available online.
DNA damage caused by F. nucleatum promoted cell proliferation with an accelerated cell cycle
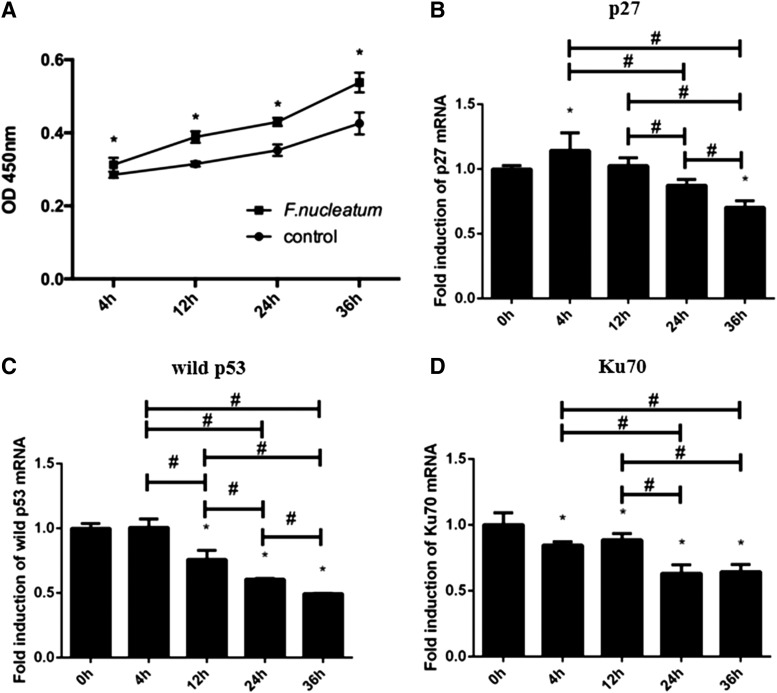
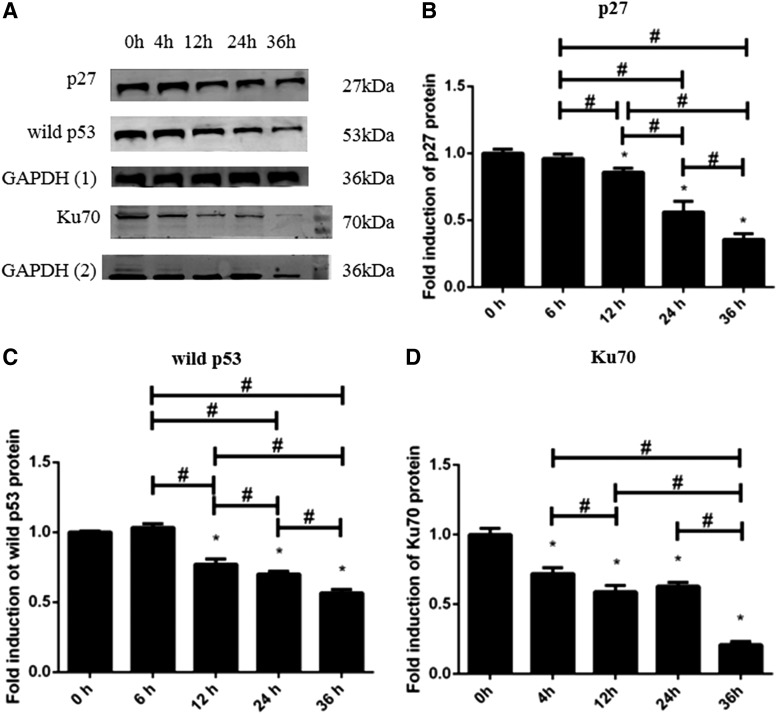
Cells infected by F. nucleatum had an increased proliferative capacity in a time-dependent manner at each time point (p < 0.05) (Fig. 2A). The proportion of cells in the G1 phase of the cell cycle was significantly decreased at the time points of 12 to 36 h while the proportion in the S phase was increased from 4 to 24 h. Of note, although no significant alteration was seen in S phase at 36 h, the G2/M phase in the infected cells was significantly higher than that in the control (Table 1). It was found that the mRNA expression of p27 was increased from 0 to 4 h and then decreased from 4 to 36 h (Fig. 2B). As shown in Figure 3A and B, the expression of p27 at the protein level was significantly decreased from 12 to 36 h (p < 0.05).
FIG. 2.
The ability of cell proliferation was increased after F. nucleatum infection from 4 to 36 h (A). The effect of F. nucleatum infection on the mRNA expression of p27 (B), wild p53 (C), and Ku70 (D) was evaluated by qPCR. The data were analyzed according to relative gene expression by 2−ΔΔCt. The data are presented as the mean ± standard deviation of three independent assays. *Significant difference (p < 0.05) compared with the control group. #Significant difference (p < 0.05) compared between the tested groups. qPCR, quantitative real-time polymerase chain reaction.
Table 1.
Tca8113 Cells Were Infected with Fusobacterium nucleatum for 4, 12, 24, and 36 h
| Time (h) | G0/G1 (%) |
S (%) |
G2/M (%) |
||||
|---|---|---|---|---|---|---|---|
| Control | F. nucleatum | Control | F. nucleatum | Control | F. nucleatum | ||
| 0 | 74.41 ± 0.96 | 74.41 ± 0.96 | 20.26 ± 1.05 | 20.26 ± 1.05 | 5.33 ± 0.83 | 5.33 ± 0.83 | |
| 4 | 53.79 ± 0.97 | 52.75 ± 1.73 | 32.70 ± 0.64 | 36.23 ± 0.71* | 11.02 ± 0.72 | 13.51 ± 1.03* | |
| 12 | 52.30 ± 0.37 | 47.33 ± 2.67* | 27.29 ± 1.74 | 34.97 ± 2.47* | 20.41 ± 1.29 | 17.69 ± 0.55* | |
| 24 | 67.18 ± 1.36 | 57.09 ± 0.13* | 21.97 ± 0.38 | 26.90 ± 0.72* | 10.86 ± 1.23 | 16.02 ± 0.69* | |
| 36 | 73.21 ± 1.61 | 61.36 ± 1.23* | 25.54 ± 1.25 | 23.70 ± 1.20 | 1.25 ± 0.75 | 14.95 ± 0.48* | |
The cell cycle distribution is shown as the cell numbers in each phase (%). The data are presented as the mean ± standard deviation of three independent assays. *Significant difference (p < 0.05) compared with the control group.
FIG. 3.
The protein expression of p27, wild p53, and Ku70 in response to F. nucleatum infection (A). The expression of p27, wild p53, and Ku70 was normalized to GAPDH and the relative fold change was compared with the control group (B–D). The fold inductions are presented as the mean ± standard deviation of three independent assays. *Significant difference (p < 0.05) compared with the control group. #Significant difference (p < 0.05) compared between the tested groups.
Expression of Ku70 and p53
We checked the expression of Ku70 (Figs. 2D and 3A, D). Interestingly, the mRNA and protein expression of Ku70 were continuously downregulated from 4 to 36 h (p < 0.05), which suggested cell repair ability might be weakened within the infection time. We also checked the expression of p53. As shown in Figures 2 and 3, the mRNA and protein expression of wild p53 (Figs. 2C and 3A, C) was continuously downregulated from 12 to 36 h (p < 0.05).
Overexpression of Ku70 weakened the cell proliferation ability by regulating the expression of Ku/p53
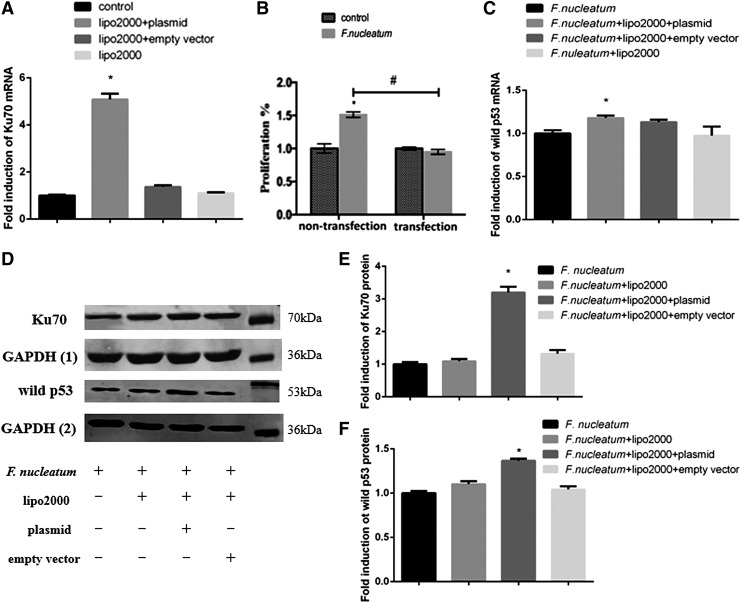
As shown in Figure 4A, the mRNA expression of Ku70 was significantly increased compared to the control groups (p < 0.05). After infection with F. nucleatum, the expression of Ku was still overexpressed in the transfected group (Fig. 4D, E). Accordingly, the previously increased cell proliferation ability was balanced out (Fig. 4B) with the upregulation of wild p53 (Fig. 4C, D, F).
FIG. 4.
The efficiency of transfection was confirmed by qPCR (A). The effect of Ku70 overexpression on cell proliferation was tested (B). After overexpression with Ku70, the cells were infected with F. nucleatum again and the mRNA expression of wild-type p53 is shown (C). The fold inductions are presented as the mean ± standard deviation of three independent assays. *Significant difference (p < 0.05) compared with the control group. #Significant difference (p < 0.05) compared between the tested groups. After transfected by Ku70, the expression of Ku70 (D, E) and wild-type p53 (D, F) in response to F. nucleatum was explored at the protein levels. The fold inductions are presented as the mean ± standard deviation of three independent assays.
Discussion
The role of F. nucleatum in cancer has been revealed in recent years. As investigated, F. nucleatum is abundantly detected in colorectal cancer with the detection rate from 10% to 90% and is considered as the significant contributor to colorectal cancer (Rubinstein et al., 2019). F. nucleatum expresses high levels of fusobacterium adhesion A (FadA) and activates oncogenic signaling to promote colorectal cancer (Rubinstein et al., 2019). Evidences above implicate the possible tumorigenic role of F. nucleatum in other types of cancer.
As one of major periodontal pathogens in oral cavity, F. nucleatum appears to be reasonably associated with oral cancer (Whitmore and Lamont, 2014). Coinfection with Porphyromonas gingivalis and F. nucleatum was found to promote OSCC development by regulating the TLR2-IL-6-STAT3 axis in a chemical-induced tumorigenic mice model. Similar to our findings, tongue epithelium-derived oral cavity SCC cell lines gained significantly increased proliferation ability after coincubation with periodontal pathogens including F. nucleatum (Binder Gallimidi et al., 2015).
We presently found that F. nucleatum caused cellular DNA damage with increasing upregulation of γH2AX. As known, the quantity of F. nucleatum can be associated with the presence of cancer. In this study, F. nucleatum infection at a higher MOI of 500 induced significant increased expression of γH2AX than at an MOI of 200 as well as noninfected control (shown in Supplementary Fig. S1). Similarly, higher amount of F. nucleatum in saliva was found in patients with colorectal cancer (Guven et al., 2019). Interestingly, we have also recently reported that F. nucleatum was highly detected in OSCC tissues comparing to normal tissues and its presence was correlated with subgingival plaques (Chang et al., 2018).
DNA damage has recently been critically reviewed and found to be closely related to the hallmarks of cancer (such as sustained proliferative signals) and is sustained during each developmental stage of cancer (Gorgoulis et al., 2018). Thus, we chose a cell line of tongue squamous cell carcinoma to initially explore the effect of DNA damage caused by F. nucleatum in OSCC. For further study, normal cells (immortalized cells and primary cells) and other OSCC cell lines should be included to better understand the role of DNA damage in OSCC initiation and development.
The continuously increased expression of γH2AX and decreased expression of Ku indicated enhanced DNA damage with insufficient DNA repair. Meanwhile, we suggested that the increased cell growth was to some extent attributed to the dampened repair mechanism. To our knowledge, this is the first time to demonstrate that the serious DNA damage caused by F. nucleatum without timely repair causes an abnormal proliferation ability in OSCC cells.
In the present study, generally, the percentage of cells in the S phase was significantly increased in response to DNA damage. The downregulation of p27 was seen in this study. In accordance with our results, several studies have also revealed that downregulation of p27 causes cell cycle arrest in S phase and an enhanced cell proliferation ability (Keles et al., 2003; Menchon et al., 2011; Li et al., 2012).
We explored the expression of Ku70 and p53, the major proteins involved in regulating NHEJ repair. Once cells are damaged, the DNA-binding domain of Ku70 is acetylated while the Ku70-dependent suppression of p53 expression is abrogated. That is, the expression of Ku70 is reduced while the expression of p53 is increased (Lamaa et al., 2016). However, contrary results were shown in the present study in that the expression of Ku70 and wild p53 were synchronously decreased in association with enhanced cell proliferation ability. We hypothesize that the DNA damage caused by F. nucleatum at the MOI of 500 is so serious that the intracellular Ku70 is insufficient to provide prompt repair. Furthermore, without the normal regulation of Ku70, wild p53 was aberrantly expressed from upregulation to downregulation along with a possible mutation. Similar to our findings, other researchers have found a positive correlation between Ku70 and p53 changes in colon cancer (Komuro et al., 2002; Lu et al., 2015). At the same time, the results of the present study support the hypothesis that deletion of Ku70 or Ku80 increases the underlying damages and limits the postinjury response mechanisms mediated by p53 (Khanna and Jackson, 2001; Podhorecka et al., 2010).
To further verify the pathway, we overexpressed Ku70 by transfection. As expected, the increased cell proliferation ability was inhibited with upregulation of wild p53. Thus, combined with the results above, we concluded that the possible promoting role of F. nucleatum in OSCC relied on the Ku70/p53 pathway after causing DNA damage. Ku70 is known as the key component of classic NHEJ that plays a critical role in detecting DSBs (Puebla-Osorio et al., 2014). Thus, in the present study, we focused the specific role of Ku70 in DNA damage caused by F. nucleatum infection. However, another model is clearly needed to explore the role of Ku70/80 heterodimer under F. nucleatum infection.
In general, several deficiencies remain that need further investigation. F. nucleatum ATCC 25586 is the strain of F. nucleatum that was applied in the present study. Other subspecies of F. nucleatum and nonpathogens should be included in future studies, which will facilitate the understanding of the specific role of F. nucleatum in OSCC (Henne et al., 2018). Also, to complete the mechanism of the Ku70/p53 pathway, the upstream and downstream molecules need further exploration. Moreover, in addition to cell proliferation, F. nucleatum infection is involved in the regulation of cell invasion and metastasis by affecting proteinase production through the activation of p38 mitogen-activated protein kinase (Uitto et al., 2005). The possible effect of DNA damage caused by F. nucleatum on cell invasion and metastasis is needed to be further explored. Nevertheless, we demonstrated a positive role of F. nucleatum in causing DNA damage that promoted cell growth and enriched the mechanisms of OSCC development.
Supplementary Material
Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by the National Natural Science Foundation of China (81670997 and 81870771).
Supplementary Material
References
- Bach M., Savini C., Krufczik M., Cremer C., Rosl F., and Hausmann M. (2017). Super-resolution localization microscopy of gamma-H2AX and heterochromatin after folate deficiency. Int J Mol Sci 18, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder Gallimidi A., Fischman S., Revach B., Bulvik R., Maliutina A., Rubinstein A.M., et al. (2015). Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 6, 22613–22623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenstein A., Akhmedov A., Camici G.G., Luscher T.F., and Tanner F.C. (2013). p27(Kip1) inhibits tissue factor expression. Biochem Biophys Res Commun 439, 559–563 [DOI] [PubMed] [Google Scholar]
- Chang C., Geng F., Shi X., Li Y., Zhang X., Zhao X., et al. (2019). The prevalence rate of periodontal pathogens and its association with oral squamous cell carcinoma. Appl Microbiol Biotechnol 103, 1393–1404 [DOI] [PubMed] [Google Scholar]
- Gorgoulis V.G., Pefani D.E., Pateras I.S., and Trougakos I.P. (2018). Integrating the DNA damage and protein stress responses during cancer development and treatment. J Pathol 246, 12–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten F.R., and Grivennikov S.I. (2019). Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51, 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Greten F.R., and Karin M. (2010). Immunity, inflammation, and cancer. Cell 140, 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke K., Hernandez P.P., Zimmermann J., Klose C.S.N., Kofoed-Branzk M., Guendel F., et al. (2019). Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven D.C., Dizdar O., Alp A., Akdogan Kittana F.N., Karakoc D., Hamaloglu E., et al. (2019). Analysis of Fusobacterium nucleatum and Streptococcus gallolyticus in saliva of colorectal cancer patients. Biomark Med 13, 725–735 [DOI] [PubMed] [Google Scholar]
- Henne K., Schilling H., Stoneking M., Conrads G., and Horz H.P. (2018). Sex-specific differences in the occurrence of Fusobacterium nucleatum subspecies and Fusobacterium periodonticum in the oral cavity. Oncotarget 9, 20631–20639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao J.R., Chang C.C., Lee W.T., Huang C.C., Ou C.Y., Tsai S.T., et al. (2018). The interplay between oral microbiome, lifestyle factors and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogenesis 39, 778–787 [DOI] [PubMed] [Google Scholar]
- Kastan M.B., and Bartek J. (2004). Cell-cycle checkpoints and cancer. Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- Keles N., Erdamar B., Kaur A., and Deger K. (2003). p21, p53, and p27 Kip1 alterations in benign and malignant tumors of sinonasal epithelium. Otolaryngol Head Neck Surg 129, 77–84 [DOI] [PubMed] [Google Scholar]
- Khanna K.K., and Jackson S.P. (2001). DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27, 247–254 [DOI] [PubMed] [Google Scholar]
- Komuro Y., Watanabe T., Hosoi Y., Matsumoto Y., Nakagawa K., Tsuno N., et al. (2002). The expression pattern of Ku correlates with tumor radiosensitivity and disease free survival in patients with rectal carcinoma. Cancer 95, 1199–1205 [DOI] [PubMed] [Google Scholar]
- Lamaa A., Le Bras M., Skuli N., Britton S., Frit P., Calsou P., et al. (2016). A novel cytoprotective function for the DNA repair protein Ku in regulating p53 mRNA translation and function. EMBO Rep 17, 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li V.C., Ballabeni A., and Kirschner M.W. (2012). Gap 1 phase length and mouse embryonic stem cell self-renewal. Proc Natl Acad Sci U S A 109, 12550–12555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Tang X., Li C., Pan C., Li Q., Geng F., et al. (2015). Porphyromonas gingivalis promotes the cell cycle and inflammatory cytokine production in periodontal ligament fibroblasts. Arch Oral Biol 60, 1153–1161 [DOI] [PubMed] [Google Scholar]
- Lu Y., Gao J., and Lu Y. (2015). Down-expression pattern of Ku70 and p53 coexisted in colorectal cancer. Med Oncol 32, 98. [DOI] [PubMed] [Google Scholar]
- Malumbres M., and Barbacid M. (2009). Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9, 153–166 [DOI] [PubMed] [Google Scholar]
- Mari P.O., Florea B.I., Persengiev S.P., Verkaik N.S., Bruggenwirth H.T., Modesti M., et al. (2006). Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A 103, 18597–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menchon C., Edel M.J., and Izpisua Belmonte J.C. (2011). The cell cycle inhibitor p27Kip(1) controls self-renewal and pluripotency of human embryonic stem cells by regulating the cell cycle, Brachyury and Twist. Cell Cycle 10, 1435–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moergel M., Kammerer P., Kasaj A., Armouti E., Alshihri A., Weyer V., et al. (2013). Chronic periodontitis and its possible association with oral squamous cell carcinoma - a retrospective case control study. Head Face Med 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obst B., Wagner S., Sewing K.F., and Beil W. (2000). Helicobacter pylori causes DNA damage in gastric epithelial cells. Carcinogenesis 21, 1111–1115 [PubMed] [Google Scholar]
- Parkin D.M. (2006). The global health burden of infection-associated cancers in the year 2002. Int J Cancer 118, 3030–3044 [DOI] [PubMed] [Google Scholar]
- Pereira S.S., Morais T., Costa M.M., Monteiro M.P., and Pignatelli D. (2013). The emerging role of the molecular marker p27 in the differential diagnosis of adrenocortical tumors. Endocr Connect 2, 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera M., Al-Hebshi N.N., Perera I., Ipe D., Ulett G.C., Speicher D.J., et al. (2018). Inflammatory bacteriome and oral squamous cell carcinoma. J Dent Res 97, 725–732 [DOI] [PubMed] [Google Scholar]
- Podhorecka M., Skladanowski A., and Bozko P. (2010). H2AX Phosphorylation: its role in DNA damage response and cancer therapy. J Nucleic Acids 2010, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla-Osorio N., Kim J., Ojeda S., Zhang H., Tavana O., Li S., et al. (2014). A novel Ku70 function in colorectal homeostasis separate from nonhomologous end joining. Oncogene 33, 2748–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai P., He F., Kwang J., Engelward B.P., and Chow V.T. (2016). Pneumococcal pneumolysin induces DNA damage and cell cycle arrest. Sci Rep 6, 22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., and Bonner W.M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- Rubinstein M.R., Baik J.E., Lagana S.M., Han R.P., Raab W.J., Sahoo D., et al. (2019). Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep 20, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., and Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang P., Guo Y., Liang X., Li Y., and Ding S. (2019). Helicobacter pylori-induced DNA damage is a potential driver for human gastric cancer AGS cells. DNA Cell Biol 38, 272–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto V.J., Baillie D., Wu Q., Gendron R., Grenier D., Putnins E.E., et al. (2005). Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells. Infect Immun 73, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore S.E., and Lamont R.J. (2014). Oral bacteria and cancer. PLoS Pathog 10, e1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wu J., Chen T., Li Q., Peng W., Li H., et al. (2018). Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a Toll-like receptor 4/p21-activated kinase 1 cascade. Dig Dis Sci 63, 1210–1218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.