Abstract
Carcass quality traits such as back fat (BF), loin depth (LD), and ADG are of extreme economic importance for the swine industry. This study aimed to (i) estimate the genetic parameters for such traits and (ii) conduct a single-step genome-wide association study (ssGWAS) to identify genomic regions that affect carcass quality and growth traits in purebred (PB) and three-way crossbred (CB) pigs. A total of 28,497 PBs and 135,768 CBs pigs were phenotyped for BF, LD, and ADG. Of these, 4,857 and 3,532 were genotyped using the Illumina PorcineSNP60K Beadchip. After quality control, 36,328 SNPs were available and were used to perform an ssGWAS. A bootstrap analysis (n = 1,000) and a signal enrichment analysis were performed to declare SNP significance. Genome regions were based on the variance explained by significant 10-SNP sliding windows. Estimates of PB heritability (SE) were 0.42 (0.019) for BF, 0.39 (0.020) for LD, and 0.35 (0.021) for ADG. Estimates of CB heritability were 0.49 (0.042) for BF, 0.27 (0.029) for LD, and 0.12 (0.021) for ADG. Genetic correlations (SE) across the two populations were 0.81 (0.02), 0.79 (0.04), and 0.56 (0.05), for BF, LD, and ADG, respectively. The variance explained by significant regions for each trait in PBs ranged from 1.51% to 1.35% for BF, from 4.02% to 3.18% for LD, and from 2.26% to 1.45% for ADG. In CBs, the variance explained by significant regions ranged from 1.88% to 1.37% for BF, from 1.29% to 1.23% for LD, and from 1.54% to 1.32% for ADG. In this study, we have described regions of the genome that determine carcass quality and growth traits of PB and CB pigs. These results provide evidence that there are overlapping and nonoverlapping regions in the genome influencing carcass quality and growth traits in PBs and three-way CB pigs.
Keywords: breeding, genetics, heritability, swine
Introduction
Carcass quality traits are important breeding goals in swine because of their high economic value. In many breeding schemes, animals from the purebred (PB) parental lines are reared and selected in nucleus herds, with the objective to improve crossbred (CB) performance (Wientjes and Calus, 2017). However, several authors have reported genetic correlations between PB and CB carcass quality traits lower than one (Wientjes and Calus, 2017; Godinho et al., 2018). Therefore, using the performance of PB individuals is less efficient in improving the performance of their CB commercial progeny due to genotype by environment interactions and nonadditive genetic effects (Zumbach et al., 2007; Lopes et al., 2017). Different methods have been proposed as an alternative to overcome those limitations such as combined PB and CB selection (Bijma and van Arendonk, 1998) and genomic selection (Guo et al., 2017; Chen et al., 2019). After the development of high-density SNPs, some studies have reported that genomic selection can be applied to various livestock species including swine (Jiao et al., 2014; Yang et al., 2017; Palombo et al., 2018). Many genome-wide association studies (GWAS) have been performed in different pig populations to identify associations between candidate genes and important production traits, including carcass and meat quality traits (Sanchez et al., 2014; Sevillano et al., 2018), growth, feed intake, and behavior (Howard et al., 2015; Lu et al., 2017), muscle tissue (Steibel et al., 2011), and reproductive traits (Wang et al., 2018). Nevertheless, in pig breeding programs, the description of genomic regions associated with economically important traits is done separately for PBs (Meng et al., 2017) and CBs (Guo et al., 2017; Yang et al., 2017).
Thus, this study aimed to (i) estimate the genetic parameters and heritability of back fat (BF), loin depth (LD), and ADG and (ii) conduct a single-step genome-wide association study (ssGWAS) to compare genomic regions that affect these traits measured in PB and three-way CB pigs.
Material And Methods
Phenotypic Data
Animal use approval was not needed for this study because the data were from an existing database and were provided by The Maschhoffs, LLC (Carlyle, IL). The dataset included two populations: a terminal Duroc nucleus PB population and a commercial CB population sired by the same Duroc boars from a terminal line.
Individual piglet information (identification, gender, and cross-fostering) and litter description (litter identification, sow identification, and sow parity) were collected within 24 h of birth for both populations. The PB population consisted of 28,497 Duroc individuals (gilts and boars). Individuals were raised under controlled conditions typical of nucleus farms in a fixed-time system. Individuals were put on test at 88.5 ± 9.92 d of age and off-tested at 178.4 ±7.96 d of age. The CB population consisted of 135,768 individuals (gilts and barrows) generated by crossing the PB Duroc line with commercial maternal F1 lines. CB commercial individuals were raised in two commercial fixed-weight testing flows (A = 40,876 animals and B = 94,892 animals), harvested at an average carcass weight of 98.8 ±10.19 and 97.9 ±7.63 kg, respectively. In both commercial testing flows, a contemporary group was defined as the group of animals that entered a given facility at the same time. In system A, individuals were allocated in single-sire, single-gender groups of 20 head and housed in the same pen. The whole group was harvested on the same day upon reaching market requirements. The previously defined groups will be referred to as batch hereinafter. In flow B, individuals were not allocated and housed with the same criteria, but gender and harvest date were available. Individuals were harvested when reaching market target weight on an individual or group basis. In this case, batch was defined as the group of individuals raised in the same facility and having same sire, gender, and harvest date. Since the batch definition may not be equivalent between systems A and B, cross-validation was used to evaluate the prediction accuracy of statistical models with and without the inclusion of system B individuals (data not shown). Previous results showed an advantage in including flow B data (data not shown), and thus these were retained for further analyses. The corresponding phenotypes were measured differently in the two populations, which is representative of typical swine breeding schemes. On PB, BF (pBF) and loin eye area (pLD) were ultrasonically (Biotronics, Inc., Ames, IA) measured at the end of the testing/finishing period. For CB, BF (cBF) and loin depth (cLD) were measured using a Fat-O-Meter system (Frontmatec A/S, Kolding, DK) at harvest. In PB, ADG (pADG) was obtained as the BW at the end of the performance test period minus birth BW, divided by the age at the end of the performance test. In CB, ADG (cADG) was obtained as the hot carcass weight minus BW, divided by the age of the animal at harvest.
To assess whether postmortem and in vivo measures are genetically dissimilar, we estimated genetic correlations between the two measures collected on a subset of CB individuals (n = 5,124). High genetic correlations were observed between ultrasound BF and carcass BF (0.93 ± 0.02) and between loin eye area and carcass LD (0.90 ± 0.03). As expected, the highest genetic correlation was observed between ADG and hot carcass ADG (0.99 ± 0.01). These results suggest that it is appropriate to consider postmortem and in vivo traits as the same.
Data editing was performed in the R environment (Version 3.6.0, http://www.R-project.org/). Dam line, batch, and contemporary group were required to have at least 100, 10, and 100 pigs with records for BF, LD, and ADG, respectively. The phenotypic values for carcass quality traits were subjected to standardization to achieve a standard deviation of 10 before further statistical analysis. After data editing, 28,497 and 135,768 individual records were available for the three traits on PB and CB, respectively. Descriptive statistics are reported in Table 1.
Table 1.
Descriptive statistics for growth and carcass quality traits in purebred and crossbred pigs
| Mean | Min | Max | SD | |
|---|---|---|---|---|
| Purebred (n = 28,497) | ||||
| Back fat, mm | 14.9 | 4.8 | 39.1 | 3.8 |
| Loin depth, mm | 81.4 | 16.9 | 122.0 | 10.1 |
| ADG, kg/d | 0.493 | 0.184 | 0.807 | 0.058 |
| Crossbred (n = 135,768) | ||||
| Back fat, mm | 18.7 | 6.0 | 39.0 | 4.1 |
| Loin depth, mm | 67.2 | 31.0 | 92.0 | 7.0 |
| ADG, kg/d | 0.541 | 0.228 | 0.794 | 0.056 |
Pedigree and Genomic Data
After tracing nine generations back for animals with phenotypes and/or genotypes, pedigrees for 179,054 animals were available for the combined PB and CB populations. A total of 4,857 PB and 3,532 CB individuals from flow A were genotyped using the PorcineSNP60 BeadChip (Illumina, Inc., San Diego, CA) according to the manufacturer’s instructions. Within-population marker quality control was performed, and only markers meeting the requirements for both populations were kept for the association analysis. Markers with a call rate <0.98 and minor allele frequency <0.05 were excluded from the genotype data set. Moreover, animals with call rate <0.90 were removed from the data set. A panel of 36,328 SNPs in common among the two populations was available for downstream analyses after quality control.
Statistical Analysis
Data and pedigree were processed with the RENUMF90 software (Misztal et al., 2014), variance components and heritabilities were estimated through Gibbs Sampling using the software GIBBS3F90 and POSTGIBBSF90, whereas SNP effects were computed using the software POSTGSF90 (Misztal et al., 2002). A total of 60,000 iterations were run with the first 10,000 discarded as burn-in and thinning every five rounds. Variance components were estimated using single trait animal models. The statistical model adopted for pBF, pLD, and pADG was:
where y is the vector for the trait under investigation; b is the vector of fixed effects (sow parity, sex, and contemporary group); a is the vector of additive genetic values of the animals assumed N(0,H) with H being a realized relationship matrix that combines genomic and pedigree information (Legarra et al., 2009); the inverse of H () was constructed by blending the inverse of the SNP-derived genomic matrix () and the inverse of the numerator relationship matrix for the genotyped individuals (); the inbreeding was considered in the computation of A; p is the vector of random effect of litter assumed N(0,I), where I is an identity matrix with dimensions equal to the number of elements in p; e is the vector of residuals assumed to be normally distributed, N(0, I); and X, Z, and W were the corresponding incidence matrix.
The model of analysis adopted for cBF, cLD, and cADG is as follows:
where y is the vector of the trait under investigation; b is the vector of fixed effects (sow parity, sex, cross-fostering, and contemporary group); with H constructed as described above; p is the vector of random effect of the biological litter, assumed N(0,I); s is the vector of random effect of harvest batch assumed N(0,I); and X, Z, W, and K are the corresponding incidence matrices. Heritabilities were calculated as the ratio of additive genetic variance divided by the sum of additive genetic, litter, batch, and residual effects for CB, and of additive genetic, litter and residual variances for PB. Genetic correlations among populations were estimated using bivariate models. The model specifications were the same as those used to estimate the variance components. The additive genetic and residual variances were expressed as:
where A is the numerator relationship matrix, and G0 is a 2 × 2 covariance matrix with the PB and CB variances in the diagonals and the covariances in the off-diagonals. Residual effects were assumed to be uncorrelated and normally distributed N(0, I).
Genome-Wide Association Study
The GWAS was performed using the single-step genomic-BLUP approach (Aguilar et al., 2010; Christensen and Lund, 2010). This method was already employed for GWAS by Wang et al. (2012), Tiezzi et al. (2015), and Aguilar et al. (2019). The G matrix was constructed using the second method reported by VanRaden (2008) and proposed by Leutenegger et al. (2003) and Amin et al. (2007) weighting each marker contribution by its expected variance:
where Z is the marker incidence matrix containing genotypes centered by allele frequency, and D is a diagonal matrix with elements containing the inverse of the expected marker variance (VanRaden, 2008). After solving the ssGBLUP model, genomic breeding values of genotyped individuals (âg) were back-solved to obtain marker effects (û) accounting for their shared genomic variance, as described by the formula:
where Z is the marker incidence matrix, and D is a diagonal matrix constructed as described above.
Individual marker effects were then obtained by solving:
Similarly to Wang et al. (2014), marker effects were used to calculate direct genomic values (DGV) for all individuals based on 10 SNPs overlapping windows:
where DGVi was a vector of DGV, for all animals, for the ith genome window; was the jth column of matrix S corresponding to marker j across individuals, S was obtained as S = ZD, which scales the columns of Z by the marker variance contained in the diagonal of D. The genome windows at the beginning and at the end of each chromosome with <10 SNPs were removed. The variance of the vector DGVi was computed for each overlapping window. This value was then expressed as the proportion of total genomic variance by dividing it by the variance of DGV computed based on all 36,328 SNPs. The percentage of genome covered by the windows within the first quartile of genomic variance was calculated as the length of all windows divided by the length of the genome (Mb). The former was obtained as the difference between the end and start positions of each window, then summed across windows. The latter was calculated as the sum of the position of the last marker in the Sus Scrofa chromosomes (10.2 assembly). The windows within the first quartile of genomic variance were investigated for significance. The significance value of QTL window was assessed using a nonparametric bootstrap analysis with 1,000 replicates as described in detail by Howard et al. (2015). A GWAS significance value of P < 0.05 was adopted for the bootstrap analysis. We then arbitrarily selected the 15 windows explaining the largest amount of genomic variance for subsequent functional annotation. Based on the starting and ending coordinates of each window, gene annotations were obtained using the Biomart platform on Ensemble (Flicek et al., 2013) through the ‘Biomart’ R package (http://www.bioconductor.org). A gene ontology analysis was carried out using DAVID Bioinformatics Resources version 6.7 (http://david.abcc.ncifcrf.gov/) (Huang et al., 2009). A gene network analysis, using the names of the candidate genes, was performed in GeneMANIA (http://genemania.org/). The genetic interaction networks were analyzed using the Cytoscape software (Shannon, 2003). In addition, we performed a signal enrichment analysis using the method proposed by Fang et al. (2019) to determine whether the GWAS signals were enriched in a predefined functional category.
Results And Discussion
Genetic Variation Between PB and CB
Genetic parameters and heritability of carcass quality traits and growth are presented in Table 2. Heritability values were moderate to high for carcass quality traits (0.39 to 0.42 for PB and 0.27 to 0.49 for CB) and low to moderate for ADG (0.35 for PB and 0.12 for CB). The PB LD heritability (0.39 ±0.01) found in this study was on average higher than those reported by Miar et al. (2014) in Landrace × Large White and Duroc CB (0.22 ± 0.08). Among the two ADG, pADG showed the highest heritability (0.35 ± 0.02). The values of heritability estimates for ADG presented in this study were lower than those presented by Suzuki et al. (2005) and Cabling et al. (2015) in Duroc pigs.
Table 2.
Variance components and heritability of back fat, loin depth, and ADG1
| σ a | σ litter | σ batch | σ e | h 2 | SE | |
|---|---|---|---|---|---|---|
| Purebred | ||||||
| Back fat, mm | 83.58 (74.43;92.39) | 11.15 (9.25;13.00) | 106.82 (101.80;111.80) | 0.415 | 0.019 | |
| Loin depth, mm | 33.38 (29.31;37.29) | 5.82 (4.95;6.72) | 47.26 (44.97;49.36) | 0.386 | 0.020 | |
| ADG, kg/d | 4.80 (4.14;5.50) | 1.40 (1.15;1.47) | 7.79 (7.42;8.16) | 0.345 | 0.021 | |
| Crossbred | ||||||
| Back fat, mm | 83.13 (67.01;99.33) | 13.07 (10.57;15.46) | 7.27 (6.18;8.39) | 66.89 (58.98;75.54) | 0.487 | 0.042 |
| Loin depth, mm | 142.30 (107.20;177.90) | 32.45 (26.57;38.79) | 29.79 (25.50;33.85) | 325.72 (307.10;343.60) | 0.268 | 0.029 |
| ADG, kg/d | 2.04 (1.30;2.80) | 2.76 (2.53;2.98) | 0.18 (0.11;0.24) | 11.80 (11.38;12.20) | 0.121 | 0.021 |
1 95% highest probability density (HPD) intervals are reported in brackets.
Estimates of additive genetic variances for BF and ADG were relatively similar to those of others pig populations (Godinho et al., 2018; Davoli et al., 2019). In contrast, estimates of additive genetic variance for cLD were higher than those obtained by van Wijk et al. (2005). This could be attributed to differences in management and farming conditions.
Estimates of genetic correlations (rpc) among carcass quality traits in PB and CB and their relative standard error are shown in Table 3. The range of rpc was similar for the three traits, with moderately higher values for carcass quality traits. The estimated values ranged from 0.79 to 0.81 for carcass quality traits and 0.59 for growth performance, in line with the values reported in the literature. Godinho et al. (2018) obtained rpc estimates (SE) for BF, LD, and ADG equal to 0.82 (0.03), 0.71 (0.07), and 0.61 (0.06). Lutaaya et al. (2001) also presented rpc estimates for PB and CB over an 8-years period testing. In their study, rpc was 0.81 for ADG and 0.51 for BF.
Table 3.
Genetic correlations between purebred and crossbred for the three traits undergoing study
| Trait | Correlation (SE) |
|---|---|
| Back fat, mm | 0.81 (0.02) |
| Loin depth, mm | 0.79 (0.04) |
| ADG, kg/d | 0.56 (0.05) |
In a crossbreeding scheme, when the goal of selection is to improve the CB performances at the production level, the importance of having CB phenotype increases when the genetic correlation between PB and CB is lower than 0.80 (Wei and van der Werf, 1994). For example, the estimate of rpc for BF of 0.81 indicates that adding CB information may contribute to the improvement of the CB performance for this carcass trait, but this contribution would be limited.
GWAS Study Results
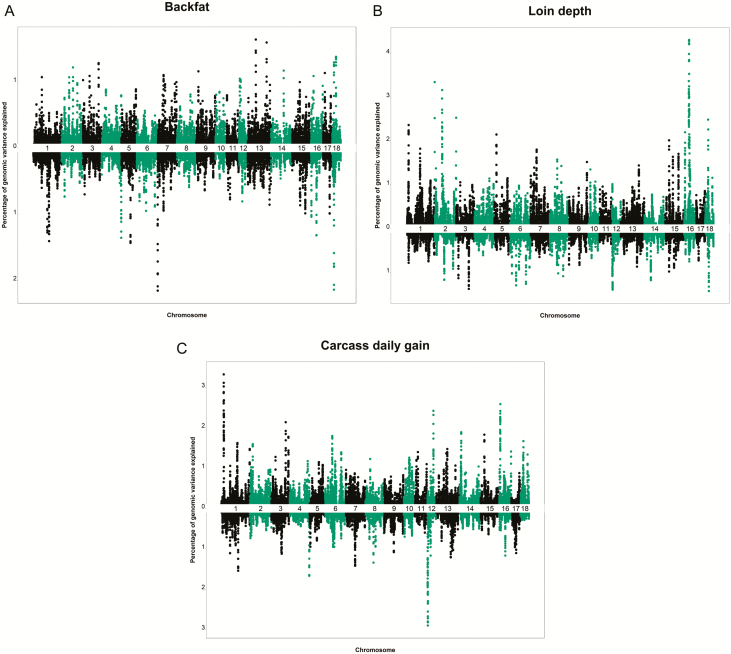
The present study allowed us to relate carcass quality traits to SNP polymorphisms across the genome of PB and CB swine. The Miami plots of SNP additive genetic variance explained by 10 SNP moving windows are reported in Figure 1a–c and a summary of the windows explaining the higher proportion of variance is reported in Table 4 and Supplementary Table S1, while the significance level of each SNP is given in Supplementary Table S2. The percentage of the genome covered by the top 25% windows were 2.46%, 2.01%, and 2.03%, while the genetic variances of these windows were 13.3%, 14.5%, and 14.3% for pBF, pLD, and pADG, respectively (Figure 1a–c). Of these, the regions of higher impact on the pBF were located on Sus Scrofa chromosome (SSC) 13 from 47.6 to 48.6 Mb and from 19.8 to 19.9 Mb, and SSC18 from 26.8 to 27.6 Mb. The first and second top SNPs rs80885388 (P = 5.66 × 10–6) and rs81229953 (P = 6.88 × 10–6) located on SSC13, while the third top SNP rs81468050 (P = 8.68 × 10–6) was located on SSC18. Our results are in in the same region of a previously identified QTL influencing carcass quality traits (Wang et al., 2015) and BF (Edwards et al., 2008b) in Duroc pigs. The regions of higher impact on the pLD were located on SSC16 from 32.5 to 35.5 Mb, SSC2 at around 0.46 Mb, and SSC16 from 133.5 to 133.8 Mb. Within these regions, the top SNP rs81479214 (P = 3.76 × 10–6) was located on SSC16, the second top SNP rs343026684 (P = 7.39 × 10–6) was located on SSC2, while the third top SNP rs80986934 (P = 3.72 × 10–6) was located on SSC16. A QTL affecting loin weight was reported to be located at about 6 Mb on SSC2 (Russo et al., 2008).
Figure 1.
Miami plot for the percentage of genetic variance explained by the 10-SNP moving windows for back fat (a), loin depth (b), and ADG (c). The results reported above and below the x-axis are for purebred and crossbred, respectively.
Table 4.
Summary of the windows within the three chromosomes and three genomic regions that explained the higher genomic variance for carcass quality traits in purebred pigs with a list of annotated genes in the proximity of each window1
| Trait | Windows | Chr | σ 2, % | Start, Mb | Stop, Mb | Genes | SNP | σ 2 PB, % | σ 2 CB, % | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Back fat | 3 | 13 | 1.51 | 47.6 | 48.6 | SLC25A26, LRIG1, ENSSSCG00000020125, KBTBD8 | rs80885388 | 0.022 | 0.000019 | 5.66 × 10–6 |
| 3 | 13 | 1.41 | 198.2 | 199.1 | RUNX1, CLIC6, ENSSSCG00000025700, ENSSSCG00000012052 | rs81229953 | 0.020 | 0.00074 | 6.88 × 10–6 | |
| 4 | 18 | 1.35 | 26.9 | 27.5 | KIAA1549, ATP6V0A4, ENSSSCG00000035963, ENSSSCG00000018723, SVOPL, TRIM24 | rs81468050 | 0.0017 | 0.0012 | 8.68 × 10–6 | |
| Loin depth | 15 | 16 | 4.02 | 32.4 | 35.4 | ENSSSCG00000016901, ENSSSCG00000016902, ENSSSCG00000016903, GPX8, MIR449A, MIR449B, MCIDAS, CCNO, DHX29, MTREX, ENSSSCG00000031138, ENSSSCG00000037130, PLPP1, SLC38A9, DDX4, ENSSSCG00000032013, IL31RA, IL6ST | rs81479214 | 0.043 | 0.0070 | 3.76 × 10–6 |
| 1 | 2 | 3.34 | 0.006 | 0.466 | ENSSSCG00000014554, ODF3, BET1L, RIC8A, SIRT3, PSMD13, ENSSSCG00000014554, NLRP6, PGGHG, IFITM5, ENSSSCG00000014565, ENSSSCG00000038912, ENSSSCG00000032436, ENSSSCG00000032591, ENSSSCG00000038005, B4GALNT4, PKP3, ANO9, ENSSSCG00000024389, PTDSS2, RNH1, HRAS, LRRC56, LMNTD2, RASSF7, ENSSSCG00000024389, PHRF1, IRF7, CDHR5, ENSSSCG00000040951, DRD4, DEAF1, ENSSSCG00000012849, EPS8L2, ENSSSCG00000012847 | rs343026684 | 0.072 | 0.018 | 7.39 × 10–6 | |
| 4 | 16 | 3.18 | 33.5 | 33.7 | IL31RA, IL6ST ANKRD55, ENSSSCG00000038692, ENSSSCG00000036369, NDUFS4, ENSSSCG00000034649, ARL15, ENSSSCG00000016892 | rs80986934 | 0.044 | 0.0064 | 3.72 × 10–6 | |
| ADG | 8 | 1 | 2.26 | 10.2 | 10.9 | rs80916931 | 0.031 | 0.0013 | 5.59 × 10–6 | |
| 4 | 16 | 2.26 | 5.8 | 6.3 | MYO10, ENSSSCG00000034022, ENSSSCG00000021807 | rs81344589 | 0.039 | 0.00085 | 6.98 × 10–6 | |
| 4 | 18 | 1.45 | 18.8 | 18.9 | UBE2H, ENSSSCG00000040437, ENSSSCG00000018417, ENSSSCG00000018762, ENSSSCG00000018739, NRF1, SMKR1, STRIP2, AHCYL2, ENSSSCG00000019583, SMO, ENSSSCG00000019904 | rs81472316 | 0.034 | 0.00012 | 5.81 × 10–6 |
1SNP = marker with largest variance within windows.
2Genetic variance of the marker with largest impact within window for purebred (σ2PB) and crossbred (σ2CB), expressed as percentage of total genetic variance, respectively.
The regions with the largest effect on pADG were located on SSC1 from 10.3 to 10.9 Mb, SSC16 from 5.7 to 6.4 Mb, and SSC18 from 18.7 to 18.9 Mb. The first top SNP rs80916931 (P = 5.59 × 10–6) was located on SSC1, the second top SNP rs81344589 (P = 6.98 × 10–6) was located on SSC16, while the third top SNP rs81472316 (P = 5.81 × 10–6) was located on SSC18. Our findings agree, with previous QTL mapping and GWAS studies in swine. For instance, a QTL associated with ADG on SCC1 spanning 10.7 to 16.1 Mb was found by de Koning et al. (2001). Similarly, a QTL associated with ADG in Duroc at 20.5 Mb was reported by Wang et al. (2015).
In the present study, the genetic variances explained by the first quartile most important windows were 9.9%, 10.0%, and 10.4% for cBF, cLD, and cADG, respectively (Figure 1a–c). The regions of higher impact on cBF were located on SSC7 from 1.1 to 1.6 Mb, SSC18 from 10.6 to 11.4 Mb, and SSC5 from 71.9 to 72.6 Mb. For this trait, the top SNP rs320534160 (P = 1.45 × 10–4) was located on SSC7, the second top SNP rs346337753 (P = 2.94 × 10–4) was located on SSC18, while the third top SNP rs81385196 (P = 1.23 × 10–4) was located on SSC5. In Meishan × Yorkshire CB, Paszek et al. (2001) found QTLs on SSC7 (4.3 to 11.6 Mb), while Harmegnies et al. (2006) found QTLs on SSC5 (63.1 to 90.1 Mb) both related to BF thickness.
The regions of higher impact on cLD were located on SSC18 from 25.4 to 26.1 Mb, SSC3 from 11.1 to 11.2 Mb, and SSC6 from 133.5 to 133.8 Mb. The top SNP rs81467885 (P = 1.79 × 10–4) was located on SSC18, the second top SNP rs81374918 (P = 1.33 × 10–4) was located on SSC3, while the third top SNP rs318606636 (P = 1.52 × 10–4) was located on SSC6. A QTL associated with loin area on SCC18 spanning 5.7 to 27.6 Mb was previously reported in Duroc × Pietrain CB (Edwards et al., 2008a).
The most significant regions for cADG were located on SSC4 from 140.2 to 140.7 Mb, SSC1 from 191.9 to 193.8 Mb, and SSC7 from 139.9 to 140.7 Mb. The top SNP rs318606636 (P = 1.20 × 10–6) was located on SSC4, the second most significant SNP rs81349654 (P = 1.04 × 10–6) was located on SSC1, while the third most significant SNP rs80885610 (P = 7.22 × 10–7) was located on SSC7. Regions affecting ADG on SSC1 and SSC4 were found in many studies, in agreement with the current study (Hernández-Sánchez et al., 2003; Qiao et al., 2015).
Genes Identified by the GWAS and Their Functions
The genes located in correspondence of the windows with the largest variance are reported in Tables 4 and 5, Supplementary Tables S3 and S4. We identified one genomic region that showed genome-wide significant association in PB and CB for BF on SSC18 between (10.6 to 27.5 Mb). Several other studies reported QTLs related to BF in the same region (Edwards et al., 2008a; Sanchez et al., 2014).
Table 5.
Summary of the windows within the three chromosomes and three genomic regions that explained the higher genomic variance for carcass quality traits in crossbred pigs with a list of annotated genes in the proximity of each window1
| Trait | Windows | Chr | σ 2, % | Start, Mb | Stop, Mb | Genes | SNP | σ 2 CB, % | σ 2 PB, % | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Back fat | 5 | 7 | 1.88 | 1.07 | 1.64 | GMDS, MYLK4 | rs320534160 | 0.035 | 0.00088 | 1.45 × 10–4 |
| 6 | 18 | 1.85 | 10.6 | 11.4 | KIAA1549, ATP6V0A4, ENSSSCG00000035963, ENSSSCG00000018723, SVOPL, RIM24, TRIM24, ZC3HAV1L | rs346337753 | 0.012 | 0.0021 | 2.94 × 10–4 | |
| 4 | 5 | 1.37 | 71.9 | 72.5 | ENSSSCG00000000784, ENSSSCG00000027998, CNTN1, ENSSSCG00000039178 | rs81385196 | 0.022 | 0.000065 | 1.23 × 10–4 | |
| Loin depth | 4 | 18 | 1.29 | 25.3 | 26.1 | FAM3C, WNT16, CPED1, TSPAN12, ING3 | rs81467885 | 0.024 | 0.0021 | 1.79 × 10–4 |
| 5 | 3 | 1.28 | 111.1 | 111.8 | RBKS, ENSSSCG00000034129, ENSSSCG00000033248, SLC4A1AP, SUPT7L, GPN1, ENSSSCG00000039695, ZNF512, C2orf16, GCKR, FNDC4, IFT172, KRTCAP3, NRBP1, PPM1G, ZNF513, SNX17, EIF2B4, GTF3C2 | rs81374918 | 0.024 | 0.0026 | 1.33 × 10–4 | |
| 2 | 6 | 1.23 | 133.5 | 133.8 | TOX3, ENSSSCG00000027421 | rs318606636 | 0.036 | 0.011 | 1.52 × 10–4 | |
| ADG | 4 | 4 | 1.54 | 140.2 | 140.6 | rs80915915 | 0.0074 | 0.0032 | 1.20 × 10–6 | |
| 6 | 1 | 1.40 | 191.9 | 193.8 | AGBL1, ENSSSCG00000034903, ESR2 | rs81349654 | 0.0011 | 0.00074 | 1.04 × 10–6 | |
| 9 | 7 | 1.32 | 139.8 | 140.7 | TMEM266, ETFA, ISL2, SCAPER, ENSSSCG00000039452, ENSSSCG00000032875, NRG4 | rs80885610 | 0.0078 | 0.00007 | 7.22 × 10–7 |
1SNP = marker with largest variance within windows.
2Genetic variance of the marker with largest impact within window for crossbred (σ2CB) and purebred (σ2PB) expressed as percentage of total genetic variance, respectively.
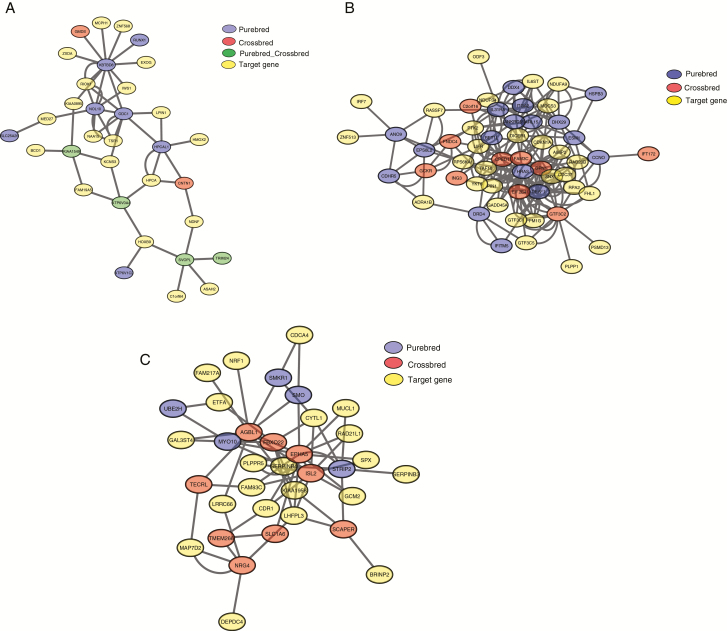
The results from the gene network analysis performed using GeneMania suggest a dense co-expression network between the candidate genes of PB and CB (Figure 2a–c). Most of the related genes in the network are also associated with functions affecting carcass quality traits. For example, phospholipid phosphatase 1 (PLPP1) plays an important role in lipid metabolism in the adipose tissue (Revilla et al., 2018). In this study, a region on SSC16 (34.3 to 34.4 Mb), with a significant effect on the LD, was in proximity of the GPX8 gene. The GPX8 has been previously associated with loin muscle tissue in an F2 population of Duroc × Pietrain (Steibel et al., 2011), consistent with our results. The region on SSC3 (126.12 to 126.13 Mb) was associated with the ODC1 gene, which is involved in the polyamine biosynthesis pathway (Sollero et al., 2011). Sollero et al. (2011) observed higher expression of ODC1 in LD of PB lines when compared with Yorkshire × Landrace CB pigs. Another potential candidate gene for LD is FAM3C (SSC18 25.5 to 25.6 Mb). The FAM3C is implicated in muscle cell growth and differentiation of the cells and was previously reported as candidate gene for the loin muscle thickness in Landrace pigs (Chen et al., 2019).
Figure 2.
Gene network for (a) back fat, the network consists of 33 genes (circles) connected by 56 interactions (edges), (b) loin depth, the network consists of 56 genes connected by 199 interactions and (c) ADG, the network consists of 35 genes connected by 86 interactions.
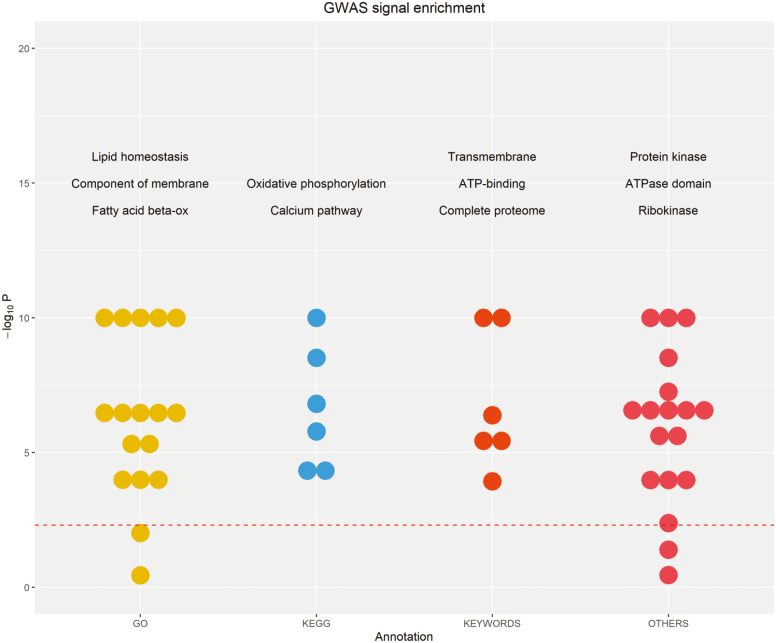
The enrichment analysis indicated that most of the genes annotated in the 15 windows per each trait with the highest explained variance were involved in fatty acid metabolism, oxidative phosphorylation, component of membrane, calcium pathway, ATP binding, and ribokynase (Figure 3). Here, we focus on two of these terms (fatty acid metabolic process and oxidative phosphorylation), which are more likely related to carcass quality traits due to their biological activity. Expressed genes in these two terms are reported in Supplementary Table S4. Among these genes, acyl-coenzyme A dehydrogenase (ACADS) and ATPase isoform (ATP6V0A4) are significant genes for growth in pigs. ACADS encodes a tetrameric mitochondrial flavoprotein, which is a member of the acyl-CoA dehydrogenase family. Yang et al. (2012) reported that ACADS was involved in the fatty acid metabolic pathway with a significant effect on feed conversion. ATP6V0A4 is an essential component of the vacuolar proton pump (V-ATPase), which plays an important function in the translocation of protons across the membranes. Kim et al. (2008) found a different expression of ATP6V0A4 among pig breeds that affect meat quality.
Figure 3.
GWAS signal enrichment on the basis of four gene annotation sources: gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, keywords, and others annotation sources. The dots are the top items with the highest enrichments in each annotation source, while the dashed red line is the significance level (P < 0.05 in original scale). The name of the major functions of these items are reported in the figure.
Genomic Regions for Carcass Quality Traits in PB and CB Pigs
The comparison of the results from the ssGWAS for PB and CB obtained in current study highlighted similarities among the genetic architecture of the two population (as expected) but also sizable differences (Tables 4 and 5; Figure 1a–c). The genetic variances explained by the first 100 most significant windows were 5.8%, 6.2%, 5.2%, 4.4%, 3.7%, and 4.3% for pBF, pLD, pADG, cBF, cLD, and cADG, respectively (Supplementary Table S1). Among these windows, 12 located on SSC18 at about 10 Mb were identified within both pBF and cBF. In particular, the 10.6 to 27.5 Mb extended region explained on average the 1.35% and 1.85% of genetic variance for pBF and cBF, respectively. Previous GWAS studies in swine have reported QTLs located on SSC18 affecting BF in different PB and CB populations (Fontanesi et al., 2012; Sanchez et al., 2014).
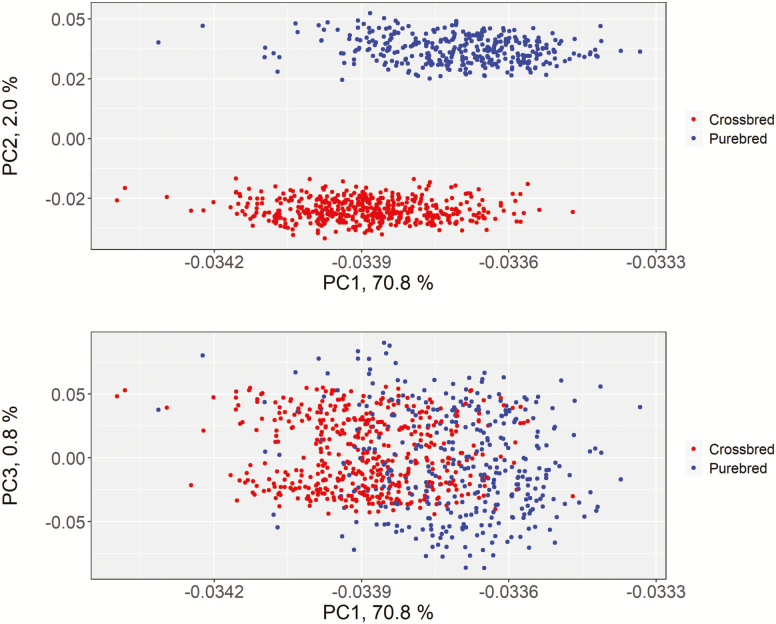
Differences were also highlighted for regions affecting LD and ADG in PB and CB pigs. One window located on SSC15 at about 22 Mb was in common for pLD and cLD, while the most significant windows were located on SSC16 and SSC18 for pLD and cLD, respectively (see Tables 4 and 5, and Supplementary Table S1). Moreover, the windows that explained the higher genetic variance for pADG (2.26%) were located on SSC1, while those that explained that largest genetic variance for cADG (1.54%) was located on SSC14. These results may be due to higher level of genetic differentiation within population with differences in terms of allele frequency (Figure 4 and Supplementary Figure S1) which could alter the power of association favoring different variants in one compared with the other population. However, these results could also be explained partially by different experimental conditions and size in the two populations.
Figure 4.
Scores plot from the principal component analysis (PC1 vs. PC2 above and PC1 vs. PC3 below) of the marker effect for back fat, loin depth, and ADG in purebred and crossbred.
We performed a principal component analysis and obtained the eigenvalue decomposition of the G-matrix. Figure 4 illustrates the result of the principal component analysis. Though the first three principal components comprised 73.6% of total variance, we observed a clear genetic differentiation between PB and CB. In our study, carcass quality traits were affected by different genomic regions in PB and CB.
Conclusions
This study investigated the genetic structure underlying the carcass quality traits in PB and CB pigs. ssGWAS allowed the identification of genomic regions associated with BF, LD, and ADG. For BF, we discovered an overlap of 12 regions within both PB and CB. For BF, LD, and ADG, we found associations between significant markers and genes in the genomic regions that include these markers. The present study also provides biological information about these genes such as fatty acid metabolism, providing further information toward improving our knowledge of the genetic mechanisms determining carcass quality traits. Our results show that the carcass quality traits are likely controlled by a large number of different genes in PB and CB pigs with a different genetic structure in the populations examined.
Supporting Information
Table S1. Percentage of genetic variance explained by the 10-SNP moving windows for back fat, loin depth, and average carcass daily gain in purebred and crossbred pigs.
Table S2. Summary of significant SNP associated with carcass quality traits in purebred and crossbred swine. The table reports, SNP ID, chromosome (chr), position (pos) (in base pairs), SNP effect (α), genetic variance, and P-values.
Table S3. List of annotated genes in the proximity of the first 15 windows that explained the largest genetic variance. The table reports trait, chromosome (chr), SNP (start, stop), position SNP (start, stop) in base pairs, gene, and position gene (start, stop).
Table S4. List of pathway-enrichment analysis performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID). The table reports trait, gene name, pathway, and functions.
Figure S1. Allele frequency of significant SNPs for back fat, loin depth, and ADG in the two population: purebred and crossbred. Diff. α is the difference between SNP effect in purebred and crossbred.
Conflict of Interest StatementNone declared.
Literature Cited
- Aguilar I., Legarra A., Cardoso F., Masuda Y., Lourenco D., and Misztal I.. . 2019. Frequentist p-values for large-scale-single step genome-wide association, with an application to birth weight in American Angus cattle. Genet. Sel. Evol. 51:28. doi: 10.1186/s12711-019-0469-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar I., Misztal I., Johnson D. L., Legarra A., Tsuruta S., and Lawlor T. J.. . 2010. Hot topic: a unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 93:743–752. doi:10.3168/jds.2009–2730 [DOI] [PubMed] [Google Scholar]
- Amin N., van Duijn C. M., and Aulchenko Y. S.. . 2007. A genomic background based method for association analysis in related individuals. PLoS One 2:e1274. doi:10.1371/journal.pone.0001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma P., and van Arendonk J. A. M.. . 1998. Maximizing genetic gain for the sire line of a crossbreeding scheme utilizing both purebred and crossbred information. Anim. Sci. 66:529–542. doi:10.1017/S135772980000970X [Google Scholar]
- Cabling M. M., Kang H. S., Lopez B. M., Jang M., Kim H. S., Nam K. C., Choi J. G., and Seo K. S.. . 2015. Estimation of genetic associations between production and meat quality traits in duroc pigs. Asian-Australas. J. Anim. Sci. 28:1061–1065. doi:10.5713/ajas.14.0783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Wu P., Yang Q., Wang K., Zhou J., Yang X., et al. . 2019. Genome-wide association study for backfat thickness at 100 kg and loin muscle thickness in domestic pigs based on genotyping by sequencing. Physiol Genomics. 51:261–266. doi:10.1152/physiolgenomics.00008.2019 [DOI] [PubMed] [Google Scholar]
- Christensen O. F., and Lund M. S.. . 2010. Genomic prediction when some animals are not genotyped. Gen. Sel. Evol. 42:2. doi:10.1186/1297-9686-42-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli R., Catillo G., Serra A., Zappaterra M., Zambonelli P., Zilio D. M., Steri R., Mele M., Buttazzoni L., and Russo V.. . 2019. Genetic parameters of backfat fatty acids and carcass traits in Large White pigs. Animal 13:924–932. doi:10.1017/S1751731118002082 [DOI] [PubMed] [Google Scholar]
- Edwards D. B., Ernst C. W., Raney N. E., Doumit M. E., Hoge M. D., and Bates R. O.. . 2008a. Quantitative trait locus mapping in an F2 Duroc × Pietrain resource population: II. Carcass and meat quality traits1. J. Anim. Sci. 86:254–266. doi:10.2527/jas.2006–626 [DOI] [PubMed] [Google Scholar]
- Edwards D. B., Ernst C. W., Tempelman R. J., Rosa G. J., Raney N. E., Hoge M. D., and Bates R. O.. . 2008b. Quantitative trait loci mapping in an F2 Duroc x Pietrain resource population: I. Growth traits. J. Anim. Sci. 86:241–253. doi:10.2527/jas.2006-625 [DOI] [PubMed] [Google Scholar]
- Fang L., Jiang J., Li B., Zhou Y., Freebern E., Vanraden P. M., Cole J. B., Liu G. E., and Ma L.. . 2019. Genetic and epigenetic architecture of paternal origin contribute to gestation length in cattle. Commun. Biol. 2:100. doi:10.1038/s42003-019-0341-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., Ahmed I., Amode M. R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., . et al. 2013. Ensembl 2013. Nucleic Acids Res. 41(Database issue):D48–D55. doi:10.1093/nar/gks1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi L., Schiavo G., Galimberti G., Calò D. G., Scotti E., Martelli P. L., Buttazzoni L., Casadio R., and Russo V.. . 2012. A genome wide association study for backfat thickness in Italian Large White pigs highlights new regions affecting fat deposition including neuronal genes. BMC Genomics 13:583. doi:10.1186/1471-2164-13-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho R. M., Bergsma R., Silva F. F., Sevillano C. A., Knol E. F., Lopes M. S., Lopes P. S., Bastiaansen J. W. M., and Guimarães S. E. F.. . 2018. Genetic correlations between feed efficiency traits, and growth performance and carcass traits in purebred and crossbred pigs. J. Anim. Sci. 96:817–829. doi:10.1093/jas/skx011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Huang Y., Hou L., Ma J., Chen C., Ai H., Huang L., and Ren J.. . 2017. Genome-wide detection of genetic markers associated with growth and fatness in four pig populations using four approaches. Gen. Sel. Evol. 49:21. doi:10.1186/s12711-017-0295-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmegnies N., Davin F., De Smet S., Buys N., Georges M., and Coppieters W.. . 2006. Results of a whole-genome quantitative trait locus scan for growth, carcass composition and meat quality in a porcine four-way cross. Anim. Genet. 37:543–553. doi:10.1111/j.1365-2052.2006.01523.x [DOI] [PubMed] [Google Scholar]
- Hernández-Sánchez J., Visscher P., Plastow G., and Haley C.. . 2003. Candidate gene analysis for quantitative traits using the transmission disequilibrium test: the example of the melanocortin 4-receptor in pigs. Genetics 164:637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. T., Jiao S., Tiezzi F., Huang Y., Gray K. A., and Maltecca C.. . 2015. Genome-wide association study on legendre random regression coefficients for the growth and feed intake trajectory on Duroc Boars. BMC Genet. 16:59. doi:10.1186/s12863-015-0218-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang d. a. W., Sherman B. T., and Lempicki R. A.. . 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. doi:10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jiao S., Maltecca C., Gray K. A., and Cassady J. P.. . 2014. Feed intake, average daily gain, feed efficiency, and real-time ultrasound traits in Duroc pigs: II. Genomewide association. J. Anim. Sci. 92:2846–2860. doi:10.2527/jas.2014-7337 [DOI] [PubMed] [Google Scholar]
- Kim N. K., Lim J. H., Song M. J., Kim O. H., Park B. Y., Kim M. J., Hwang I. H., and Lee C. S.. . 2008. Comparisons of longissimus muscle metabolic enzymes and muscle fiber types in Korean and western pig breeds. Meat Sci. 78:455–460. doi:10.1016/j.meatsci.2007.07.014 [DOI] [PubMed] [Google Scholar]
- de Koning D. J., Rattink A. P., Harlizius B., Groenen M. A. M., Brascamp E. W., and van Arendonk J. A. M.. . 2001. Detection and characterization of quantitative trait loci for growth and reproduction traits in pigs. Livest. Sci. 72:185–198. doi:10.1016/S0301-6226(01)00226-3 [DOI] [PubMed] [Google Scholar]
- Legarra A., Aguilar I., and Misztal I.. . 2009. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 92:4656–4663. doi:10.3168/jds.2009-2061 [DOI] [PubMed] [Google Scholar]
- Leutenegger A. L., Prum B., Génin E., Verny C., Lemainque A., Clerget-Darpoux F., and Thompson E. A.. . 2003. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 73:516–523. doi:10.1086/378207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M. S., Bovenhuis H., Hidalgo A. M., van Arendonk J. A. M., Knol E. F., and Bastiaansen J. W. M.. . 2017. Genomic selection for crossbred performance accounting for breed-specific effects. Genet. Sel. Evol. 49:51. doi:10.1186/s12711-017-0328-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Jiao S., Tiezzi F., Knauer M., Huang Y., Gray K. A., and Maltecca C.. . 2017. The relationship between different measures of feed efficiency and feeding behavior traits in Duroc pigs. J. Anim. Sci. 95:3370–3380. doi:10.2527/jas.2017.1509 [DOI] [PubMed] [Google Scholar]
- Lutaaya E., Misztal I., Mabry J. W., Short T., Timm H. H., and Holzbauer R.. . 2001. Genetic parameter estimates from joint evaluation of purebreds and crossbreds in swine using the crossbred model. J. Anim. Sci. 79:3002–3007. doi:10.2527/2001.79123002x [DOI] [PubMed] [Google Scholar]
- Meng Q., Wang K., Liu X., Zhou H., Xu L., Wang Z., and Fang M.. . 2017. Identification of growth trait related genes in a Yorkshire purebred pig population by genome-wide association studies. Asian-Australas. J. Anim. Sci. 30:462–469. doi:10.5713/ajas.16.0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miar Y., Plastow G., Bruce H., Moore S., Manafiazar G., Kemp R., Charagu P., Huisman A., van Haandel B., Zhang C., . et al. 2014. Genetic and phenotypic correlations between performance traits with meat quality and carcass characteristics in commercial crossbred pigs. PLoS One 9:e110105. doi:10.1371/journal.pone.0110105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misztal I., Tsuruta S., Lourenco D. A. L., Masuda Y., Aguilar I., Legarra A., and Vitezica Z.. . 2014. Manual for BLUPF90 family of programs. Athens:University of Georgia. [Google Scholar]
- Misztal I., Tsuruta S., Strabel T., Auvray B., Druet T., and Lee D.. . 2002. BLUPF90 and related programs (BGF90). Proc. 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France. [Google Scholar]
- Palombo V., Milanesi M., Sgorlon S., Capomaccio S., Mele M., Nicolazzi E., Ajmone-Marsan P., Pilla F., Stefanon B., and D’Andrea M.. . 2018. Genome-wide association study of milk fatty acid composition in Italian Simmental and Italian Holstein cows using single nucleotide polymorphism arrays. J. Dairy Sci. 101:11004–11019. doi:10.3168/jds.2018-14413 [DOI] [PubMed] [Google Scholar]
- Paszek A. A., Wilkie P. J., Flickinger G. H., Miller L. M., Louis C. F., Rohrer G. A., Alexander L. J., Beattie C. W., and Schook L. B.. . 2001. Interval mapping of carcass and meat quality traits in a divergent swine cross. Anim. Biotechnol. 12:155–165. doi:10.1081/ABIO-100108342 [DOI] [PubMed] [Google Scholar]
- Qiao R., Gao J., Zhang Z., Li L., Xie X., Fan Y., Cui L., Ma J., Ai H., Ren J., . et al. 2015. Genome-wide association analyses reveal significant loci and strong candidate genes for growth and fatness traits in two pig populations. Genet. Sel. Evol. 47:17. doi:10.1186/s12711-015-0089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla M., Puig-Oliveras A., Crespo-Piazuelo D., Criado-Mesas L., Castelló A., Fernández A. I., Ballester M., and Folch J. M.. . 2018. Expression analysis of candidate genes for fatty acid composition in adipose tissue and identification of regulatory regions. Sci. Rep. 8:2045. doi:10.1038/s41598-018-20473-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V., Fontanesi L., Scotti E., Beretti F., Davoli R., Nanni Costa L., Virgili R., and Buttazzoni L.. . 2008. Single nucleotide polymorphisms in several porcine cathepsin genes are associated with growth, carcass, and production traits in Italian Large White pigs. J. Anim. Sci. 86:3300–3314. doi:10.2527/jas.2008-0920 [DOI] [PubMed] [Google Scholar]
- Sanchez M. P., Tribout T., Iannuccelli N., Bouffaud M., Servin B., Tenghe A., Dehais P., Muller N., Del Schneider M. P., Mercat M. J., . et al. 2014. A genome-wide association study of production traits in a commercial population of Large White pigs: evidence of haplotypes affecting meat quality. Genet. Sel. Evol. 46:12. doi:10.1186/1297-9686-46-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevillano C. A., Ten Napel J., Guimarães S. E. F., Silva F. F., and Calus M. P. L.. . 2018. Effects of alleles in crossbred pigs estimated for genomic prediction depend on their breed-of-origin. BMC Genomics 19:740. doi:10.1186/s12864-018-5126-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T.. . 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. doi:10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollero B. P., Guimarães S. E., Rilington V. D., Tempelman R. J., Raney N. E., Steibel J. P., Guimarães J. D., Lopes P. S., Lopes M. S., and Ernst C. W.. . 2011. Transcriptional profiling during foetal skeletal muscle development of Piau and Yorkshire-Landrace cross-bred pigs. Anim. Genet. 42:600–612. doi:10.1111/j.1365-2052.2011.02186.x [DOI] [PubMed] [Google Scholar]
- Steibel J. P., Bates R. O., Rosa G. J., Tempelman R. J., Rilington V. D., Ragavendran A., Raney N. E., Ramos A. M., Cardoso F. F., Edwards D. B., . et al. 2011. Genome-wide linkage analysis of global gene expression in loin muscle tissue identifies candidate genes in pigs. PLoS One 6:e16766. doi:10.1371/journal.pone.0016766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Irie M., Kadowaki H., Shibata T., Kumagai M., and Nishida A.. . 2005. Genetic parameter estimates of meat quality traits in Duroc pigs selected for average daily gain, longissimus muscle area, backfat thickness, and intramuscular fat content. J. Anim. Sci. 83:2058–2065. doi:10.2527/2005.8392058x [DOI] [PubMed] [Google Scholar]
- Tiezzi F., Parker-Gaddis K. L., Cole J. B., Clay J. S., and Maltecca C.. . 2015. A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS One 10:e0114919. doi:10.1371/journal.pone.0114919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi:10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ding X., Tan Z., Xing K., Yang T., Wang Y., Sun D., and Wang C.. . 2018. Genome-wide association study for reproductive traits in a Large White pig population. Anim. Genet. 49:127–131. doi:10.1111/age.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liu D., Hernandez-Sanchez J., Chen J., Liu C., Wu Z., Fang M., and Li N.. . 2015. Genome wide association analysis reveals new production trait genes in a male Duroc population. J.-F. Liu, editor. PLoS One. 10:e0139207. doi:10.1371/journal.pone.0139207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Misztal I., Aguilar I., Legarra A., Fernando R. L., Vitezica Z., Okimoto R., Wing T., Hawken R., and Muir W. M.. . 2014. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front. Genet. 5:134. doi:10.3389/fgene.2014.00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Misztal I., Aguilar I., Legarra A., and Muir W. M.. . 2012. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. (Camb.). 94:73–83. doi:10.1017/S0016672312000274 [DOI] [PubMed] [Google Scholar]
- Wei M., and van der Werf J. H. J.. . 1994. Maximizing genetic response in crossbreds using both purebred and crossbred information. Anim. Sci. 59:401–413. doi:10.1017/S0003356100007923 [Google Scholar]
- Wientjes Y. C. J., and Calus M. P. L.. . 2017. Board invited review: the purebred-crossbred correlation in pigs: a review of theory, estimates, and implications. J. Anim. Sci. 95:3467–3478. doi:10.2527/jas.2017.1669 [DOI] [PubMed] [Google Scholar]
- van Wijk H. J., Arts D. J., Matthews J. O., Webster M., Ducro B. J., and Knol E. F.. . 2005. Genetic parameters for carcass composition and pork quality estimated in a commercial production chain. J. Anim. Sci. 83:324–333. doi:10.2527/2005.832324x [DOI] [PubMed] [Google Scholar]
- Yang T., Wang Z., Miar Y., Bruce H., Zhang C., and Plastow G.. . 2017. A genome-wide association study of meat colour in commercial crossbred pigs. Can. J. Anim. Sci. 97: 721–733. [Google Scholar]
- Yang F., Wang Q., Wang M., He K., and Pan Y.. . 2012. Associations between gene polymorphisms in two crucial metabolic pathways and growth traits in pigs. Chin. Sci. Bull. 57:2733–2740. doi: 10.1007/s11434-012-5328-3 [Google Scholar]
- Zumbach B., Misztal I., Tsuruta S., Holl J., Herring W., and Long T.. . 2007. Genetic correlations between two strains of Durocs and crossbreds from differing production environments for slaughter traits. J. Anim. Sci. 85:901–908. doi:10.2527/jas.2006-499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.