Abstract
The ability to control antibody activity by pH has important applications in diagnostics, therapeutic antibody targeting, and antibody-guided imaging. Here, we report the rational design of bivalent peptide–DNA ligands that allow pH-dependent control of antibody activity. Our strategy uses a pH-responsive DNA triple helix to control switching from a tight-binding bivalent peptide–DNA lock into a weaker-binding monovalent ligand. Different designs are introduced that allow antibody activation at both basic and acidic pHs, either autonomously or in the presence of an additional oligonucleotide trigger. The pH of antibody activation could be precisely tuned by changing the DNA triple helix sequence. The peptide–DNA locks allowed pH-dependent antibody targeting of tumor cells both in bulk and for single cells confined in water-in-oil microdroplets. The latter approach enables high-throughput antibody-mediated detection of single tumor cells based on their distinctive metabolic activity.
Short abstract
Bivalent, peptide−DNA ligands conditionally block antibody activity. Formation of a triple helix structure triggered by extracellular acidification results in antibody activation and tumor targeting.
Introduction
Antibodies play a pivotal role in many areas of life sciences, ranging from molecular diagnostics and imaging to targeted drug delivery and immunotherapy.1−5 Their omnipresence results from the ability to generate high-affinity antibodies for virtually any molecular target. Nonetheless, molecular diagnostics and antibody-based therapies suffer from false-positives and side-effects resulting from background expression of target antigens by healthy cells.6 Bispecific antibodies, whose binding relies on the simultaneous interaction with two different receptors, have been developed to increase specificity but do not effectively avoid background binding.7 Instead of increasing the specificity of molecular recognition, several strategies have been reported that block antibody binding such that antibody activity can only be restored in the presence of a specific molecular cue. Church and co-workers used DNA origami to construct nanocontainers (NanoRobots) to control the accessibility of antibody fragments using aptamer-based molecular locks. Activation of the aptamers by binding to cell-surface receptors and growth factors resulted in the opening of the DNA container, allowing the antibody Fab fragments to bind cell-surface receptors on cancer cells.8,9 Protease-activatable antibodies have been constructed by employing a prodrug design in which the antibody’s antigen binding sites are masked by fusing a ligand with attenuated affinity via a protease susceptible peptide linker.10−12 These prodrug antibodies have shown promising results in in vivo mouse models, but the approach is restricted to protease-based triggers and requires extensive protein engineering for each specific antibody.
A common feature of cancer cells is that—even at saturating oxygen concentrations—their energy metabolism is dominated by glycolysis, a phenomenon known as the Warburg effect. The resulting increased production of lactate acidifies the extracellular environment from pH 7.3–7.4 in healthy tissue to pH 6.2–6.9 in tumors.13−16 The Warburg effect has been the basis for the development of a variety of pH-dependent imaging strategies, diagnostic assays, and drug carriers.17−20 Typically, these strategies rely on pH-dependent imaging agents or pH-dependent disassembly of nanoparticles; e.g., turn-on fluorescent probes conjugated to tumor specific antibodies have been used for dual-specific tumor imaging, and pH-sensitive fluorescent probes have been applied for the detection of single circulating tumor cells in microfluidic droplets.21 Surprisingly, the development of targeting ligands that display increased affinity at low pH remains largely unexplored. Some examples of pH-responsive aptamers have been reported, either by introducing pH-dependent DNA structures such as i-motifs or triple helices or by including pH-dependency as a variable during aptamer selection.19,22−27 Directed evolution has also been used to develop pH-dependent antibodies, typically resulting in the introduction of histidines to modulate the surface charge of the antigen binding pocket. However, such directed evolution approaches are time-consuming and need to be repeated for each new antibody–antigen interaction.28−30 Moreover, in almost all examples the interaction is attenuated at low pH, which is impractical for tumor targeting.
Our group recently introduced bivalent peptide–DNA ligands as highly effective and generic molecular locks to reversibly control antibody activity. These self-assembling bivalent ligands can effectively bridge the two antigen binding sites present in monoclonal antibodies, allowing control of antibody activity by proteases, light, and toehold-mediated displacement of the dsDNA linker using oligonucleotide triggers.31−33 Here, we show that introduction of pH-responsive DNA triple helix structures in the linker of these ligands allows activation of antibody activity by either an increase or a decrease of the solution’s pH. Harnessing the programmable pH-dependency of the DNA triple helix, the pH sensitivity of these ligands can be rationally tuned, providing a generic strategy that can be applied to any antibody and operate in a user-defined pH range. pH-dependent antibody-based targeting is demonstrated for tumor cells, both in ensemble assays and on single cells confined in water-in-oil microdroplets generated using a microfluidic device. The latter approach provides an attractive approach for high-throughput antibody-based targeting of single tumor cells based on their distinctive metabolic activity.
Results
Antibody Activation at High pH
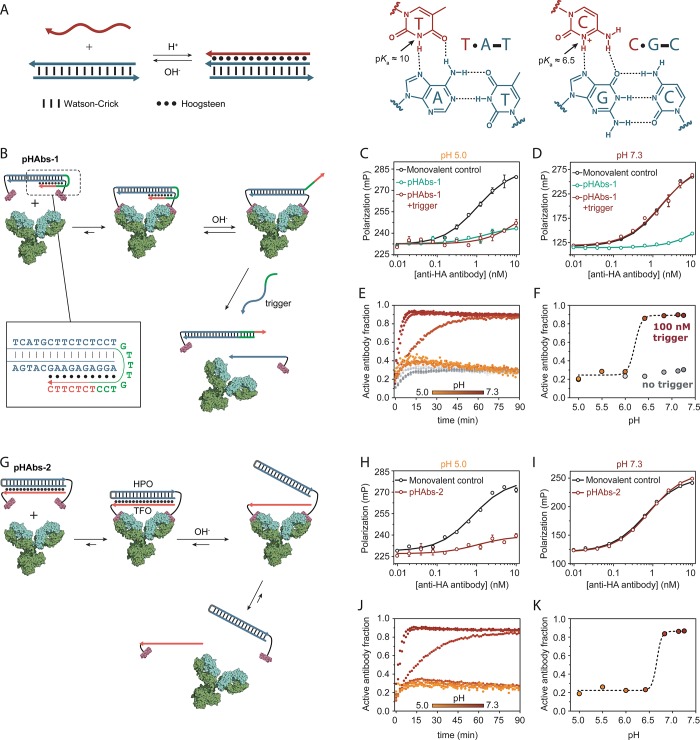
In this work we used the extensively studied DNA triple helix as a pH-responsive domain to construct a variety of bivalent peptide–DNA ligand architectures.34,35 DNA triple helix formation is governed by both TAT and CGC triplets which are composed of standard, antiparallel Watson–Crick interactions and additional parallel Hoogsteen interactions (Figure 1A). As the Hoogsteen interactions in TAT and CGC triplets rely on protonation of the N3 of, respectively, thymine (pKa ∼ 10) and cytosine (pKa ∼ 6.5), DNA triple helices are strongly pH dependent.36 Their difference in pKa, CGC triplets requiring a lower pH than TAT triplets to form, allows the pH response of a DNA triple helix to be tuned over more than 5 units of pH by sequence design.37 Because of these favorable properties, DNA triple helices have already been used to introduce pH control in toehold-mediated strand displacement reactions, hybridization chain reactions, DNA tile assembly, and plasmonic nanoassemblies.38−41
Figure 1.
Antibody activation at high pH. (A) DNA-based triple helices are formed by standard antiparallel Watson–Crick base pairing (dash) and pH-sensitive parallel Hoogsteen interactions (dots). Triple helix formation is governed by protonation of the N3 of thymine (pKa ∼ 10) or cytosine (pKa ∼ 6.5). (B) Design of bivalent antibody ligand pHAbs-1, assembled via a 20 base pair DNA duplex containing a 15 nucleotide overhang that folds back on the duplex to form a 10 nucleotide triple helix. (C, D) Competition assays between a fluorescently labeled HA peptide epitope and pHAbs-1 or a monovalent control in absence and presence of 100 nM trigger oligonucleotide at pH 5.0 and 7.3, respectively. (E) Fraction of activated antibody in time when blocked by pHAbs-1 in absence (gray symbols) and presence of 100 nM trigger oligonucleotide (colored symbols) at various pH values. (F) Steady state fraction of activated antibody at 90 min as a function of pH. (G) Design of bivalent antibody ligand pHAbs-2, assembled through the formation of a 20 nucleotide intermolecular triple helix between an epitope-functionalized DNA hairpin (HPO) and an epitope-functionalized triplex forming oligonucleotide (TFO), each conjugated to a HA-tag peptide epitope. (H, I) Competition assays between a fluorescently labeled HA peptide epitope and pHAbs-2 or a monovalent control at pH 5.0 and 7.3, respectively. (J) Fraction of activated antibody in time when blocked by pHAbs-2 at various pH values. (K) Steady state fraction of activated antibody at 90 min as a function of pH. Error bars represent standard error of the mean of duplicate measurements.
Two strategies were explored that allow antibody activation by an increase in pH. The first approach employs pH-dependent toehold-mediated strand displacement by a trigger oligonucleotide to control the integrity of a bivalent peptide–DNA lock.32 This bivalent ligand, termed pHAbs-1, harbors an 8 nucleotide long toehold domain on one of the oligonucleotides, which at low pH is sequestered within an intramolecular triple helix that is formed on part of the dsDNA linker (Figure 1B). At high pH the intramolecular triple helix is destabilized, thus exposing the toehold domain and allowing a trigger oligonucleotide to invade the ligand, yielding two weaker-binding, monovalent ligands that spontaneously dissociate to activate the antibody. A toehold of 8 nucleotides was chosen to ensure fast strand displacement kinetics and to provide sufficient thermodynamic driving force (i.e., base pairing) to disassemble the bivalent ligand via toehold-mediated strand displacement. As proof-of-principle a monoclonal antibody was used that targets the HA-tag peptide, a well-known peptide epitope derived from the human influenza virus hemagglutinin protein. Importantly, we verified that the interaction between the antibody and the HA peptide epitope by itself is not pH-sensitive (Kd = 0.46 nM, Figure S1).32,42 To construct the peptide–DNA chimera, oligonucleotides modified with a primary amine at either their 5′- or 3′-end were conjugated to cysteine-functionalized HA peptide epitopes via a heterobifunctional Sulfo-SMCC cross-linker and subsequently purified by reversed phase HPLC (Figure S2). To evaluate the efficiency of pHAbs-1 to block and concurrently activate the anti-HA antibody upon a change in pH, competition assays were performed with a fluorescently labeled HA peptide. To this end, a mixture of anti-HA antibody and pHAbs-1 that was preassembled at pH 5.0 was titrated to the competing fluorescently labeled HA peptide at either pH 5.0 or pH 7.3. A monovalent control containing only a single peptide–DNA conjugate was included to serve as reference for a fully activated antibody. Figure 1C shows that pHAbs-1 efficiently blocks binding of the antibody to the competing peptide at pH 5.0 both in the absence and presence of 100 nM of trigger, as fluorescence polarization of the competing peptide does not increase with increasing antibody concentrations. At pH 7.3 the antibody remains blocked in the absence of the trigger oligonucleotide, while complete restoration of antibody activity is observed in the presence of 100 nM trigger oligonucleotide (Figure 1D). To analyze the pH-dependency of oligonucleotide-triggered antibody activation in more detail, we monitored the fraction of activated antibody following rapid dilution of antibody-pHAbs-1 complex preassembled at pH 5.0 in buffers ranging in pH from 5.0 to 7.3. Figure 1E shows a strong increase in the fraction of activated antibody at pH > 6.5, reaching full equilibrium within 15 min. A small increase is also observed at low pH, which may be due to competition between the bivalent ligand and the fluorescent peptide. Figure 1F shows that the amount of activated antibody rises sharply between pH 6.0 and 6.5, which is in accordance with the apparent pKa that was reported previously for a triple helix with the same TAT/CGC content as the one used in our lock.37 The sharp transition between blocked and activated antibody indicates a high degree of cooperativity, which probably originates from the pH-dependent formation of a DNA triple helix involving the simultaneous protonation of multiple pyrimidine bases.
The second ligand design, termed pHAbs-2, consists of an intermolecular DNA triple helix formed by the interaction between a hairpin-forming oligonucleotide–peptide conjugate (HPO) and a triplex-forming oligonucleotide–peptide conjugate (TFO, Figure 1G). At low pH the bivalent ligand assembles through the formation of an intermolecular, 20 nucleotide long triple helix and tightly binds to the antibody. Due to the pH-dependency of the triple helix, the bivalent ligand unfolds at neutral/basic pH and separates into two monovalent peptide–DNA ligands that readily dissociate to restore antibody activity. As the integrity of the bivalent ligand is governed by a DNA triple helix only, antibody activation is solely controlled by pH, without the requirement for an external trigger. Figure 1H shows that in the presence of pHAbs-2 the competing peptide does not bind to the antibody at pH 5.0, while the antibody is fully activated at pH 7.3, showing a binding curve that is identical to that of the monovalent control (Figure 1I). To analyze the kinetics of antibody activation by pHAbs-2 as a function of pH, the anti-HA antibody blocked by pHAbs-2 was rapidly diluted in buffer with a pH ranging from 5.0 to 7.3 (Figure 1J,K). Similar to pHAbs-1, robust inhibition of antibody activity is observed at low pH, while fast antibody activation is observed over a narrow pH range between pH 6.5 and 7.0.
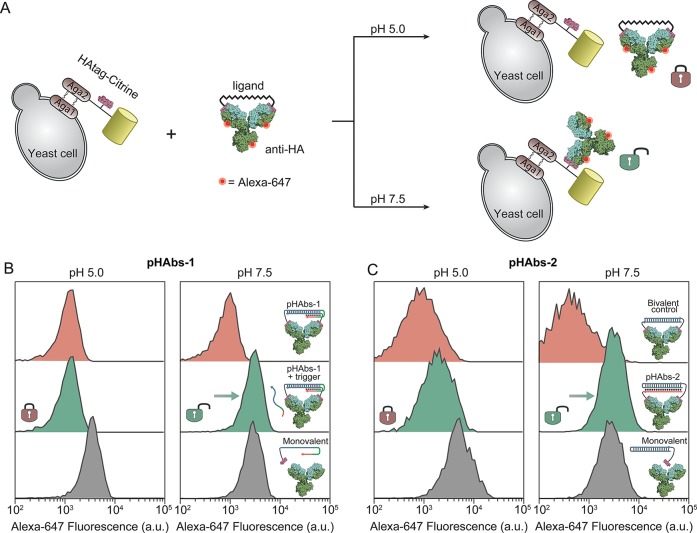
Having confirmed efficient antibody blocking by pHAbs-1 and pHAbs-2 at acidic pH and restoration of antibody activity at neutral/basic pH, we next explored whether these noncovalent antibody locks can be used for pH-dependent antibody targeting to cell-surface markers. As a model target, yeast cells were cultured that display the HA peptide epitope genetically fused to a yellow fluorescent protein (Citrine) on their surface (Figure 2A). Figure 2B shows flow cytometry analysis of yeast cells incubated at pH 5.0 and pH 7.5 with an Alexa647-labeled anti-HA antibody blocked by pHAbs-1. In the presence of pHAbs-1, low cell labeling is observed at both pHs. However, a 5-fold higher labeling efficiency is observed upon addition of the trigger oligonucleotide at pH 7.5, while cell labeling remains blocked in the absence of trigger. The labeling efficiency at pH 7.5 is identical to the labeling efficiency of a monovalent control, confirming that the toehold domain in pHAbs-1 is robustly sequestered in the triple helix conformation at low pH but is fully activated at a higher pH. Figure 2C shows similar cell targeting experiments with the anti-HA antibody blocked by pHAbs-2. When the antibody is incubated with a bivalent control that does not harbor a triple helix domain, low cell targeting is observed at both the tested pHs. When incubated with pHAbs-2 minimal targeting of the antibody is observed at pH 5.0, while increasing the pH results in binding of the antibody to the yeast cells with similar efficiency as the monovalent control. Taken together, these results confirm the feasibility of rationally implementing triple helix domains in the bivalent peptide–DNA locks to introduce pH-dependent control over antibody-based cell targeting.
Figure 2.
pH-dependent targeting of the anti-HA antibody to yeast cells. (A) Schematic representation of a yeast cell displaying the HA peptide epitope as a fusion protein with Aga2 and the yellow fluorescent protein Citrine. Governed by an increase in pH the bivalent ligands activate the Alexa-647-labeled anti-HA antibody, resulting in targeting of the antibody to the HA peptide displaying yeast cells. (B) Flow cytometry analysis of yeast cells after incubating for 1 h at room temperature with 2 nM anti-HA antibody preincubated with pHAbs-1 and control ligands at pH 5.0 (left) and pH 7.5 (right). (C) Similar as in part B, but for the anti-HA antibody preincubated with pHAbs-2 and appropriate control ligands at pH 5.0 (left) and pH 7.5 (right). Histograms were constructed from the Alexa-647 fluorescence intensity of 3000 Citrine-positive yeast cells.
Antibody Activation at Low pH
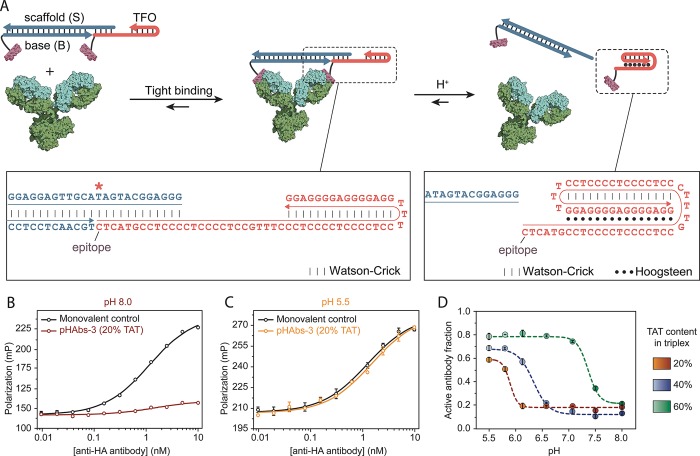
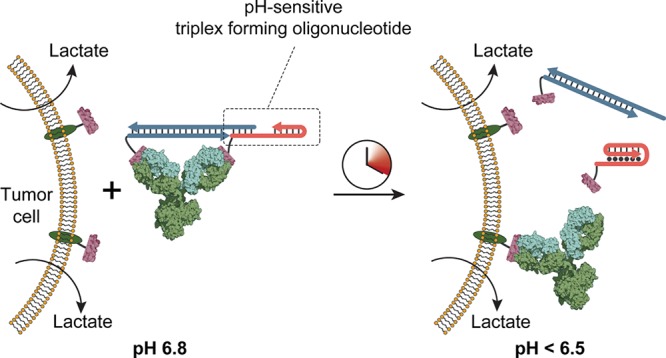
Since tumor cells exhibit a distinct metabolic activity that results in acidification of the extracellular environment, antibody-based cancer diagnostics and treatment might greatly benefit from conditional activation of antibodies at low pH. Therefore, a third antibody-binding ligand, termed pHAbs-3, was designed to form a stable antibody lock at neutral/basic pH and dissociate at low pH (pH 6.0–6.5). Since this implies that the formation of a triple helix disrupts the bivalent character of the ligand, a more complex ligand design is required. The pHAbs-3 design is based on the mutually exclusive formation of a duplex domain that governs the assembly of the bivalent ligand and an intramolecular triple helix domain that invades this duplex (Figure 3A).43 One peptide epitope is conjugated to a base strand (B), which forms a DNA duplex with the scaffold strand (S) that harbors a 12-nucleotide overhang on its 5′ end. To this overhang the 5′-terminal domain of a triplex-forming DNA–peptide conjugate (TFO) hybridizes to assemble the bivalent ligand. At low pH the TFO forms an intramolecular triple helix (TAT content of 20%) that partially invades the duplex between the scaffold and TFO strands. Based on previous observations, the 6 base pairs that remain after triple helix formation are expected to be insufficient to retain the bivalent character of the ligand, inducing its dissociation into two monovalent peptide–DNA ligands.32 The pH-induced activation of the antibody relies on a subtle balance between the stability of the S:TFO duplex and triple helix formation. Therefore, the competition assay with the fluorescently labeled HA peptide epitope was used to screen for optimal ligand design. An initial design with fully complementary S:TFO hybridization efficiently blocked the antibody at pH 8.0 but showed only moderate activation at low pH (Figure S3). To optimize antibody activation at low pH, the stability of the S:TFO duplex was systematically attenuated by introducing mismatches in the scaffold strand, while keeping the stability of the triple helix domain unaltered. Ligands with one or two base pair mismatches performed optimally, showing efficient inhibition of the antibody at pH 8.0, while ensuring antibody activation at pH 5.5. Introducing four mismatches destabilized the DNA duplex too much, resulting in inefficient blocking of the antibody at pH 8.0. In all subsequent experiments a ligand with a single base pair mismatch was used (Figure 3B,C and Figure S3, mismatch indicated by asterisk).
Figure 3.
Antibody activation at low pH. (A) Design of bivalent antibody ligand pHAbs-3, based on a mutually exclusive DNA duplex and triple helix formation. The epitope-functionalized base strand (B) hybridizes to a scaffold strand (S) that harbors a 12 nucleotide overhang. To this overhang a peptide-functionalized, triplex-forming oligonucleotide (TFO) hybridizes to assemble the bivalent ligand at neutral pH. (B, C) Competition assay between a fluorescently labeled HA peptide and pHAbs-3, with S harboring a single mismatch (indicated by asterisk) and TFO with a 20% TAT content at pH 8.0 and 5.5, respectively. A serial dilution of anti-HA antibody preincubated with 2 equiv of ligand was diluted 10-fold in buffer containing 2 nM of FITC-labeled HA peptide epitope. A monovalent control is included as reference for fully activated antibody. (D) Fraction of activated antibody as a function of pH when blocked by pHAbs-3 with a TAT content of 20%, 40%, and 60% after 90 min of incubation. Fitting of eq 2 to the experimental data yielded an apparent pKa of 5.9, 6.3, and 7.3 for the 20%, 40%, and 60% TAT ligands, respectively. Error bars represent standard error of the mean of duplicate measurements.
Next, the pH-dependency of pHAbs-3 with a 20% TAT triple helix domain was evaluated by measuring the fraction of activated antibody as a function of pH. Figure 3D shows that at pH > 6.0 the antibody is efficiently blocked, while decreasing the pH below 6.0 results in disassembly of the bivalent ligand and concurrent activation of the antibody. Previous work has shown that the pKa of triple helix formation is encoded in the relative TAT content of the primary nucleotide sequence. To explore whether we can use this property to tune the pH at which the bivalent ligand disassembles and the antibody is activated, we synthesized two additional ligands with triple helices harboring a relative TAT content of 40% and 60%. Notably, all the ligands contain the single mismatch that was optimized for 20% TAT pHAbs-3. The 40% and 60% TAT ligands efficiently blocked the antibody at high pH, but as expected antibody activation was shifted to higher pH. Fitting of Equation S2 to the experimental data yielded an apparent pKa,app of 5.9, 6.3, and 7.3 for pHAbs-3 with a TAT content of 20%, 40%, and 60% respectively. These results show that the pH response of the lock can be accurately controlled by sequence design over at least 1.5 pH units. Interestingly, ligands with increasing TAT content are more efficiently activated at pH 5.5, which is most likely due to the attenuated stability of the S:TFO duplex as a result of increasing AT content.
pH-Dependent Targeting of Human Carcinoma Cells
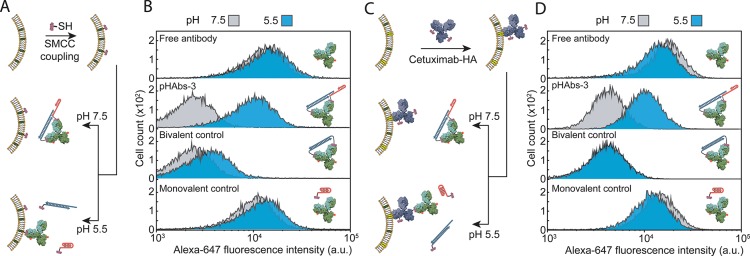
Having established a robust strategy to activate a monoclonal antibody by a decrease in pH, we next investigated whether the pHAbs-3 ligand can be used to specifically target antibodies to mammalian cancer cells solely based on the acidity of the cellular environment. As a first proof-of-principle, A431 epidermoid carcinoma cells were covalently labeled with HA peptide epitopes. Exposed primary amines on the cell surface were functionalized with maleimide moieties by reacting with the bifunctional Sulfo-SMCC cross-linker. Next, the maleimide-activated cell surface was reacted with HA peptides modified with a C-terminal cysteine, yielding A431 cells covalently decorated with HA peptide epitopes (Figure 4A). Figure 4B shows flow cytometry analysis of cells that were incubated with free anti-HA antibody at either pH 5.5 or pH 7.5, revealing efficient, pH-independent cell labeling. When the anti-HA antibody was preincubated with the pHAbs-3 ligand it was efficiently inhibited at pH 7.5, showing similar cell labeling as an antibody incubated with a permanently closed bivalent peptide–DNA ligand. In contrast, a significant increase in cell labeling was observed at pH 5.5, with antibody labeling similar to a control in which the anti-HA antibody was incubated with the monovalent TFO conjugate. These results show that pHAbs-3 allows efficient, pH-induced control of antibody-mediated cell targeting.
Figure 4.
pH-dependent targeting of antibodies to human carcinoma cells. (A) Schematic representation of direct low pH triggered targeting of mammalian cells. First, A431 epidermoid carcinoma cells are covalently prelabeled with HA peptide epitopes via a Sulfo-SMCC cross-linker. The prelabeled cells are subsequently resuspended in PBS at pH 7.5 or 5.5 containing 2 nM anti-HA antibody blocked by the pHAbs-3 ligand (20% TAT). (B) Flow cytometry analysis showing Alexa-647 fluorescence intensities for A431 cells incubated for 30 min at pH 7.5 or 5.5 with 2 nM anti-HA antibody in absence of ligand (free antibody), in the presence of the 2 equiv of pHAbs-3 ligand (20% TAT), 2 equiv of a pH-insensitive bivalent ligand (bivalent control), or 4 equiv of an incomplete ligand (monovalent control). (C) Schematic representation of a dual specific strategy for the pH-controlled targeting of the overexpressed EGFR on A431 cells. First, the EGFR overexpressing A431 cells were incubated with 2 nM cetuximab-HA to specifically label EGFR overexpressing cells with the HA peptide epitope. Next, the cells were resuspended in PBS at pH 7.5 or 5.5 containing 2 nM anti-HA antibody blocked by the 20% TAT pHAbs-3 ligand. (D) Flow cytometry analysis showing Alexa-647 fluorescence intensities for A431 cells incubated for 30 min at pH 7.5 or 5.5 with 2 nM anti-HA antibody in absence of ligand (free antibody), in the presence of the 2 equiv of 20% TAT pHAbs-3 ligand, 2 equiv of a pH-insensitive bivalent ligand (bivalent control), or 4 equiv of an incomplete ligand (monovalent control). Histograms were reconstructed from the Alexa-647 fluorescence intensity of 10 000 individual A431 cells.
Covalently decorating cells with the HA peptide provides a generally applicable strategy to allow antibody targeting that does not rely on expression of specific cell-surface markers, but solely on the pH of the extracellular environment. We also explored a second approach that relies on HA peptide labeling of cells mediated by a primary antibody. This approach represents dual specific targeting, as it requires the expression of a specific cell-surface marker in combination with a pH decrease of the extracellular environment. To this end, an anti-EGFR antibody (Cetuximab) was covalently labeled with HA peptide epitopes. After incubating EGFR overexpressing A431 cells for 20 min with HA-labeled Cetuximab, the cells were washed and resuspended in PBS at pH 7.5 or pH 5.5 containing 2 nM of the Alexa647-labeled anti-HA antibody and analyzed by flow cytometry (Figure 4C,D). When incubated with free anti-HA antibody, the A431 cells show strong fluorescence intensity at both high and low pH, which confirms successful prelabeling with HA peptide epitopes mediated by the primary antibody. When the cells were incubated with the anti-HA antibody blocked by the pHAbs-3 ligand (20% TAT), low cell labeling was observed at pH 7.5, while efficient targeting was observed at pH 5.5. Again, no labeling was observed when the anti-HA antibody was blocked with the permanently closed bivalent peptide–DNA ligand at both pHs, whereas incubation of the anti-HA antibody with a monovalent control showed efficient cell labeling at both pH 5.5 and 7.5. Together these results show that the developed pHAbs-3 ligand efficiently blocks the activity of an antibody at high pH, while robustly activating the antibody at low pH. This strategy thus represents an attractive approach to increase the specificity of tumor targeting by allowing binding to EGFR-overexpressing cells (or another cell-surface marker) only by the acidic environment of tumor tissue.
Targeting Tumor Cells Based on Single-Cell Metabolic Activity
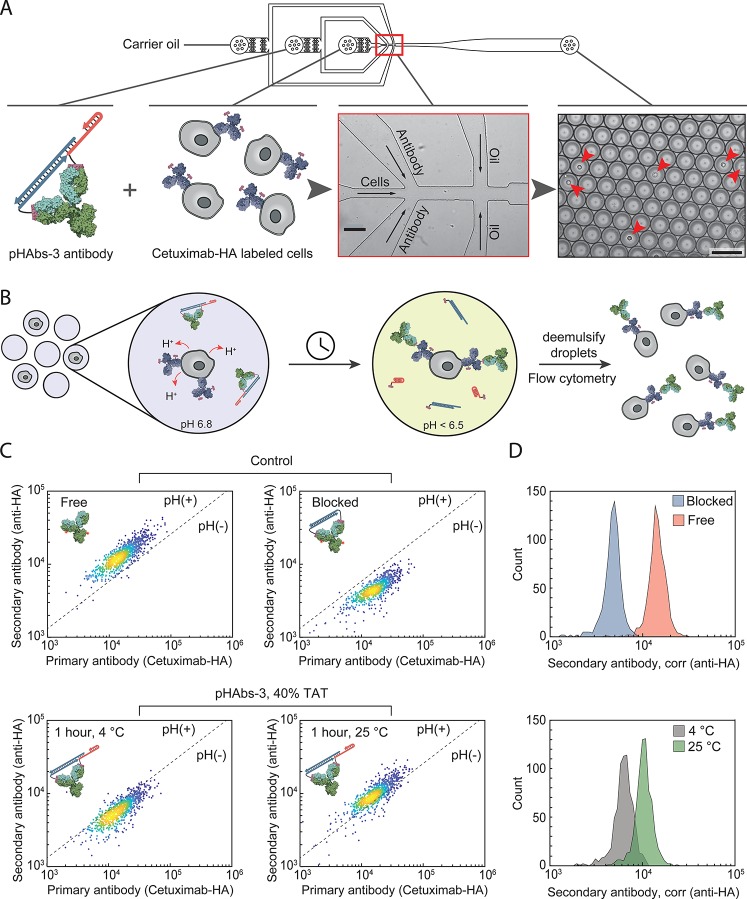
Having established the feasibility of using pHAbs-3 to control antibody targeting of mammalian cells triggered by a decrease in pH of the extracellular environment, we next set out to explore the application of this pH-dependent labeling strategy for the evaluation of the metabolic activity of single mammalian cells. Del Ben et al. recently showed that, by confining individual cells in picoliter-sized water-in-oil droplets, the lactate produced by malignant cells accumulates inside the constrained volume of the droplet. The resulting pH decrease of the droplet could subsequently be detected by a pH-sensitive fluorescent dye present in the water phase.21 Consequently, circulating tumor cells could be identified in the blood of metastatic patients solely based on their metabolic activity, while not depending on the heterogeneous presence of specific cell-surface markers. Using pH-induced antibody activation instead of a pH-sensitive dye will allow the encapsulated tumor cells to be labeled, making it no longer necessary to interrogate all the droplets. Instead, the droplets can be de-emulsified to retrieve all encapsulated cells after labeling, and the cells can be analyzed and sorted in high throughput using standardized flow cytometry equipment. Figure 5A,B shows the microfluidic chip design and experimental workflow for coencapsulating mammalian cells with the anti-HA antibody in complex with the pHAbs-3 ligand. The microfluidic chip contains three inlets: one inlet for the carrier oil, a second inlet for the pHAbs-3 ligand–antibody complex, and a third inlet for cells prelabeled with HA-labeled cetuximab. Upon injection in the microfluidic chip, the prelabeled cells are first mixed with the anti-HA antibody blocked by the pHAbs-3 ligand. Next, monodisperse picoliter-sized water-in-oil droplets are generated via flow-focusing at the junction where the water and oil channels merge. As proof-of-concept, A431 cells were prelabeled with 2 nM HA-labeled cetuximab and 2 nM Cy3-labeled cetuximab. The cells were injected in the microfluidic device and mixed on chip with the Alexa647-labeled anti-HA antibody blocked by the pHAbs-3 ligand. Importantly, the concentration of injected cells was chosen such that cells are individually encapsulated (ergo, most droplets are empty). To allow fast activation of the antibody by acidification of cell-containing droplets, experiments were performed with pHAbs-3 harboring a 40% TAT ligand, which was shown to be activated at pH < 6.5. The generated droplets were collected in a tube that was prechilled on ice to slow down cell metabolism. After all droplets were produced and collected, half of the droplets were incubated for 1 h at room temperature to restore cell metabolism, while the other half was kept at 4 °C. Finally, the droplets were de-emulsified to retrieve the cells after which they were analyzed by flow cytometry. The buffer contained 2 g L–1 glucose, which was found necessary to ensure full metabolic activity of the cells. In addition, to avoid degradation of the DNA linker by secreted nucleases, the buffer was supplemented with 0.1 mg mL–1 dsDNA from salmon testes to scavenge nuclease activity. Figure 5C shows flow cytometry analysis of primary antibody labeling (as detected by cetuximab-Cy3) versus secondary antibody binding (anti-HA antibody) of A431 cells after single-cell encapsulation. Similar to the ensemble experiments, a 5-fold decrease in secondary antibody labeling was observed when the anti-HA antibody was blocked by the nonactivatable bivalent ligand, showing effective antibody inhibition under these conditions and efficient scavenging of excreted nucleases by the supplemented dsDNA. When human carcinoma cells were encapsulated together with the anti-HA antibody blocked by the pHAbs-3 ligand, only a minor increase in antibody binding was observed compared to the nonactivatable control when the droplets were immediately stored at 4 °C. However, when the same droplets were incubated for 1 h at room temperature to stimulate metabolic activity, pH-dependent anti-HA labeling of the entire population was observed. To ensure that the activation of the antibody is not due to degradation of the bivalent ligand or temperature-dependent endocytosis, a set of control experiments was performed in a pH buffer with high buffer capacity. In this case no increase in anti-HA labeling was observed for cells that were incubated at 25 °C (Figure S4). A strong correlation was observed between secondary and primary antibody labeling, since more efficient primary antibody labeling will result in a higher quantity of HA peptides displayed on the cell to which the secondary antibody can bind. Figure 5D shows histograms of (pH-responsive) secondary antibody labeling corrected for the primary antibody labeling heterogeneity, revealing a distinct shift to higher pH-responsive labeling when the cells are incubated at 25 °C compared to 4 °C. Interestingly, the heterogeneity in pH-responsive secondary antibody labeling is smaller than the heterogeneity in primary antibody labeling, as the activation of the pH-responsive ligand by itself is independent of heterogeneous cell-surface marker expression. Together these results show that single tumor cells can sufficiently acidify the lumen of picoliter-sized droplets to disassemble the pHAbs-3 ligand, making pH-induced antibody activation an attractive orthogonal marker for the high-throughput evaluation of single-cell metabolic activity.
Figure 5.
Antibody labeling of carcinoma cells based on single-cell metabolic activity. (A) Design of the microfluidic droplet device containing inlets for the oil with surfactant, the pH-responsive antibody–ligand complex, and the prelabeled cells. Cells were mixed with the antibody–ligand complex after which monodisperse water-in-oil droplets are generated. Scale bars represent 100 μm. (B) After tumor cells are encapsulated in the picoliter-sized droplets their distinct metabolic activity will result in accumulation of lactate and a decrease in pH of the droplets containing tumor cells. This pH decrease results in activation of the antibody, which can be measured in high throughput by demulsifying the droplets and analyzing the cells by flow cytometry. (C) A431 human epidermoid carcinoma cells were encapsulated in droplets together with either a pH+ control (free antibody), a pH– control (permanently blocked antibody), or the anti-HA antibody blocked with the 40% TAT pHAbs-3 ligand for 1 h at either 4 °C to inhibit cell metabolism or 25 °C to promote cell metabolism. After 1 h of incubation the droplets were demulsified, and the degree of (pH-responsive) secondary antibody labeling versus primary antibody labeling via Cetuximab-HA of the retrieved cells was analyzed by flow cytometry. (D) Histograms showing the degree of pH-responsive secondary antibody derived from the scatter plots shown in (C). Scatter plots and histograms were constructed from ∼1000 individual cells per experiment.
Discussion
In this work pH-responsive peptide–DNA ligands were developed that act as robust molecular locks to noncovalently control the activity of monoclonal antibodies based on changes in pH. The general design of the antibody-blocking ligands exploits a bivalent interaction between the antigen binding sites of the antibody and two identical peptide epitopes that are linked via a DNA-based spacer. By integrating pH-responsive triple helix structures in the bivalent ligand, three types of pH-sensitive molecular locks were developed that allow switching from bivalent to monovalent interactions in response to pH changes. Two ligand designs were introduced that enable efficient antibody activation upon a pH increase, either autonomously or in combination with a specific trigger oligonucleotide. The third ligand design efficiently blocked antibody activity at neutral pH and allowed antibody activation at lower pH. The pH response of this ligand could be encoded in the sequence of the triple helix, allowing the pH at which the antibody is activated to be tuned between 5.5 and 7.5. Using these ligands, pH-dependent antibody-mediated tumor cell targeting was successfully demonstrated, both in ensemble experiments and at the single-cell level.
Expanding on the approach developed by Del Ben and co-workers, we employed a pH-activatable anti-HA antibody to detect the acidification of tumor cells in water-in-oil microfluidic droplets. An important advantage of this approach compared to detecting the pH inside the droplets is that the cells themselves are labeled, making it no longer necessary to interrogate all the droplets. Instead the droplets can be de-emulsified to retrieve all encapsulated cells after labeling, and the cells can be analyzed and sorted in high throughput using standard flow cytometry equipment. In principle, pH-dependent antibody targeting could be combined with a panel of other antibody labels that target specific cell-surface markers, introducing metabolic activity as an orthogonal label for tumor cell detection, which could be particularly useful for detecting and isolating circulating tumor cells (CTCs). While we have demonstrated the ability of pH-activatable antibodies to label a tumor cell based on the Warburg effect, the efficiency of the system to distinguish tumor cells among a large background of healthy cells remains to be established, and the stability of the current system against nucleases could be further improved. If successful, the method should be further validated by comparing it with established methods for CTC detection, such as the FDA approved CellSearch technology. Since the latter relies on targeting of the epithelial cell adhesion molecule (EpCAM), selection based on acidification may be able to detect a broader set of CTCs, including those that have undergone mesenchymal-to-epithelial transition and are most likely responsible for initiation of metastasis.44 Finally, the ability to efficiently retrieve (potential) tumor cells using cell sorting is essential to provide a much more detailed insight into tumor heterogeneity and tumor progression, e.g., by allowing single-cell sequencing.
In contrast to protein engineering approaches that target a specific antibody–antigen interaction, the pH-switchable bivalent peptide–DNA ligands introduced here represent a generic mechanism to install pH sensitivity that can be applied to in principle any monoclonal antibody, without the requirement for genetic or chemical modification. Since antibody activation is based on the difference in affinity between bivalent and monovalent binding, the only requirement is the availability of epitope or mimitope peptides of sufficient affinity (Kd ≤ 10 nM).31 The ability to bind and release antibodies by subtle changes in pH has many potential applications, e.g., as a biotechnological strategy to purify antibodies under mild conditions or to trigger protein activation following endocytosis.45,46 Activating a monoclonal antibody upon a decrease in pH is particularly useful in cancer associated diagnostics and therapeutics, as demonstrated here by high-throughput evaluation of single tumor cell metabolism. The ability to activate therapeutic antibodies at low pH might also provide attractive opportunities to increase the specificity of immunotherapy, allowing antibody activation only at the tumor site and reducing background targeting of healthy cells.
Acknowledgments
We thank M. Rosmalen and S. Wouters for support with the flow cytometry equipment and titration experiments and N. Sinha for support with the microfluidic setup. We are grateful to F. del Ben for fruitful discussions regarding metabolic profiling of tumor cells in microdroplets. This work was supported by funding from by European Research Council (ERC) Starting Grant (280255) and the Ministry of Education, Culture and Science (Gravity program, 024.001.035).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b00964.
Experimental procedures, safety statement, HA epitope–antibody dissociation constant at various pHs, HPLC traces of purified oligonucleotide–peptide conjugates, mismatch screening of pHAbs-3, antibody activation in microdroplets at high buffer capacity, flow cytometry gating strategy for doublet discrimination, oligonucleotide sequences, and MS analysis of oligonucleotide–peptide conjugates (PDF)
Author Contributions
W.E., J.T., and M.M. designed the experiments. A.I. and F.R. designed the pH-dependent sequences. W.E., K.Z., and N.S. performed experiments and analyzed data. W.E. and M.M. wrote the manuscript. All authors discussed the data and commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Weiner L. M.; Dhodapkar M. V.; Ferrone S. Monoclonal antibodies for cancer immunotherapy. Lancet 2009, 373, 1033–1040. 10.1016/S0140-6736(09)60251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrebaeck C. A. K. Antibodies in diagnostics – from immunoassays to protein chips. Immunol. Today 2000, 21, 379–382. 10.1016/S0167-5699(00)01683-2. [DOI] [PubMed] [Google Scholar]
- Mestel R. Cancer: imaging with antibodies. Nature 2017, 543, 743. 10.1038/543743a. [DOI] [PubMed] [Google Scholar]
- Warram J. M.; et al. Antibody-based imaging strategies for cancer. Cancer Metastasis Rev. 2014, 33, 809–822. 10.1007/s10555-014-9505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan D. J.; O’Connor D. P.; Rexhepaj E.; Ponten F.; Gallagher W. M. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nat. Rev. Cancer 2010, 10, 605. 10.1038/nrc2902. [DOI] [PubMed] [Google Scholar]
- Busam K. j.; et al. Cutaneous side-effects in cancer patients treated with the antiepidermal growth factor receptor antibody C225. Br. J. Dermatol. 2001, 144, 1169–1176. 10.1046/j.1365-2133.2001.04226.x. [DOI] [PubMed] [Google Scholar]
- Kontermann R. E.; Brinkmann U. Bispecific antibodies. Drug Discovery Today 2015, 20, 838–847. 10.1016/j.drudis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Douglas S. M.; Bachelet I.; Church G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Amir Y.; et al. Universal computing by DNA origami robots in a living animal. Nat. Nanotechnol. 2014, 9, 353–357. 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. M.; Kari C.; Fragoso R. C.; Rodeck U.; Williams J. C. Design and development of masked therapeutic antibodies to limit off-target effects: Application to anti-EGFR antibodies. Cancer Biol. Ther. 2009, 8, 2147–2152. 10.4161/cbt.8.22.9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster O.; et al. Site-specific targeting of antibody activity in vivo mediated by disease-associated proteases. J. Controlled Release 2012, 161, 804–812. 10.1016/j.jconrel.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers L. R.; et al. Tumor-specific activation of an EGFR-targeting probody enhances therapeutic index. Sci. Transl. Med. 2013, 5, 207ra144–207ra144. 10.1126/scitranslmed.3006682. [DOI] [PubMed] [Google Scholar]
- Webb B. A.; Chimenti M.; Jacobson M. P.; Barber D. L. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 2011, 11, 671. 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- Warburg O.; Wind F.; Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande rHeiden M. G.; Cantley L. C.; Thompson C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haji-Michael P. G.; Ladrière L.; Sener A.; Vincent J.-L.; Malaisse W. J. Leukocyte glycolysis and lactate output in animal sepsis and ex vivo human blood. Metab., Clin. Exp. 1999, 48, 779–785. 10.1016/S0026-0495(99)90179-8. [DOI] [PubMed] [Google Scholar]
- Gillies R. J.; Raghunand N.; Garcia-Martin M. L.; Gatenby R. A. pH imaging. A review of pH measurement methods and applications in cancers. IEEE Eng. Med. Biol. Mag. Q. Mag. Eng. Med. Biol. Soc. 2004, 23, 57–64. 10.1109/MEMB.2004.1360409. [DOI] [PubMed] [Google Scholar]
- Tian Y.; et al. pH-dependent transmembrane activity of peptide-functionalized gold nanostars for computed tomography/photoacoustic imaging and photothermal therapy. ACS Appl. Mater. Interfaces 2017, 9, 2114–2122. 10.1021/acsami.6b13237. [DOI] [PubMed] [Google Scholar]
- Lee E. S.; Na K.; Bae Y. H. Polymeric micelle for tumor pH and folate-mediated targeting. J. Controlled Release 2003, 91, 103–113. 10.1016/S0168-3659(03)00239-6. [DOI] [PubMed] [Google Scholar]
- Urano Y.; et al. Selective molecular imaging of viable cancer cells with pH-activatable fluorescence probes. Nat. Med. 2009, 15, 104–109. 10.1038/nm.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Ben F.; et al. A method for detecting circulating tumor cells based on the measurement of single-cell metabolism in droplet-based microfluidics. Angew. Chem., Int. Ed. 2016, 55, 8581–8584. 10.1002/anie.201602328. [DOI] [PubMed] [Google Scholar]
- Porchetta A.; Idili A.; Vallée-Bélisle A.; Ricci F. General strategy to introduce pH-induced allostery in DNA-based ceceptors to achieve controlled release of ligands. Nano Lett. 2015, 15, 4467–4471. 10.1021/acs.nanolett.5b00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell E. M.; et al. pHAST (pH-driven aptamer switch for thrombin) Catch-and-release of target protein. Bioconjugate Chem. 2016, 27, 1493–1499. 10.1021/acs.bioconjchem.6b00124. [DOI] [PubMed] [Google Scholar]
- Del Grosso E.; Idili A.; Porchetta A.; Ricci F. A modular clamp-like mechanism to regulate the activity of nucleic-acid target-responsive nanoswitches with external activators. Nanoscale 2016, 8, 18057–18061. 10.1039/C6NR06026A. [DOI] [PubMed] [Google Scholar]
- Li L.; et al. Modulating aptamer specificity with pH-responsive DNA bonds. J. Am. Chem. Soc. 2018, 140, 13335–13339. 10.1021/jacs.8b08047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. K. L.; Eisenstein M.; Soh H. T. Direct selection strategy for isolating aptamers with pH-sensitive binding activity. ACS Sens. 2018, 3, 2574–2580. 10.1021/acssensors.8b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Liu X.; Willner I. DNA switches: from principles to applications. Angew. Chem., Int. Ed. 2015, 54, 1098–1129. 10.1002/anie.201404652. [DOI] [PubMed] [Google Scholar]
- Schröter C.; et al. A generic approach to engineer antibody pH-switches using combinatorial histidine scanning libraries and yeast display. mAbs 2015, 7, 138–151. 10.4161/19420862.2014.985993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter C.et al. Isolation of pH-sensitive antibody fragments by fluorescence activated cell sorting and yeast surface display. In Protein Eng.; Humana Press: New York, NY, 2018; pp 311–331. [DOI] [PubMed] [Google Scholar]
- Strauch E.-M.; Fleishman S. J.; Baker D. Computational design of a pH-sensitive IgG binding protein. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 675–680. 10.1073/pnas.1313605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B. M. G.; et al. Reversible blocking of antibodies using bivalent peptide–DNA conjugates allows protease-activatable targeting. Chem. Sci. 2013, 4, 1442–1450. 10.1039/c3sc22033h. [DOI] [Google Scholar]
- Janssen B. M. G.; van Rosmalen M.; van Beek L.; Merkx M. Antibody activation using DNA-based logic gates. Angew. Chem., Int. Ed. 2015, 54, 2530–2533. 10.1002/anie.201410779. [DOI] [PubMed] [Google Scholar]
- Wouters S. F. A.; Wijker E.; Merkx M. Optical control of antibody activity by using photocleavable bivalent peptide–DNA locks. ChemBioChem 2019, 20, 2463–2466. 10.1002/cbic.201900241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucale M.; Zuccheri G.; Samorì B. The dynamic properties of an intramolecular transition from DNA duplex to cytosine – thymine motif triplex. Org. Biomol. Chem. 2005, 3, 575–577. 10.1039/B418353N. [DOI] [PubMed] [Google Scholar]
- Völker J.; Botes D. P.; Lindsey G. G.; Klump H. H. Energetics of a stable intramolecular DNA triple helix formation. J. Mol. Biol. 1993, 230, 1278–1290. 10.1006/jmbi.1993.1241. [DOI] [PubMed] [Google Scholar]
- Leitner D.; Schröder W.; Weisz K. Influence of sequence-dependent cytosine protonation and methylation on DNA triplex stability. Biochemistry 2000, 39, 5886–5892. 10.1021/bi992630n. [DOI] [PubMed] [Google Scholar]
- Idili A.; Vallée-Bélisle A.; Ricci F. Programmable pH-triggered DNA nanoswitches. J. Am. Chem. Soc. 2014, 136, 5836–5839. 10.1021/ja500619w. [DOI] [PubMed] [Google Scholar]
- Amodio A.; et al. Rational design of pH-controlled DNA strand displacement. J. Am. Chem. Soc. 2014, 136, 16469–16472. 10.1021/ja508213d. [DOI] [PubMed] [Google Scholar]
- Idili A.; Porchetta A.; Amodio A.; Vallée-Bélisle A.; Ricci F. Controlling hybridization chain reactions with pH. Nano Lett. 2015, 15, 5539–5544. 10.1021/acs.nanolett.5b02123. [DOI] [PubMed] [Google Scholar]
- Amodio A.; Adedeji A. F.; Castronovo M.; Franco E.; Ricci F. pH-Controlled assembly of DNA tiles. J. Am. Chem. Soc. 2016, 138, 12735–12738. 10.1021/jacs.6b07676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A.; Urban M. J.; Idili A.; Ricci F.; Liu N. Selective control of reconfigurable chiral plasmonic metamolecules. Sci. Adv. 2017, 3, e1602803. 10.1126/sciadv.1602803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen W.; Meijer L. H. H.; Somers B.; Greef T. F. A. de; Merkx M. Antibody-controlled actuation of DNA-based molecular circuits. Nat. Commun. 2017, 8, 14473. 10.1038/ncomms14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; et al. Triplex DNA nanoswitch for pH-sensitive release of multiple cancer drugs. ACS Nano 2019, 13, 7333–7344. 10.1021/acsnano.9b03846. [DOI] [PubMed] [Google Scholar]
- Mani S. A.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low D.; O’Leary R.; Pujar N. S. Future of antibody purification. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2007, 848, 48–63. 10.1016/j.jchromb.2006.10.033. [DOI] [PubMed] [Google Scholar]
- Boyken S. E.; et al. De novo design of tunable, pH-driven conformational changes. Science 2019, 364, 658–664. 10.1126/science.aav7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.