Abstract
Recently, it has been proposed that strains of Propionibacterium acnes from the type III genetic division should be classified as P. acnes subsp. elongatum subsp. nov., with strains from the type I and II divisions collectively classified as P. acnes subsp. acnes subsp. nov. Under such a taxonomic re-appraisal, we believe that types I and II should also have their own separate rank of subspecies. In support of this, we describe a polyphasic taxonomic study based on the analysis of publicly available multilocus and whole-genome sequence datasets, alongside a systematic review of previously published phylogenetic, genomic, phenotypic and clinical data. Strains of types I and II form highly distinct clades on the basis of multilocus sequence analysis (MLSA) and whole-genome phylogenetic reconstructions. In silico or digital DNA–DNA similarity values also fall within the 70–80 % boundary recommended for bacterial subspecies. Furthermore, we see important differences in genome content, including the presence of an active CRISPR/Cas system in type II strains, but not type I, and evidence for increasing linkage equilibrium within the separate divisions. Key biochemical differences include positive test results for β-haemolytic, neuraminidase and sorbitol fermentation activities with type I strains, but not type II. We now propose that type I strains should be classified as P. acnes subsp. acnes subsp. nov., and type II as P. acnes subsp. defendens subsp. nov. The type strain of P. acnes subsp. acnes subsp. nov. is NCTC 737T (=ATCC 6919T=JCM 6425T=DSM 1897T=CCUG 1794T), while the type strain of P. acnes subsp. defendens subsp. nov. is ATCC 11828 (=JCM 6473=CCUG 6369).
Keywords: Propionibacterium acnes subsp. defendens subsp. nov., type I, type II, Multilocus Sequence Analysis, Genomics, Phenotype
Propionibacterium acnes is a Gram-stain-positive anaerobic bacterium and a member of the ‘cutaneous’ group of human propionibacteria along with Propionibacterium granulosum , Propionibacterium avidum and ‘ Propionibacterium humerusii'. Although found predominately on the skin, it can also be isolated from the oral cavity and the genitourinary and gastrointestinal tracts (Patrick & McDowell, 2011). While the bacterium is most noted for its association with the inflammatory skin condition acne vulgaris (Lomholt & Kilian, 2010; McDowell et al., 2012; Fitz-Gibbon et al., 2013), there is now a growing recognition that the spectrum of opportunistic infections and clinical conditions with which it may be associated has been underestimated (Tunney et al., 1999; Cohen et al., 2005; Cavalcanti et al., 2011; Eishi, 2013; Barnard et al., 2016).
In the last 10 years, significant advances in our understanding of this bacterium at the population genetic level have been made using single, multilocus and whole-genome sequence analyses (McDowell et al., 2005, 2008; Lomholt & Kilian, 2010; McDowell et al., 2012; Fitz-Gibbon et al., 2013; Tomida et al., 2013; Scholz et al., 2014). Such work has demonstrated the phylogenetically distinct nature of the originally described serotypes of P. acnes , designated types I and II, and identified a third type, designated type III, which displays an ability to form long filamentous cell structures not seen with types I and II (McDowell et al., 2005, 2008). These studies have also identified further phylogenetic subdivisions within the type I clade (IA1, IA2, IB, IC) which differ in genome content, inflammatory potential, association with disease, production of putative virulence determinants and resistance to antibiotics used in the treatment of acne, as well as biochemical and aggregative properties (Valanne et al., 2005; McDowell et al., 2013; Tomida et al., 2013; Johnson et al., 2016; Scholz et al., 2016).
Very recently, Dekio et al. (2015) proposed that P. acnes type III be reclassified as P. acnes subsp. elongatum subsp. nov. based on phylogenetic, genomic and phenotypic differences, with strains of type I and II classified as P. acnes subsp. acnes subsp. nov. (Dekio et al., 2015). In bacterial taxonomy, there are currently no clear guidelines for the establishment of subspecies, and the proposal of such essentially remains at the discretion of the researcher. Nevertheless, the proposal of a novel bacterial subspecies is normally based on consistent phylogenetic differences and phenotypic variations between groups of strains within a species (Brenner et al., 2000). If the major phylogroups of P. acnes are now to be reclassified within a subspecies framework, then strains of types I and II also deserve their own taxonomic rank of subspecies. In this paper, we describe a polyphasic taxonomic study based on the analysis of publicly available multilocus sequence and whole-genome datasets, alongside a review of published phylogenetic, genomic, phenotypic and clinical data, to support the reclassification of P. acnes types I and II as distinct subspecies. We propose type I as P. acnes subsp. acnes subsp. nov. as it contains the type strain (ATCC6919T), and type II as P. acnes subsp. defendens subsp. nov. Type III strains remain as P. acnes subsp. elongatum subsp. nov. (hereafter described as type III) as previously proposed (Dekio et al., 2015).
In 2005, we demonstrated that the P. acnes serotypes known as types I and II represented highly distinct phylogenetic groups based on sequence analysis of the recA housekeeping gene, as well as the putative haemolysin/ FtsJ-like methyltransferase gene tly (McDowell et al., 2005). Application of recA typing was also central in the identification of strains representing the type III phylogenetic division (McDowell et al., 2008). Since then, two key multi-locus sequence analysis (MLSA) methods based on eight (MLSA8) and nine protein-encoding genes (MLSA9) have been described for this bacterium, both based on completely different sets of genetic loci (Lomholt & Kilian, 2010; McDowell et al., 2012). With both independent MLSA schemes we find that types I, II and III form highly distinct clades consistent with the original recA and tly analysis, and supported by high bootstrap values (Fig. 1). This phylogenetic clustering is also highly congruent with that obtained upon whole-genome analysis of 124 731 SNPs in shared or ‘core’ regions of 85 P . acnes genomes spanning all the major phylogenetic divisions (Fig. S1, available in the online Supplementary Material); the average P-distance between each of the types based on core region analysis is 0.444 for types I and II, 0.487 for types I and III, and 0.470 for types II and III (Table 1). Of the core region SNPs, 26 % are unique to type I, with 22 % unique to type II and 24 % unique to type III (Table 1). The genetic distance between types I and II is therefore similar to the distance between type I and type III, and type II and type III. In addition, even though the 16S rRNA gene of P. acnes demonstrates a high degree of intra-species sequence identity, the observation of distinct and non-overlapping ribotypes for type I (RT1; RT3; RT4; RT5; RT8; RT16; RT532), type II (RT2; RT6), and type III (RT9) provides further evidence for their different phylogenies (Fitz-Gibbon et al., 2013; Barnard et al., 2016) (Fig. S1).
Fig. 1.
Minimum evolution phylogenetic tree (mega v7.0) (Kumar et al., 2016) of concatenated gene sequences (4253 bp) from all STs currently represented in the MLST8 database (http://pubmlst.org/pacnes/), and covering all major genetic divisions. Sequence input order was randomized, and bootstrapping resampling statistics were performed using 500 data sets. Bootstrap values (≥70 %) are shown on the arms of the tree. Horizontal bar represents genetic distance. CC, Clonal complex.
Table 1.
Genetic characteristics of P. acnes phylogroups
| Genetic Grouping | P-distance (core SNPs) | ds* | Percentage unique core region SNPs | ||
|---|---|---|---|---|---|
| Type I | Type II | Type III | |||
| Type I | – | 0.444 | 0.487 | 0.006±0.001 | 26 |
| Type II | 0.444 | – | 0.470 | 0.005±0.001 | 22 |
| Type III | 0.487 | 0.470 | – | 0.002±0.001 | 24 |
| Type I, II, III | – | – | – | 0.024±0.003 | – |
*Based on the analysis of concatenated MLSA8 sequence data using the Nei–Gojobori method (Jukes–Cantor) in mega v5.0.
Alongside phylogenetic analyses, previous whole-genome typing patterns based on methods such as random amplification of polymorphic DNA (RAPD) and non-coding repeat sequences, as well as the analysis of non-core regions, also support types I and II as highly distinct divisions at the genome level (Perry et al., 2003; Tomida et al., 2013; Hauck et al., 2015). While digital or in silico DNA–DNA hybridization values (GGDC 2.0 algorithm) between types I, II and III are above the 70 % cut-off value currently used for bacterial species demarcation, thus confirming their membership of the same species, the whole-genome relatedness values are consistent with the proposal that types I and II are also placed in distinct taxonomic ranks in line with that recently proposed for type III (Dekio et al., 2015). Strains representing the different phylogroups within type I (IA1, IA2, IB, IC) share high in silico DNA–DNA hybridization values of 91–100 %, but this drops to 74.1–78.5 % when analysed against the type II strains ATCC11828 and JCM18920, and 72.0–72.8 % with the type III strain JCM18909 (Dekio et al., 2015). Strains of type II and III share relatedness values of 72.9–73.2 % (Dekio et al., 2015). These hybridization values between the major divisions are within the 70–80 % similarity boundary recently recommended for bacterial subspecies (Meier-Kolthoff et al., 2014).
Detailed comparative analysis of type I and II whole-genome sequences also reveals some salient differences between the divisions. These include specific genomic inversions and insertions present in type II strains, but not type I, which encode genes related to carbohydrate processing and modification, ABC transporters, nickel import, bacitracin resistance and hypothetical proteins (Fig. S2) (McDowell et al., 2013; Scholz et al., 2016). One of the most striking differences relates to the presence in type II strains of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas locus (Brüggemann et al., 2012b; Fitz-Gibbon et al., 2013). In contrast, type I and type III strains contain CRISPR/Cas gene remnants within their genome, indicating deletion of the locus during the evolutionary history of these phylogroups; the deletions are more extensive in type I strains compared with type III. The deletion of the CRISPR/Cas system in type I and type III strains makes these divisions more susceptible to horizontal gene transfer (HGT) and the acquisition of fitness or virulence traits. The observation of such CRISPR/Cas gene remnants has led to the suggestion that the type I and III divisions may constitute younger subpopulations than type II strains which are descended from a more ancient lineage (Brüggemann et al., 2012a, b). Since age = ds/(clock rate ×2), where ds is the mean number of synonymous substitutions per site and clock rate is the synonymous molecular clock rate, calculation of the ds values for strains currently representing the major type I, II and III divisions may give deeper insights into their relative ages. Interestingly, using the Nei–Gojobori method (Jukes–Cantor) (Nei & Gojobori, 1986) in mega v7.0 (Kumar et al., 2016), we observed that the ds value for the entire type I division was slightly higher than that for type II based on an initial analysis of concatenated MLSA8 sequence data, while values for type III were lower (Table 1). To investigate this further, we examined the shared core-coding regions of 85 P. acnes genomes currently available. Multiple sequence alignments were performed using MUSCLE (Edgar, 2004) and the Jukes–Cantor ds values calculated for each pair of sequences in the alignment using the Nei–Gojobori method as implemented in the Bioperl package Bio::Align::DNAStatistics (Stajich et al., 2002). As before, the resulting ds values obtained for type I (0.008), type II (0.005) and type III (0.001) revealed higher synonymous nucleotide diversity within the large type I clade compared with type II and type III, indicative of an older age. Further studies are therefore required to provide clarity on the series of evolutionary events that have given rise to the emergence and diversity of the current P. acnes clades now proposed as subspecies, including the possible diversity-purging effects of periodic selection (Cohan, 2001).
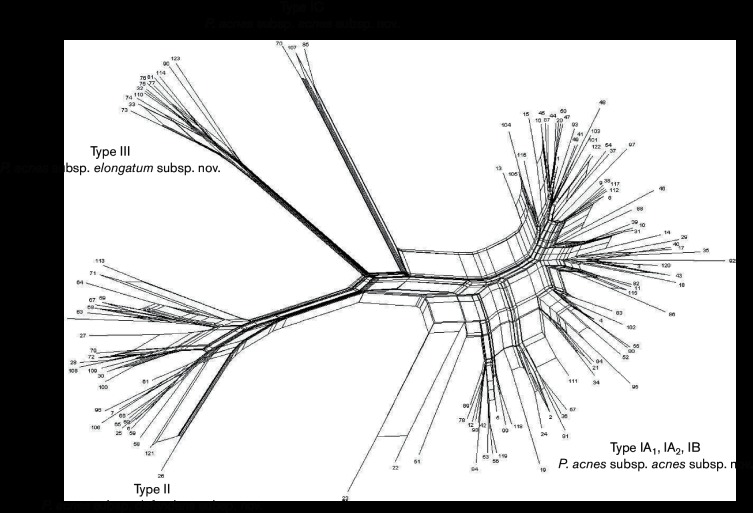
P. acnes has a clonal, epidemic population structure and is in linkage disequilibrium, though rates of HGT within the population as a whole are statistically significant (Lomholt & Kilian, 2010; McDowell et al., 2012, 2013). Previous studies have, however, found that rates of recombination appear to differ throughout the population, and that the association of alleles is less significant when distinct phylogroup populations are considered (McDowell et al., 2012, 2013). In particular, we see a drop in the index of association value (IA) when strains from the type I and II divisions are considered separately, indicating increasing linkage equilibrium within these distinct clusters (McDowell et al., 2013); this can also be observed on a Neighbour-Net split graph based on MLST8 allelic profile data (Fig. 2). Detailed inspection of MLSA8 datasets also indicates conjugal transfer and replacement of unusually large chromosomal segments in the genome dynamics of the type I clade, particularly between types IA2 and IB (Lomholt & Kilian, 2010; McDowell et al., 2012, 2013). The idea that rates of genetic interchange are more frequent within, but not between, the major divisions is indicative of increasing sexual isolation which occurs with more genetically divergent organisms (Majewski, 2001). Reduced rates of recombination may also indicate ecological differences since members of the same habitat are more likely to undergo recombination events (sympatric speciation); such population subdivisions can introduce linkage disequilibrium into an analysis if isolates from different niches (Ecotypes) are included (Spratt & Maiden, 1999). Comprehensive analysis of genome differences between the major types does indeed provide potential evidence for distinct environmental challenges within the human host.
Fig. 2.
Neighbour-net split graph (SplitsTree v4.14.4) of allelic profiles from all STs currently represented in the MLST8 database (http://pubmlst.org/pacnes/), and covering all major genetic divisions (Huson & Bryant, 2006). A distance matrix was generated from the allelic profile data and saved in NEXUS format for input to SplitsTree. Parallelogram formations indicative of recombination/ reticulation events are evident within the major type I and II divisions.
Studies by Johnson & Cummins (1972) first revealed types I and II as distinct phenotypes of P. acnes based on serological agglutination tests and cell wall sugar analysis; type I strains contain galactose in their cell wall, but this sugar is absent in type II strains which occasionally also contain meso-diaminopimelic acid (DAP) (Table 2). The development of more recent monoclonal antibody typing methods for P. acnes have further highlighted differences between the cell wall structures of type I and II, as well as type III, based on the expression of unique antigenic determinants, including those in lipoteichoic acid and adhesin proteins (Holland et al., 2010; McDowell et al., 2011; Bae et al., 2014). Differences in cell surface hydrophobicity have also been described for types I and II, and upon growth in liquid media, such as protease peptone yeast (PPY) or brain–heart infusion (BHI) broth, type II strains form a turbid solution with a slight fine sediment, while strains of type IA and IC can form a large granular sediment or auto-aggregate with a clear solution (Cohen et al., 2005); type IB strains behave as type II with respect to this characteristic. Types I and II can be differentiated from one another and type III upon analysis of bacterial whole-cell proteins by matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) fingerprinting, highlighting further variation at the phenotype level (Nagy et al., 2013; Dekio et al., 2015). Furthermore, differences in the susceptibility of types I and II to bacteriophage infection have also been known for some time (Webster & Cummins, 1978; Liu et al., 2015). The main phylogroups of P. acnes share a high degree of similarity with regard to their biochemical phenotype, including traditional tests used to differentiate the bacterium from other ‘cutaneous’ propionibacteria (Table 2). Notable phylogroup differences, however, include β-haemolytic and neuraminidase activity, as well as sorbitol fermentation, all of which are essentially restricted to the type I division (McDowell et al., 2008; Lomholt & Kilian, 2010; Niazi et al., 2010) (Table 2). The production of lipase also appears to be much lower amongst type II strains versus those from the type I and III divisions (McDowell et al., 2008; Niazi et al., 2010) (Table 2); we have previously described how type II strains have deletions in the TATA box and open reading frame of two candidate lipase genes which may explain this reduced activity (Tomida et al., 2013).
Table 2.
Key phenotypic similarities and differences between type I, II and III strains
+, At least 90 % of isolates are positive; −, at least 90 % of isolates are negative; d+, 40–89 % of isolates are positive; d−, 11–39 % of isolates are positive; nd, not determined.
| Characteristic* | Type I | Type II | Type III |
|---|---|---|---|
| Indole production | + | d+ | + |
| Catalase activity | + | + | + |
| Nitrate reduction | + | + | d+ |
| Gelatin liquefaction | + | + | − |
| Aesculin hydrolysis | − | − | − |
| β-Haemolysis (5 days at 37 °C) | d+ | − | − |
| Neuraminidase | d+ | − | − |
| Lipase | d+ | d− | d+ |
| l-Pyrrolydonyl arylamidase | d+ | d− | − |
| Pyruvate | d+ | + | − |
| Fermentation of: | |||
| Sorbitol | d+ | − | − |
| Maltose | − | − | − |
| Sucrose | − | − | − |
| Glycerol | d+ | d+ | + |
| Ribose | d− | d+ | − |
| Cell wall components | |||
| Dermatan sulphate-binding adhesins | d+ | − | − |
| DAP isomer | ll- | ll- (meso) | LL- |
| Amino acids | Ala, Gly, Glu |
Ala, Gly, Glu | nd |
| Sugars | Galactose, Glucose, Mannose | Glucose Mannose | nd |
*Key phenotypic characteristics were compiled from data from one or more of the following publications: McDowell et al. (2005, 2008, 2011); Lomholt & Kilian (2010); Niazi et al. (2010); Patrick & McDowell (2011); Dekio et al. (2015).
One defining difference between the type I and II phylogroups rests on their association with acne vulgaris. On the basis of both culture and metagenomic analyses, widely disseminated clonal lineages from the type I division have been described in association with acneic skin, but not those from the type II or type III divisions, which appear to be associated more with blood, medical devices and soft tissue infections (Lomholt & Kilian, 2010; McDowell et al., 2011, 2012; Fitz-Gibbon et al., 2013; Rollason et al., 2013). Recently, type III strains have also been linked with the depigmenting skin condition progressive macular hypomelanosis (Petersen et al., 2015, Barnard et al., 2016). Interrogation of the P. acnes MLST8 isolate database, which contains information on a large collection of geographically widespread isolates and their clinical sources, reveals a statistically significant enrichment overall for strains from the type I clade in acneic versus healthy skin (P<0.001; Fishers exact test, two tailed), while those from the type II clade appear to show no association overall (P=0.213; Fishers exact test). More specifically, associations are found between acneic skin and strains from the type IA1 clonal complexes CC1 (RT1 and RT532) (P<0.01; Fishers exact test), CC3 (RT1, RT4 and RT5) (P=0.043; Fishers exact test) and CC4 (RT8) (P=0.021; Fishers exact test) (Figs 1 and S1). In a previous study, we found that a globally disseminated clonal lineage with the MLST genotype ST6 (Warwick MLST7 scheme analysis) or ST1 (MLST8 analysis) strikingly represented the majority of type IA1 isolates we analysed from a cohort of patients with acne (McDowell et al., 2011). In contrast, specific type II lineages (RT2 and RT6) belonging to CC72 (MLST8) appear to be associated with healthy skin on the basis of metagenomic and culture-based detection (McDowell et al., 2012; Fitz-Gibbon et al., 2013; Johnson et al., 2016). The observation that type II strains, but not those from the type I clade, encode CRISPR/Cas elements may be important in this context, thus preventing the acquisition of genetic loci that may contribute to virulence and acne pathophysiology (Fitz-Gibbon et al., 2013). For example, key type I lineages from CC3 (MLST8; Fig. 1), believed to be associated with acne contain a novel plasmid with a tight adhesion (Tad) locus and two unique genomic islands, known as loci 1 and 2, that contain genes proposed to enhance virulence via increased bacterial adhesion and host immune response (Fitz-Gibbon et al., 2013; Tomida et al., 2013; Kasimatis et al., 2013).
To conclude, we now propose P. acnes type I and II as distinct subspecies based on a polyphasic taxonomy approach. The growing number of genomes now becoming available for other propionibacteria will also provide an important opportunity to reexamine the genus and the place of the ‘cutaneous’ group within it.
Description of Propionibacterium acnes subsp. acnes subsp. nov.
P. acnes subsp. acnes (ac′nes Gr. n. acme a point; incorrectly transliterated as N.L. n. acne acne; N.L. gen.n. acnes of acne). Description based on McDowell et al. (2008), Niazi et al. (2010), Patrick & McDowell (2011), and Dekio et al. (2015).
Four phylogenetically distinct type I groups have been described, known as type IA1, IA2, IB and IC; type IA2, IB and IC represent phylogenetically tight clusters compared with IA1. Cells are Gram-stain-positive, non-motile, non-spore-forming, and anaerobic-to-aerotolerant. Colonies appear as lenticular, minute-to-4.0 mm, white, and can become tan, pink or orange in 3 weeks. Growth is most rapid at 30–37 °C. Surface colonies on blood agar (horse or rabbit) are punctiform-to-0.5 mm, circular, entire-to-pulvinate, translucent-to-opaque, white-to-gray and glistening. The cell shape after anaerobic culture in broth medium ranges from small plump rods to ellipsoids which tend to occur in pairs joined at a slight angle, and the size is approximately 0.4–0.5×0.8–0.9 µm. In defined medium broth culture, type IA and IC strains form a turbid suspension, while in PPY or BHI broth they form a settled granular sediment with a clear solution. In contrast, type IB strains form a slight fine sediment and turbid solution containing suspended cells. In suitable media with good growth, the final pH is 4.5–5.0. Generally catalase-positive, cultures need to be exposed to air for 1 h before testing. All strains have an absolute requirement for pantothenate, while thiamine, biotin and nicotinamide are stimulatory. Strains are co-haemolytic and variable for β-haemolytic activity and produce a number of extracellular enzymes including ribonuclease, neuraminidase, hyaluronidase, acid phosphatase, lecithinase and lipase. Strains of type IA produce relatively low levels of the putative co-haemolytic Christie–Atkins–Munch–Peterson (CAMP) factor 1, but type IB strains produce an abundance of this protein. The total quantity of acid (especially the proportion of lactic acid) produced from fermentable carbohydrates is highly variable. Cells ferment glucose, but not sucrose or maltose. Lactate is converted to propionate by most strains but only if the initial oxidation–reduction potential of the medium is sufficiently low, or if the initial growth rate is rapid. Sorbitol fermentation is a variable but defining characteristic of type I strains. Gelatin is hydrolysed, and most strains produce indole and reduce nitrate, but aesculin is not hydrolysed. The major long-chain fatty acid produced in thioglycolate cultures is 13-methyltetradecanoic acid (32–62 %) and iso-C15 : 0 is the predominant cellular fatty acid. Prominent mass ions obtained by MALDI-TOF mass spectrometry are at 3589 and 7179 Da. Peptidoglycan contains alanine, glutamic acid, glycine and ll-DAP. Cell wall sugars are glucose, mannose and galactose. Strains have been isolated from the human skin, oral cavity and genitourinary tract. Type IA1 and IC strains are associated with acne vulgaris. The DNA G+C content is approximately 60 % based on whole-genome sequencing analysis.
This subspecies is the type subspecies of P. acnes and contains the type strain according to Rules 40a and 40b of the Bacteriological Code (Lapage, 1992). The type strain is NCTC 737T (=ATCC 6919T=JCM 6425T=DSM 1897T=CCUG 1794T), the original type strain of the species, isolated from facial acne in London, 1920 (Genbank accession number NZ_JNHS00000000).
Description of Propionibacterium acnes subsp. defendens subsp. nov.
P. acnes subsp. defendens (de.fen′dens L. part. adj. defendens, defending, guarding, protecting; referring to the fact that strains have an active CRISPR/Cas system which guards or controls against foreign mobile genetic elements). Description based on McDowell et al. (2008), Niazi et al. (2010), Patrick & McDowell (2011), and Dekio et al. (2015).
Cells are Gram-stain-positive, non-motile, non-spore forming, and anaerobic-to-aerotolerant. Their cellular and colony morphology is similar to that of type I cells, but they may appear more coccoid and are most similar to previous descriptions of ‘Corynebacterium parvum’ which is a synonym for P. acnes . In defined medium broth culture, strains form a slight fine sediment and turbid solution containing suspended cells. In addition to pantothenate, some strains require haem and vitamin K to grow. Biochemical phenotype is similar to that of type I strains but with some notable differences. Cells are negative for β-haemolysis, and neuraminidase and lipase activity is infrequently found. Abundant levels of CAMP factor 1 are produced; similar to those observed with strains of type IB. Sorbitol fermentation is negative. The predominant cellular fatty acid is iso-C15 : 0 and prominent mass ions obtained by MALDI-TOF mass spectrometry are at 3628 and 7258 Da. Peptidoglycan contains alanine, glutamic acid, glycine, ll-DAP, and occasionally meso-DAP. Cell wall sugars are mannose and glucose, but galactose is not present. Strains have been isolated from the human skin surface, oral cavity and genitourinary tract. Strains are rarely associated with acne vulgaris and some may be associated with skin health and others with opportunistic infection. The DNA G+C content is approximately 60 % based on whole-genome sequencing analysis.
The type strain of P. acnes subspecies defendens subsp. nov. is ATCC11828 (=JCM 6473=CCUG 6369) isolated from a subcutaneous abscess (Genbank accession number NC_017550).
Description of Propionibacterium acnes subsp. elongatum subsp. nov.
The description for P. acnes subsp. elongatum is given in Dekio et al. (2015).
Supplementary Data
Supplementary File 1
Acknowledgements
E. B., J. L. and H. L. are funded by the National Institutes of Health (NIH) grant R01GM099530 from the National Institute of General Medical Sciences (NIGMS) awarded to H. L. J. L. is also supported by the Ruth L. Kirschstein National Research Service Award AI007323. This work was also supported by a grant of £11.5M awarded to Professor Tony Bjourson from European Union Regional Development Fund (ERDF) EU Sustainable Competitiveness Programme for Northern Ireland; Northern Ireland Public Health Agency (HSC R and D) and Ulster University.
Footnotes
Abbreviations: CRISPR, clustered regularly interspaced short palindromic repeats; HGT, horizontal gene transfer; MLSA, multi-locus sequence analysis.
Two supplementary figures are available with the online Supplementary Material.
References
- Bae Y., Ito T., Iida T., Uchida K., Sekine M., Nakajima Y., Kumagai J., Yokoyama T., Kawachi H., et al. Intracellular Propionibacterium acnes infection in glandular epithelium and stromal macrophages of the prostate with or without cancer. PLoS One. 2014;9:e90324. doi: 10.1371/journal.pone.0090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard E., Liu J., Yankova E., Cavalcanti S. M., Magalhães M., Li H., Patrick S., McDowell A. Strains of the Propionibacterium acnes type III lineage are associated with the skin condition progressive macular hypomelanosis. Sci Rep. 2016;6:31968. doi: 10.1038/srep31968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D., Stanley J., Krieg N. Classification of prokaryotic organisms and the concept of bacterial speciation. In: Boone D. R., Castenholz W., Garrity G. M., editors. Bergey's Manual of Systematic Bacteriology. 2nd edn. New York, NY: Springer; 2000. Edited by. [Google Scholar]
- Brüggemann H., Lomholt H. B., Kilian M. The flexible gene pool of Propionibacterium acnes . Mob Genet Elements. 2012a;2:145–148. doi: 10.4161/mge.21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemann H., Lomholt H. B., Tettelin H., Kilian M. CRISPR/cas loci of type II Propionibacterium acnes confer immunity against acquisition of mobile elements present in type I P. acnes . PLoS One. 2012b;7:e34171. doi: 10.1371/journal.pone.0034171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti S. M., de França E. R., Lins A. K., Magalhães M., de Alencar E. R., Magalhães V. Investigation of Propionibacterium acnes in progressive macular hypomelanosis using real-time PCR and culture. Int J Dermatol. 2011;50:1347–1352. doi: 10.1111/j.1365-4632.2011.04978.x. [DOI] [PubMed] [Google Scholar]
- Cohan F. M. Bacterial species and speciation. Syst Biol. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- Cohen R. J., Shannon B. A., McNeal J. E., Shannon T., Garrett K. L. Propionibacterium acnes associated with inflammation in radical prostatectomy specimens: a possible link to cancer evolution? J Urol. 2005;173:1969–1974. doi: 10.1097/01.ju.0000158161.15277.78. [DOI] [PubMed] [Google Scholar]
- Dekio I., Culak R., Misra R., Gaulton T., Fang M., Sakamoto M., Ohkuma M., Oshima K., Hattori M., et al. Dissecting the taxonomic heterogeneity within Propionibacterium acnes: proposal for Propionibacterium acnes subsp. acnes subsp. nov. and Propionibacterium acnes subsp. elongatum subsp. nov. Int J Syst Evol Microbiol. 2015;65:4776–4787. doi: 10.1099/ijsem.0.000648. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eishi Y. Etiologic link between sarcoidosis and Propionibacterium acnes . Respir Investig. 2013;51:56–68. doi: 10.1016/j.resinv.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon S., Tomida S., Chiu B. H., Nguyen L., Du C., Liu M., Elashoff D., Erfe M. C., Loncaric A., et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck Y., Soler C., Gérôme P., Vong R., Macnab C., Appere G., Vergnaud G., Pourcel C. A novel multiple locus variable number of tandem repeat (VNTR) analysis (MLVA) method for Propionibacterium acnes . Infect Genet Evol. 2015;33:233–241. doi: 10.1016/j.meegid.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Holland C., Mak T. N., Zimny-Arndt U., Schmid M., Meyer T. F., Jungblut P. R., Brüggemann H. Proteomic identification of secreted proteins of Propionibacterium acnes . BMC Microbiol. 2010;10:230. doi: 10.1186/1471-2180-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Cummins C. S. Cell wall composition and deoxyribonucleic acid similarities among the anaerobic coryneforms, classical propionibacteria, and strains of Arachnia propionica . J Bacteriol. 1972;109:1047–1066. doi: 10.1128/jb.109.3.1047-1066.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., Kang D., Barnard E., Li H. Strain-level differences in porphyrin production and regulation in Propionibacterium acnes elucidate disease associations. mSphere. 2016;1:e00023-15. doi: 10.1128/mSphere.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimatis G., Fitz-Gibbon S., Tomida S., Wong M., Li H. Analysis of complete genomes of Propionibacterium acnes reveals a novel plasmid and increased pseudogenes in an acne associated strain. Biomed Res Int. 2013;2013:918320. doi: 10.1155/2013/918320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapage S. P. In: International Code of Nomenclature of Bacteria (1990 Revision) Sneath P. H. A., Lessel E. F., Skerman V. B. D., Seeliger H. P. R., Clark W. A., editors. Washington D.C: ASM Press; 1992. Edited by. [PubMed] [Google Scholar]
- Liu J., Yan R., Zhong Q., Ngo S., Bangayan N. J., Nguyen L., Lui T., Liu M., Erfe M. C., et al. The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J. 2015;9:2078–2093. doi: 10.1038/ismej.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomholt H. B., Kilian M. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One. 2010;5:e12277. doi: 10.1371/journal.pone.0012277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J. Sexual isolation in bacteria. FEMS Microbiol Lett. 2001;199:161–169. doi: 10.1111/j.1574-6968.2001.tb10668.x. [DOI] [PubMed] [Google Scholar]
- McDowell A., Valanne S., Ramage G., Tunney M. M., Glenn J. V., McLorinan G. C., Bhatia A., Maisonneuve J. F., Lodes M., et al. Propionibacterium acnes types I and II represent phylogenetically distinct groups. J Clin Microbiol. 2005;43:326–334. doi: 10.1128/JCM.43.1.326-334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell A., Perry A. L., Lambert P. A., Patrick S. A new phylogenetic group of Propionibacterium acnes . J Med Microbiol. 2008;57:218–224. doi: 10.1099/jmm.0.47489-0. [DOI] [PubMed] [Google Scholar]
- McDowell A., Gao A., Barnard E., Fink C., Murray P. I., Dowson C. G., Nagy I., Lambert P. A., Patrick S. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology. 2011;157:1990–2003. doi: 10.1099/mic.0.049676-0. [DOI] [PubMed] [Google Scholar]
- McDowell A., Barnard E., Nagy I., Gao A., Tomida S., Li H., Eady A., Cove J., Nord C. E., Patrick S. An expanded multilocus sequence typing scheme for Propionibacterium acnes: investigation of ‘pathogenic’, ‘commensal’ and antibiotic resistant strains. PLoS One. 2012;7:e41480. doi: 10.1371/journal.pone.0041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell A., Nagy I., Magyari M., Barnard E., Patrick S. The opportunistic pathogen Propionibacterium acnes: insights into typing, human disease, clonal diversification and CAMP factor evolution. PLoS One. 2013;8:e70897. doi: 10.1371/journal.pone.0070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Hahnke R. L., Petersen J., Scheuner C., Michael V., Fiebig A., Rohde C., Rohde M., Fartmann B., et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E., Urbán E., Becker S., Kostrzewa M., Vörös A., Hunyadkürti J., Nagy I. MALDI-TOF MS fingerprinting facilitates rapid discrimination of phylotypes I, II and III of Propionibacterium acnes . Anaerobe. 2013;20:20–26. doi: 10.1016/j.anaerobe.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Niazi S. A., Clarke D., Do T., Gilbert S. C., Mannocci F., Beighton D. Propionibacterium acnes and Staphylococcus epidermidis isolated from refractory endodontic lesions are opportunistic pathogens. J Clin Microbiol. 2010;48:3859–3869. doi: 10.1128/JCM.01326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick S., McDowell A. The Propionibacteriaceae. In: Goodfellow M., Kämpfer P., Busse H.-J., Trujillo M. E., Suzuki K.-I., Ludwig W., Whitman B. W. B., editors. Bergey’s Manual of Systematic Bacteriology. 2nd edn. New York: Springer; 2011. Edited by. [Google Scholar]
- Perry A. L., Worthington T., Hilton A. C., Lambert P. A., Stirling A. J., Elliott T. S. Analysis of clinical isolates of Propionibacterium acnes by optimised RAPD. FEMS Microbiol Lett. 2003;228:51–55. doi: 10.1016/S0378-1097(03)00720-1. [DOI] [PubMed] [Google Scholar]
- Petersen R., Lomholt H. B., Scholz C. F., Brüggemann H. Draft genome sequences of two Propionibacterium acnes strains isolated from progressive macular hypomelanosis lesions of human skin. Genome Announc. 2015;3:e01250-15. doi: 10.1128/genomeA.01250-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollason J., McDowell A., Albert H. B., Barnard E., Worthington T., Hilton A. C., Vernallis A., Patrick S., Elliott T., Lambert P. Genotypic and antimicrobial characterisation of Propionibacterium acnes isolates from surgically excised lumbar disc herniations. Biomed Res Int. 2013;2013:530382. doi: 10.1155/2013/530382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz C. F. P., Jensen A., Lomholt H. B., Brüggemann H., Kilian M. A novel high-resolution single locus sequence typing scheme for mixed populations of Propionibacterium acnes in vivo . PLoS One. 2014;9:e104199. doi: 10.1371/journal.pone.0104199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz C. F. P., Brüggemann H., Lomholt H. B., Tettelin H., Kilian M. Genome stability of Propionibacterium acnes: a comprehensive study of indels and homopolymeric tracts. Sci Rep. 2016;6:20662. doi: 10.1038/srep20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G., Maiden M. C. Bacterial population genetics, evolution and epidemiology. Philos Trans R Soc Lond B Biol Sci. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J. E., Block D., Boulez K., Brenner S. E., Chervitz S. A., Dagdigian C., Fuellen G., Gilbert J. G., Korf I., et al. The Bioperl toolkit: perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida S., Nguyen L., Chiu B. H., Liu J., Sodergren E., Weinstock G. M., Li H. Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio. 2013;4:e00003. doi: 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunney M. M., Patrick S., Curran M. D., Ramage G., Hanna D., Nixon J. R., Gorman S. P., Davis R. I., Anderson N. Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J Clin Microbiol. 1999;37:3281–3290. doi: 10.1128/jcm.37.10.3281-3290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanne S., McDowell A., Ramage G., Tunney M. M., Einarsson G. G., O'Hagan S., Wisdom G. B., Fairley D., Bhatia A., et al. CAMP factor homologues in Propionibacterium acnes: a new protein family differentially expressed by types I and II. Microbiology. 2005;151:1369–1379. doi: 10.1099/mic.0.27788-0. [DOI] [PubMed] [Google Scholar]
- Webster G. F., Cummins C. S. Use of bacteriophage typing to distinguish Propionibacterium acne types I and II. J Clin Microbiol. 1978;7:84–90. doi: 10.1128/jcm.7.1.84-90.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1