Abstract
Inhibition of dendritic cell maturation and activation, together with abnormal functioning of cell-mediated immunity, has been reported in chronic spinal cord injury (SCI). The development of immune-based therapies could: 1) prevent or slow down limit further tissue damage in chronic SCI, and 2) promote tissue regeneration. To identify novel candidate molecular pathways mediating SCI-induced immune changes, we performed whole–genome microarray and molecular pathway analyses. Subjects with motor complete chronic SCI (> 2 years post-injury) and uninjured controls were selected from an ongoing study. Microarray analysis was performed with RNA extracted from circulating monocytes. Partek Genomic Suite (PGS) software was used to limit the 54,000 gene list to only those genes up-regulated or down-regulated by 2-fold or more in SCI compared with control. Pathway analyses were performed with Ingenuity Systems IPA software to identify biological pathways of interest involving differentially expressed genes. Genes of interest were then confirmed by quantitative PCR (qPCR). Six SCI subjects and five uninjured controls were included in the final analyses. A molecular pathway related to immune cell trafficking was identified as being significantly upregulated in the SCI subjects. Two genes in that network, transmembrane domain protein (TMEM)176A and TMEM176B, were notable for the magnitude of overexpression. Dendritic cells have been shown to mediate recovery and/or protective autoimmunity in central nervous system injuries and have the capacity to induce neuroprotection and neurogenesis in stroke patients. High TMEM176A and TMEM176B levels have been shown to prevent dendritic cell maturation and inhibit dendritic cell activity in the general population. Here, we report overexpression of both genes in SCI compared with control subjects. Thus, we propose that TMEM176A and TMEM176B are candidate genes involved in inhibiting protective immune responses in SCI. This study may support future research aimed at developing new targets for therapies to promote immune system-mediated neuroprotection and recovery in SCI.
Keywords: autoimmunity, dendritic cells, spinal cord injury, TMEM family
Introduction
An autoimmune response is frequently observed following traumatic spinal cord injury (SCI).1 This response may produce secondary damage to the neighboring neurons that escaped the main injury. For instance, it has been reported that patients with SCI have elevated titers of antibodies against myelin basic protein in serum and cerebrospinal fluid, suggesting that trauma activates both T and B cells which are able to recognize central nervous system (CNS) proteins.2 Autoimmunity following SCI has been generally associated with poor neurological outcomes. However, it also has been suggested that an autoimmune response can have neuroprotective and/or reparative effects after trauma, including stroke and SCI, and that this neuroprotection is mediated, to a certain extent, by autoreactive T cells.3–5 Animal studies show that recovery after injury is improved in B cell-deficient mice,6 suggesting a role for B cells in neurotoxic autoimmunity. Although still a subject of debate, immune-based therapies in the acute phase of the injury may mitigate or prevent further neurological damage after SCI.5,7
Over the last decade, dendritic cells have emerged as the focus of research at the intersection of CNS trauma and autoimmunity, particularly because they are potent antigen-presenting cells that play crucial roles in host defenses connecting innate and adaptative immune responses.8 Their number, state of maturation, as well as the context in which they are activated define the resulting immune response. Maturation in vitro of dendritic cells from progenitors is significantly reduced in SCI patients and following SCI with tetraplegia associated with less mature dendritic cells than paraplegia.8 The defect in innate immunity (maturation potential of dendritic cells) observed after SCI may be caused by sympathetic nervous system dysfunction after injury.9 Injection of functional dendritic cells matured ex vivo following SCI has been shown to activate neural stem cells, causing protection from neurological damage and promoting neurological recovery.10,11
Immature dendritic cells are characterized by low expression of stimulatory molecules and inflammatory cytokines contributing to abnormal T cell function.12 Two transmembrane domain proteins (TMEM), TMEM176A and TMEM176B, are preferentially expressed in myeloid cells and are strictly associated with the immature state of dendritic cells in rats and humans. Their expression is low or absent in NK, CD4+ or CD8+ T cells and in B cells. These molecules interact with each other and can form multimers suggesting that both may be part of an intracellular complex which plays a role in the maturation of dendritic cells.13 TMEM176A and TMEM176B proteins maintain the initial immature state of dendritic cells and have no influence in mature dendritic cells.12 The exact molecular mechanisms by which these two proteins exert their role remain to be elucidated and little is known about their role in immune dysfunction after SCI. In this study, we sought to identify novel putative molecular pathways mediating SCI-induced immune changes that might warrant further investigation. We therefore performed whole-genome microarray and molecular pathway analyses of circulating white blood cells derived from men with and without chronic SCI. Given the exploratory nature of this study, we hypothesized that gene expression profiles would vary based on the presence or absence of chronic SCI.
Methods
Subjects
In this study, we evaluated a convenience sample of six male subjects with chronic motor complete SCI (more than 5 years post-injury) and five male subjects without SCI who were enrolled in the Fracture Risk after SCI (FRASCI) Study.14 Inclusion criteria for the parent study have been previously reported.14 Briefly, participants with SCI were eligible to participate if they were 18 years of age or older, 1 year or more after injury, were not ventilator dependent, and had no other neurological condition such as multiple sclerosis, stroke, and past polio. Subjects were not eligible to participate if they reported changes in their medical status or symptoms of systemic illness. Subjects without SCI were recruited from our outpatient clinics (Boston, MA) or by advertisement and were eligible to participate if they did not require an ambulatory aid, had no neurological conditions preventing independent walking, and had not been previously diagnosed with osteoporosis. Our institutional review boards approved the study and all subjects gave informed consent.
Spinal cord injury classification
SCI was confirmed by physical examination by a trained rater according to the American Spinal Injury Association Impairment Scale (AIS) as previously described.14,15
Peripheral blood mononuclear cells isolation
Approximately 30 mL of peripheral blood extracted from each participant were drawn into a heparinized tube and diluted at a ratio of 1:2 with phosphate-buffered saline (PBS) solution. The whole–blood PBS solution was layered onto one volume of Histopaque®-1077 (Sigma-Aldrich, St. Louis, MO). The layered solution was then centrifuged and white blood cells were isolated from the plasma-Histopaque interface for subsequent analysis.
Total RNA extraction and complementary DNA preparation
Total RNA was extracted from the peripheral blood mononuclear cells using the Qiagen RNeasy Mini Kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions. RNA concentration and integrity were assessed with the NanoDrop 8000 spectrophotometer (Fischer Scientific, Wilmington, DE). Two micrograms of RNA obtained from each sample were reverse transcribed into complementary DNA (cDNA) using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany).
Microarray analysis
Microarray analysis was performed by SeqWright, Inc. (Houston, TX). Complementary DNA was hybridized to HG-U133A Plus 2.0 GeneChip oligonucleotide arrays (Affymetrix, Santa Clara, CA), each of which contains 54,675 sets of oligonucleotide probes targeting approximately 38,000 human genes. Relative fluorescence of the chip region corresponding to each gene in the GeneChip was determined and the raw data were exported to CEL files for further analysis using the Partek Genomics Suite (PGS Partek Inc., St. Louis, Missouri) and Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA). PGS was used to generate a list from the raw CEL files of 1453 genes that were significantly over- or under-expressed in SCI versus control subjects (p ≤ 0.05). The significance of differentially expressed genes was evaluated based on the adjusted p values generated by the Benjamini and Hochberg16 stepwise procedure in order to control the false discovery rate.
IPA software was used for further analysis of the genes identified by PGS, including the identification of differentially expressed genetic pathways. A significance score was calculated for each pathway and that represents the possibility that the genes in the pathway are found there by chance. The significance score of a network17 is equal to the negative exponent of calculated P value of the network (i.e., a significance score of 4 equals a p value of 1 × 10−4). Each network is composed of individual genes and complexes. Out of all the genes in a particular network, only some are referred to as “focus molecules” (genes that significantly change their status). Genes included in the network that are not focus molecules are genes which have been reported to interact with the focus molecules. Thus, some genes reported in the network were not shown to have significant changes in expression but were included because of a functional relationship to genes that did. In the resulting network schematic, focus genes are shaded, while other genes are not. Genes of interest identified in the microarray analysis were confirmed by real-time polymerase chain reaction (RT-PCR).
Quantitative RT-PCR
Complementary DNA derived from peripheral blood mononuclear cells was used as a template for RT-PCR to confirm differential expression of selected genes of interest. All reactions were performed in triplicate with an iCycler Optical Module system (Bio-Rad, Philadelphia, PA) in a 96-well PCR plate (ABgene, Epson, Surray County, UK) with a final reaction volume of 20 μL/well. Each reaction sample included Sybr-Green PCR Master Mix (Bio-Rad), 50ng of template cDNA and 0.25 μM of sense and anti-sense primer mix (Invitrogen Corporation, Carlsbad, CA). Optimized thermal cycling conditions were: 95° for 7 min followed by 45 cycles at 95° for 10 sec, 55° for 15 sec, 72° for 15 sec, and a final cycle at 95° for 1 min. Differential expression of TMEM176A and TMEM176B was confirmed by qPCR using the oligonucleotides described in Table 1. Human β-actin was assayed as an endogenous control (Table 1) and all quantifications were normalized using the cycling threshold method.
Table 1.
Oligonucleotide Primers for qPCR
| 5′ – 3′ oligonucleotide forward and reverse sequences | ||
|---|---|---|
| Human ACTB (NM_001101.3) | Forward | ACCGCGAGAAGATGACCCAG |
| Reverse | GTACGGCCAGAGGCGTACAG | |
| Human TMEM176A (NM_018487.2) | Forward | CTCTTGGGTGTCTGGATTCTG |
| Reverse | TTTCCTTCTGGTCTCTTTTCCC | |
| Human TMEM176B (NM_001101314.1) | Forward | GCGAAGTCAAGAGAACCAATG |
| Reverse | CTACTCCCAAGGAAACCAAGG | |
qPCR, quantitative polymerase chain reaction; TMEM, transmembrane domain protein
Statistical analysis
To compare gene expression levels between samples from SCI subjects and uninjured controls, the relative expression of each gene was calculated using the comparative ΔΔCt method [fold of expression = 2-(ΔΔCT ± SD)]. Student's t-test or analysis of variance with Bonferroni post hoc test was used to analyze differences in gene expression between both groups.
Results
Participant characteristics have been previously reported4 and are again presented in Table 2A and Table 2B. All were male and none reported active bisphosphonate use or active heterotopic ossification. Most participants were white. There was no difference in age between the SCI and no SCI groups (p = 0.24) and their ages ranged from 31.9 to 68.7 years. For those with SCI, only one subject had AIS A tetraplegia and the remaining five subjects had AIS A paraplegia. All subjects with SCI used a wheelchair for mobility. The duration of injury ranged from 16.5 to 43.7 years.
Table 2.
A. Subject Characteristics
| Groups | Subject | Age (y) | Injury level | Injury duration (y) |
|---|---|---|---|---|
| SCI | 1 | 60.3 | T8 AIS A | 35.9 |
| 2 | 62.4 | T12 AIS A | 20.8 | |
| 3 | 46.2 | C5 AIS A | 31.2 | |
| 4 | 31.9 | T3 AIS A | 18.0 | |
| 5 | 35.1 | T8 AIS A | 16.5 | |
| 6 | 67.1 | T11 AIS A | 43.7 | |
| No SCI | 1 | 68.7 | - | - |
| 2 | 55.6 | - | - | |
| 3 | 62.5 | - | - | |
| 4 | 60.6 | - | - | |
| 5 | 50.7 | - | - | |
SCI, spinal cord injury; AIS, American Spinal Injury Association Impairment Scale.
Table 2.
B. Active Medication Use
| Drug class | SCI (n = 6) | Able-bodied (n = 5) |
|---|---|---|
| Heart burn or stomach disorder, n (%) | 2 (33.3) | 3 (60.0) |
| Inhaled or oral steroid, n (%) | 1 (16.6) | 1 (20.0) |
| Blood pressure or heart medications, n (%) | 3 (50.0) | 2 (40.0) |
| Blood thinners, n (%) | 4 (66.6) | 2 (40.0) |
| Cholesterol medication, n (%) | 3(50.0) | 3 (60.0) |
| Diuretic, n (%) | 1 (16.6) | 2 (40.0) |
| Muscle or bladder spasms, n (%) | 5 (83.3) | 1 (20.0) |
| Pain, n (%) | 2 (33.3) | 1 (20.0) |
SCI, spinal cord injury.
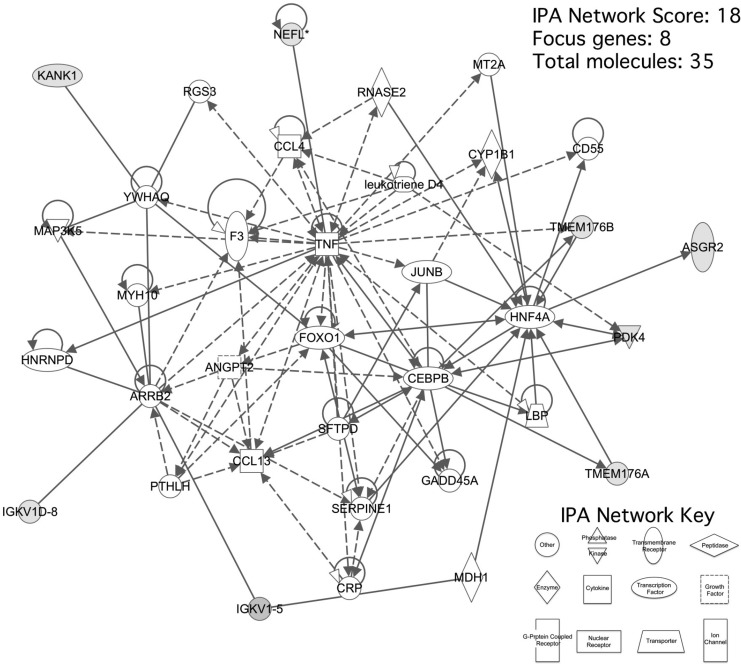
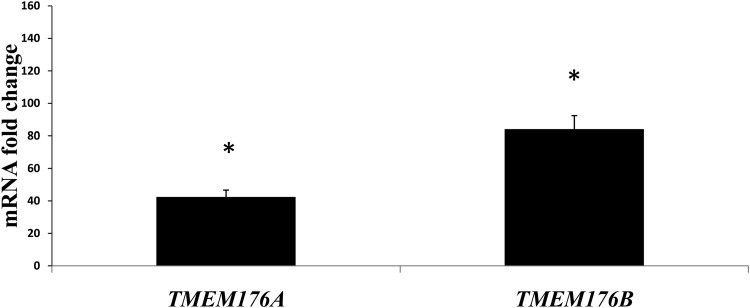
Microarray analysis revealed 1970 genes whose up- or down-regulation was statistically significant (p ≤ 0.05). IPA software identified 1453 of genes were identified that resulted to be present in 25 statistically significant molecular pathways. We selected a single pathway (Fig. 1) with an IPA score of 18, consisting of eight focus molecules within a total of 35 genes (Table 3) for further analysis based on its associated network functions of “Cell death, cellular movement and immune cell trafficking” and its inclusion of many cytokines related to inflammation and the recruitment of leukocytes. This selected pathway included TMEM176A and TMEM176B genes, which our microarray analysis had shown to be overexpressed by factors of 5.2 (p = 0.01) and 11.9 (p = 0.01), respectively in SCI patients compared with healthy controls. Quantitative real-time PCR showed that TMEM176A and TMEM176B were overexpressed 42.4 (p < 0.05) and 84.1-fold (p < 0.05) respectively (Fig. 2), validating the microarray analysis results.
FIG. 1.
Pathway analysis of peripheral blood monocytes gene expression in patients with chronic spinal cord injury compared with control healthy subjects. The network depicted is composed of genes described in the Results section. Transmembrane domain protein TMEM176A and TMEM176B are highlighted.
Table 3.
Pathway Gene List
Single selected pathway with IPA software score 18, consisting in eight focus molecules, genes identified by microarray to be significantly changed.
FIG 2.
Transmembrane domain protein TMEM176A and TMEM176B are upregulated in spinal cord injury (SCI) patients. Quantitative real-time polymerase chain reaction analysis of messenger RNA expression was performed as described under methods. Both were significantly up-regulated in SCI subjects, compared with uninjured controls (TMEM176A: 42.4-fold and TMEM176B: 84.1-fold, *p ≤ 0.05). Control values were 1,0988 and 1,09464654 for TMEM176A and TMEM176B, respectively.
Discussion
In this study, we report that TMEM176A and TMEM176B are significantly elevated in circulating monocytes from subjects with chronic SCI. During the revision of this manuscript, Segovia and colleagues published an elegant paper describing a mechanism by which TMEM176B regulates T cell-dependent immunity.18 The authors demonstrate that TMEM176B works as a cation channel and, in response to adenosine triphosphate, inhibits accumulation of calcium in the cytoplasm. Since calcium-dependent potassium influx is required for inflammasome activation, the authors conclude that TMEM176B inhibits activation of the inflammasome. They further confirm this by the following: 1) using TMEM176B deficient mice, and 2) specifically blocking TMEM176B with BayK8644, a drug that triggers activation of the inflammasome. TMEM176B is expressed on myeloid cells, including dendritic cells. Prior studies suggest that SCI may be associated with impaired dendritic cell function. Since activation of the inflammasome in dendritic cells is required for T-cell mediated immunity, increased expression of TMEM176B in subjects with SCI offers one possible explanation for immune depression observed in this population. Because dendritic cell mediated therapies have potential neuroprotective benefits in rodent models,19 it is possible that novel therapeutic approaches targeting dendritic cell suppression after SCI may warrant further investigation. While exploratory in nature, our findings may provide insight into potential molecular mechanisms by which dendritic cell function is suppressed following SCI.
TMEM176A and TMEM176B were recently characterized and little is known about their function or role in the pathophysiology of disease states. Their expression has been described in association with the immature state of dendritic cells.12 TMEM176B was first identified in 2005 in rats as a gene highly overexpressed in tolerated allografts and was initially given the name Torid (tolerance-related and induced transcript).20 Torid/TMEM176B is the rat orthologue to human LR8 gene and its expression was found to be highest in myeloid dendritic cells (DCs; mostly in CD4+ DCs), with almost no expression in the other dendritic cell subpopulations. TMEM176B expression is inversely related to the activation or maturation of dendritic cells and to the expression of MHC II and CD86. Moreover, TMEM176B gene expression strongly decreases following stimulation of dendritic cells early after activation, suggesting that it may be involved in the regulation of antigen uptake or initiation of differentiation rather than in T cell interactions.20 TMEM176A, on the other hand, was previously associated with tumor autoantigens suggesting a role in transformation, metastasis or immune evasion of hepatocellular carcinoma21 and its methylation is linked to lymph node metastasis of colorectal cancer22 and gastric tumors.23
TMEM176A and TMEM176B proteins interacts with each other to form multimers and they exhibit similar patterns of expression and regulation. Their inhibition increased the expression of costimulatory molecules, the maturation of dendritic cells and subsequent activation of allogenic T cells.12 These results strongly suggest that TMEM176A and TMEM176B play key roles in maintaining the immature, inactivated state of dendritic cells. In relation to the intracellular mechanism by which both molecules exert their action, dendritic cells express numerous ion channels and a switch of ion channel expression during maturation has been demonstrated.24 As both gene products are part of an intracellular membranous complex and interact together, these two molecules may be part of an ion channel determinant in the final adjustment of dendritic cells maturation,12 consistent with the recent article by Sergovia and colleagues.18
The present paper is the first to report overexpression of TMEM176A and TMEM176B in peripheral blood mononuclear cells derived from subjects with SCI compared with uninjured controls. We suggest that overexpression of TMEM176A and TMEM176B is a significant mediator of the functional suppression of DCs, observed after SCI. Because our findings are limited to chronic SCI, future studies focused on changes in immunophenotypes in the acute setting are needed. Accordingly, a number of therapies for use in both the acute and chronic phase of SCI should be investigated. Vaccination of subjects with acute SCI with dendritic cells pulsed with various CNS antigens has already been shown to provide neuroprotection and to promote neurological recovery.10,19,25 However, this approach requires invasive procedures to extract dendritic cells from the bone marrow for subsequent transplantation near the cord lesion. Interference with the expression or function of TMEM176A and TMEM176B could less invasively provide some or all of the benefits of dendritic cell vaccination therapy by recovering dendritic cell capacity to effectively present antigen in such a way that CD4+ T cells become activated.
To our knowledge, this is the first study reporting the association between overexpression of TMEM176A and TMEM176B in spinal cord injury. This study has a number of limitations, including the small sample size in only men with motor complete chronic SCI. The original microarray analysis was designed to include a homogenous sample to limit variability. Therefore, additional work, including more participants and women, should be undertaken to confirm these findings and to determine the generalizability of the findings in both acute and chronic SCI. Despite these limitations, our findings suggest that therapeutic approaches targeting TMEM176A and TMEM176B expression might providing neurological protection and/or promote recovery after SCI by restoring dendritic cell function.
Funding Information
The present study was funded by National Institute of Arthritis and Musculoskeletal and Skin Diseases National Institutes of Health/National Institute of Child Health and Human Development 3R21HD057030-02S1 (LRM) and R01AR064793 (RAB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Riegger T., Conrad S., Liu K., Schluesener H.J., Adibzahdeh M., and Schwab J.M. (2007). Spinal cord injury-induced immune depression syndrome (SCI-IDS). Eur. J. Neurosci. 25, 1743–1747 [DOI] [PubMed] [Google Scholar]
- 2. Ankeny D.P. and Popovich P.G. (2010). B cells and autoantibodies: complex roles in CNS injury. Trends Immunol. 31, 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ziv Y., Finkelstein A., Geffen Y., Kipnis J., Smirnov I., Shpilman S., Vertkin I., Kimron M., Lange A., Hecht T., Reyman K.G., Marder J.B., Schwartz M., and Yoles E. (2007). A novel immune-based therapy for stroke induces neuroprotection and supports neurogenesis. Stroke 38, 774–782 [DOI] [PubMed] [Google Scholar]
- 4. Saltzman J.W., Battaglino R.A., Salles L., Jha P., Sudhakar S., Garshick E., Stott H.L., Zafonte R., and Morse L.R. (2013). B-cell maturation antigen, a proliferation-inducing ligand, and B-cell activating factor are candidate mediators of spinal cord injury-induced autoimmunity. J. Neurotrauma 30, 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saltzman J.W., Battaglino R., Stott H., and Morse L.R. (2013). Neurotoxic or neuroprotective? Current controversies in SCI-induced autoimmunity. Curr. Phys. Med. Rehabil. Rep 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dekaban G.A. and Thawer S. (2009). Pathogenic antibodies are active participants in spinal cord injury. J. Clin. Invest. 119, 2881–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwartz M. and Kipnis J. (2005). Protective autoimmunity and neuroprotection in inflammatory and noninflammatory neurodegenerative diseases. J. Neurol. Sci 233, 163–166 [DOI] [PubMed] [Google Scholar]
- 8. Pan S.C., Hsieh S.M., Wang Y.H., Chiang B.L., Huang T.S,. and Chang S.C. (2005). In vitro maturation potential of monocyte-derived dendritic cells is impaired in patients with spinal cord injury: a case-control study. Arch. Phys. Med. Rehabil. 86, 974–978 [DOI] [PubMed] [Google Scholar]
- 9. Takenaka M.C., Guereschi M.G., and Basso A.S. (2017). Neuroimmune interactions: dendritic cell modulation by the sympathetic nervous system. Semin. Immunopathol. 39, 165–176 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y., Wang K., Chao R., Li J., Zhou L., Ma J., and Yan J. (2012). Neuroprotective effect of vaccination with autoantigen-pulsed dendritic cells after spinal cord injury. J. Surg. Res. 176, 281–292 [DOI] [PubMed] [Google Scholar]
- 11. Hayashi K., Ohta S., Kawakami Y., and Toda M. (2009). Activation of dendritic-like cells and neural stem/progenitor cells in injured spinal cord by GM-CSF. Neurosci. Res. 64, 96–103 [DOI] [PubMed] [Google Scholar]
- 12. Condamine T., Le Texier L., Howie D., Lavault A., Hill M., Halary F., Cobbold S., Waldmann H., Cuturi M.C., and Chiffoleau E. (2010). Tmem176B and Tmem176A are associated with the immature state of dendritic cells. J. Leukoc. Biol. 88, 507–515 [DOI] [PubMed] [Google Scholar]
- 13. Segovia M., Louvet C., Charnet P., Savina A., Tilly G., Gautreau L., Carretero-Iglesia L., Beriou G., Cebrian I., Cens T., Hepburn L., Chiffoleau E., Floto R.A., Anegon I., Amigorena S., Hill M., and Cuturi M.C. (2014). Autologous dendritic cells prolong allograft survival through Tmem176b-dependent antigen cross-presentation. Am. J. Transplant. 14, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morse L.R., Sudhakar S., Danilack V., Tun C., Lazzari A., Gagnon D.R., Garshick E., and Battaglino R.A. (2012). Association between sclerostin and bone density in chronic spinal cord injury. J. Bone Miner Res. 27, 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park A.J., Battaglino R.A., Nguyen N.M.H., and Morse L.R. (2018). Associations between lean mass and leptin in men with chronic spinal cord injury: results from the FRASCI-muscle study. PLoS One 13, e0198969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benjamini Y.and Hochberg Y (1995). Controlling the false eiscovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Series B (Methodol.) 57, 289–300 [Google Scholar]
- 17. Shanley T.P., Cvijanovich N., Lin R., Allen G.L., Thomas N.J., Doctor A., Kalyanaraman M., Tofil N.M., Penfil S., Monaco M., Odoms K., Barnes M., Sakthivel B., Aronow B.J., and Wong H.R. (2007). Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol. Med. 13, 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segovia M., Russo S., Jeldres M., Mahmoud Y.D., Perez V., Duhalde M., Charnet P., Rousset M., Victoria S., Veigas F., Louvet C., Vanhove B., Floto R.A., Anegon I., Cuturi M.C., Girotti M.R., Rabinovich G.A., and Hill M. (2019). Targeting TMEM176B enhances antitumor immunity and augments the efficacy of immune checkpoint blockers by unleashing inflammasome activation. Cancer Cell 35, 767–781 e766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hauben E., Gothilf A., Cohen A., Butovsky O., Nevo U., Smirnov I., Yoles E., Akselrod S., and Schwartz M. (2003). Vaccination with dendritic cells pulsed with peptides of myelin basic protein promotes functional recovery from spinal cord injury. J. Neurosci. 23, 8808–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Louvet C., Chiffoleau E., Heslan M., Tesson L., Heslan J.M., Brion R., Beriou G., Guillonneau C., Khalife J., Anegon I., and Cuturi M.C. (2005). Identification of a new member of the CD20/FcepsilonRIbeta family overexpressed in tolerated allografts. Am. J. Transplant. 5, 2143–2153 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y., Han K.J., Pang X.W., Vaughan H.A., Qu W., Dong X.Y., Peng J.R., Zhao H.T., Rui J.A., Leng X.S., Cebon J., Burgess A.W., and Chen W.F. (2002). Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. J. Immunol. 169, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 22. Gao D., Han Y., Yang Y., Herman J.G., Linghu E., Zhan Q., Fuks F., Lu Z.J., and Guo M. (2017). Methylation of TMEM176A is an independent prognostic marker and is involved in human colorectal cancer development. Epigenetics 12, 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun L., Zhang Y., and Zhang C. (2018). Distinct Expression and Prognostic Value of MS4A in Gastric Cancer. Open Med. (Wars.) 13, 178–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zsiros E., Kis-Toth K., Hajdu P., Gaspar R., Bielanska J., Felipe A., Rajnavolgyi E., and Panyi G. (2009). Developmental switch of the expression of ion channels in human dendritic cells. J. Immunol. 183, 4483–4492 [DOI] [PubMed] [Google Scholar]
- 25. Liu M., Zhao J., Liang H., and Bian X. (2009). Vaccination with dendritic cells pulsed with homogenate protein of spinal cord promotes functional recovery from spinal cord injury in mice. Spinal Cord 47, 360–366 [DOI] [PubMed] [Google Scholar]