Abstract
Chemoresistance has been a significant problem affecting the efficacy of drugs targeting tumors for decades. MGMT, known as O6-methylguanine-DNA methyltransferase, is a DNA repair enzyme that plays an important role in chemoresistance to alkylating agents. Hence, MGMT is considered a promising target for tumor treatment. Several methods are employed to detect MGMT, each with its own advantages and disadvantages. Some of the detection methods are; immunohistochemistry, methylation-specific PCR (MSP), pyrophosphate sequencing, MGMT activity test, and real-time quantitative PCR. Methylation of MGMT promoter is a key predictor of whether alkylating agents can effectively control glioma cells. The prognostic value of MGMT in glioma is currently being explored. The expression of MGMT gene mainly depends on epigenetic modification–methylation of CpG island of MGMT promoter. CpG island covers a length of 762 bp, with 98 CpG sites located at the 5' end of the gene, ranging from 480 to 1,480 nucleotides. The methylation sites and frequencies of CpG islands vary in MGMT-deficient tumor cell lines, xenografts of glioblastoma and in situ glioblastoma. Methylation in some regions of promoter CpG islands is particularly associated with gene expression. The change in the methylation status of the MGMT promoter after chemotherapy, radiotherapy or both is not completely understood, and results from previous studies have been controversial. Several studies have revealed that chemotherapy may enhance MGMT expression in gliomas. This could be through gene induction or selection of high MGMT-expressing cells during chemotherapy. Selective survival of glioma cells with high MGMT expression during alkylating agent therapy may change MGMT status in case of recurrence. Several strategies have been pursued to improve the anti-tumor effects of temozolomide. These include the synthesis of analogs of O6-meG such as O6-benzylguanine (O6-BG) and O6-(4-bromothenyl) guanine (O6-BTG), RNAi, and viral proteins. This review describes the regulation of MGMT expression and its role in chemotherapy, especially in glioma. Targeting MGMT seems to be a promising approach to overcome chemoresistance. Further studies exploring new agents targeting MGMT with better curative effect and less toxicity are advocated. We anticipate that these developments will improve the current poor prognosis of glioma patients.
Keywords: MGMT, methylation, alkylating agents, target therapy, chemotherapy
Introduction
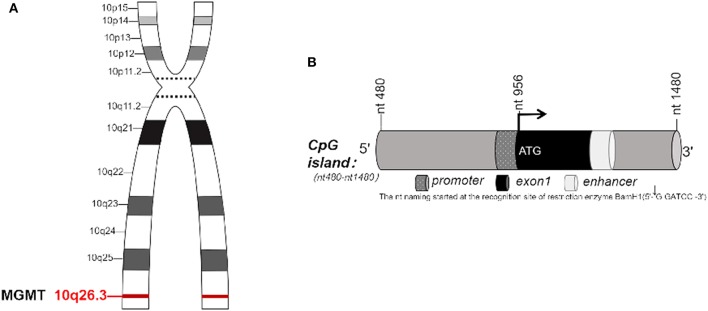
O6-methylguanine-DNA methyltransferase, known as MGMT, is a DNA “suicide” repair enzyme. It repairs damaged guanine nucleotides by transferring the methyl at O6 site of guanine to its cysteine residues, thus avoiding gene mutation, cell death and tumorigenesis caused by alkylating agents. MGMT gene is located on chromosome 10q26.3 (Figure 1A), with a total length of 300,437 bp (3, 4). The expression of MGMT gene is mainly regulated by epigenetic modification. Many studies have shown that the loss of MGMT expression is not due to gene deletion, mutation, rearrangement or unstable RNA, but due to methylation of CpG island of MGMT promoter (5–9). In 1987, Gardiner and Frommer discovered that the human MGMT gene has a CpG island with a length of 762 bp, with 98 CpG sites located at the 5' end of the gene, ranging from 480 to 1,480 nucleotides (nt) (Figure 1B). The transcription initiation site of the gene is nt 956, and the CpG island spans about 500 bp (1) at the 5' and 3' ends of the transcription initiation site. The nt naming was initiated by Harris et al. beginning from the recognition site (2) of the restriction enzyme BamH1. In the non-methylated state, the transcriptional initiation sites of MGMT adhere to four precisely located nucleosome-like structures, which fine-tunes the transcription of the gene. Methylation of CpG islands leads to heterochromatinization, accompanied by rearrangement and random localization of nucleosomes, thus obscuring the transcription initiation sites and making transcription devices unable to bind (10, 11). Other studies have shown that methylation and chromatin status modulate the transcription of MGMT gene by determining whether Sp1 and other transcription factors access the MGMT promoter (12).
Figure 1.
(A) MGMT gene is located on chromosome 10, q26.3. (B) CpG island in MGMT. (B) In 1987, Gardiner-Garden and Frommer (1) identified the CpG island with 762 bp in MGMT gene. It has 98 CpG sites, located on about 480–1,480 nucleotide (nt) at the 5' end of this gene. The transcription start site (TSS) is located at nt956, CpG island covers a length of 500 bp at both 5' end and 3' end of TSS. The name of nt was first coined by Harris et al. (2), derived from the recognition site of restriction enzyme BamH1.
Detection Methods for MGMT
Immunohistochemistry is a semi-quantitative method used to detect the expression of MGMT protein (13–15). It can distinguish between tumor cells and non-tumor cells, and thus the results are not confounded by the heterogeneity of tumors, but this method is greatly influenced by the subjectivity of the observer (16). In clinical research, many methods are used to detect the methylation status of MGMT promoter in glioma. The most frequently used method is methylation-specific PCR (MSP) (17, 18). This method was first established in 1996 for detection of promoter methylation (19). However, it can only detect the methylation of small fragments complementary to primers, and cannot determine the exact location of promoter methylation. Therefore, it is a semi-quantitative method (20). Whether conventional MSP primers (+120 toc +143, +173 to +196) bind to the key regions that regulate MGMT expression remains unclear. Pyrophosphate sequencing is a comprehensive sequencing method. In this method, methylation level of a single CpG site modified by sulfite can be quantitatively evaluated by efficient PCR amplification and sequencing. Therefore, Pyrophosphate sequencing is more reliable than MSP (21). However, the role of CpG island hypermethylation in gene silencing remains controversial (22–25). Many researchers hold the view that methylation of MGMT promoter directly inhibits gene transcription, thus detection of promoter methylation may serve as an indicator of susceptibility to alkylating agents. In fact, the most direct method used to detect the sensitivity of alkylating agents is MGMT activity test. The number of MGMT active molecules per unit protein or DNA detected by MGMT activity test can reflect the level of MGMT protein and RNA. However, because the test requires fresh or frozen tissues and in situ hybridization, it is not feasible for daily application. In contrast, quantitative detection of MGMT RNA expression by real-time quantitative PCR seems more suitable and highly sensitive, but few studies have applied it in MGMT RNA detection. Recently, Wang et al. found that a combination of immunohistochemistry and qMSP assays can provide high sensitivity and specificity for the prediction of MGMT status (26).
The Prediction and Prognostic Value of MGMT Promoter Status in Glioma
The MGMT gene encodes a DNA damage repair protein that removes alkylating agents resulting in resistance to chemotherapy. Because DNA methylation can inhibit transcription, methylation of MGMT promoter increases sensitivity to alkylating agents (27). Several studies have shown that methylation of MGMT promoter can predict whether alkylating agents can be of benefit in glioblastoma and low-grade gliomas (28–37). Two other clinical trials have revealed that methylation status of MGMT promoter can predict the prognosis of glioma patients. In these two studies, retrospective analysis of MGMT promoter methylation in elderly patients found that it could predict good prognosis in temozolomide (TMZ) group, but not in radiotherapy alone group (38, 39). The EORTC26951 clinical trial retrospectively analyzed the methylation status of MGMT promoter in anaplastic oligodendroglioma patients. It was found that methylation of MGMT promoter in anaplastic oligodendroglioma patients predicted better overall survival (OS) and PFS, whether in radiotherapy alone or in sequential radiotherapy and chemotherapy group [chemotherapy regimen: procarbacine, lomustine (CCNU), vincristine (PCV)]. But it had no prognostic value in glioblastoma patients. Elsewhere, it has been reported that methylation of MGMT promoter has no predictive value for chemosensitivity of anaplastic oligodendroglioma patients undergoing adjuvant PCV chemotherapy (40). Another phase III randomized clinical trial, NOA-04, drew a similar conclusion that methylation of MGMT promoter and IDH1 mutation reduces the risk of progression in anaplastic glioma patients, and patients with MGMT promoter methylation have a longer PFS (41) in both radiotherapy and chemotherapy groups (PVC). In addition, results from a phase III clinical trial prospectively indicate that MGMT promoter methylation status can be used as a biomarker to predict good prognosis of glioblastoma patients treated with TMZ (42) (Table 1).
Table 1.
Summary of the OS and PFS of patients receiving different treatments and characterized by non-methylated and methylated MGMT promoters in different studies.
| References | Pathology | Treatment | OS (months) | PFS (months) | ||||
|---|---|---|---|---|---|---|---|---|
| MGMTm | MGMTu | All | MGMTm | MGMTu | ALL | |||
| Criniere et al. (28) | GBM | RT+BCNU | 17.1 (14.5–26.5) | 13.1 (10.1–17.2) | NG | NG | ||
| RT | 15.1 (9.8–n.r.) | 10.2 (3.33–21.9) | NG | |||||
ALL
|
14.4 (13–16.1) | 13.6 (11.4–15.7) | 13.9 (12.5–15.3) | 7.33 (5.8–8.43) | 7.63 (6.47–8.63) | 7.37 (6.5–8.43) | ||
| Hegi et al. (29) | GBM | RT+TMZ | 21.7 (17.4–30.4) | 12.7 (11.6–14.4) | NG | 10.3 (6.5–14.0) | 5.3 (5.0–7.6) | NG |
| RT | 15.3 (13.0–20.9) | 11.8 (9.7–14.1) | 5.9 (5.3–7.7) | 4.4 (3.1–6.0) | ||||
| ALL | 18.2 (15.5–22.0) | 12.2 (11.4–13.5) | NG | NG | ||||
| Reifenberger et al. (30) | GBM (age ≥ 70) | RT+TMZ | 13.1 (11.0–15.3) | 10.4 (8.4–12.4) | 12.3(11.2–13.4) | 7.3 (6.2–8.5) | 7.2 (5.6–8.7) | 7.2 (6.3–8.0) |
| TMZ | 7.2 (5.6–8.9) | 2.6 (n.r.) | 6.8(4.8–8.8) | 6.8 (2.5–11.0) | 0.5 (n.r.) | 5.3 (0.1–10.5) | ||
| RT | 7.8 (3.4–12.2) | 8.8 (7.5–10.1) | 8.7(7.0–10.4) | 4.5 (3.5–5.4) | 5.2 (4.3–6.2) | 5.0 (4.4–5.6) | ||
| No treatment | 2.3 (0.8–3.8) | 2.0 (0.6–3.7) | 2.3(0.9–3.7) | 1.8 (1.1–2.4) | 1.7 (0.4–3.1) | 1.8 (1.0–2.5) | ||
| ALL | 8.4 (6.7–10.1) | 6.4 (3.9–8.9) | 7.7(6.3–9.0) | 5.2 (4.3–6.1) | 4.7 (3.8–5.5) | 4.8 (4.3–5.3) | ||
| Esteller et al. (31) | AA/GBM | ALL (RT+BCNU) | MGMTu/MGMTm: HR = 9.5 (95% CI: 3.0–42.7, p < 0.001) |
MGMTu/MGMTm: HR = 10.8 (95% CI: 4.4–30.8, p < 0.001) |
||||
| Hegi et al. (32) | GBM | ALL (RT+TMZ after surgery) |
MGMTu/MGMTm: The risk of death within 18 months after surgery: 92% vs. 38%; p = 0.002 | NG | ||||
| Everhard et al. (33) | LGG | ALL (TMZ) | NG | 29.5 (21.5–n.r.) | 6 (5–n.r.) | 28 (20–n.r.) | ||
| Pandith et al. (37) | Gliomas | RT+TMZ | 40.1 (29.8–50.3) | 6.8 (3.8–9.6) | 43.4 (32.5–54.1) | 23.9 (20.0–27.7) | 3.2 (0.6–5.8) | 25.8 (21.9–29.6) |
| Malmstrom et al. (38) | GBM (age≥60) | TMZ | 9.7 (8.0–11.4) | 6.8 (5.9–7.7) | 8.3 (7.1–9.5) | NG | ||
| Standard RT (60 Gy) | 8.2 (6.6–9.9)※ | 7.0 (5.7–8.3)※ | 6.0 (5.1–6.8) | |||||
| Hypofractioned RT (34 Gy) | 7.5 (6.5–8.6) | |||||||
| ALL | 9.0 (8.0–10.0) | 6.9 (5.9–7.9) | NG | |||||
| Wick et al. (39) | AA/GBM (age ≥ 65) | TMZ | n.r. (10.1–n.r.) | 7 (5.7–8.7) | 8.6 (7.3–110.2) | 8.4 (5.5–11.7) | 3.3 (3.0–3.5) | 3.3 (3.2–4.1) |
| RT | 9.6 (6.4–n.r.) | 10.4 (8–11.6) | 9.6 (8.2–10.8) | 4.6 (4.2–5.0) | 4.6 (3.7–6.3) | 4.7 (4.2–5.2) | ||
| ALL | 11.9 (9.0–n.r.) | 8.2 (7.0–10.0) | NG | 5.7 (5.0–7.4) | 3.5 (3.3–3.7) | NG | ||
| van den Bent et al. (40) | AOD/AOA (≥25% oligodendroglia elements) | RT RT+PVC | 59.3 (30.0–66.2) n.r. (n.r.) | 12.3 (11.5–28.5) 19.0 (12.3–34.5) | NG | 17.9 (11.9–43.4) 49.0 (19.1–71.2) | 7.8 (7.1–17.6) 10.5 (5.2–23.0) | NG |
| Wick et al. (41) | Anaplastic gliomas (WHO III) |
NG | MGMTu/MGMTm: | |||||
| RT | 72.1 (n.r.) | HR = 2.0 (95% CI: 1.1–3.6, p < 0.03) | 30.6 (16.3–42.8) | |||||
| TMZ/PCV | 82.6 (n.r.) | HR = 2.7 (95% CI: 1.4–5.1, p < 0.003) | 31.9 (21.1–37.3) | |||||
| Gilbert et al. (42) | GBM | Standard dose TMZ | 21.4 (17.6–29.0) | 14.6 (13.2–16.5) | 16.6 (14.9–18.0) | 6.5 (4.1–9.6) | 5.1 (4.3–5.7) | 5.5 (4.7–6.1) |
| Dense dose TMZ | 20.2 (15.4–25.1) | 13.3 (12.3–14.3) | 14.9 (13.7–16.5) | 10.1 (7.9–12.4) | 6.0 (5.5–6.5) | 6.7 (6.2–7.7) | ||
| ALL | 21.2 (17.9–24.8) | 14.0 (12.9–14.7) | NG | 8.7 (6.6–11.2) | 5.7 (5.1–6.1) | NG | ||
 , included other treatments: BCNU alone or supportive care; ※, standard RT and hypofractioned RT is grouped together; GBM, glioblastoma; LGG, low-grade gliomas; AA, anaplastic astrocytoma; AOD, anaplastic oligodendroglia tumors; AOA, anaplastic oligoastrocytoma; RT, radiotherapy; TMZ, temozolomide; BCNU, carmustine; PCV, procarbazine, lomustine, and vincristine; OS, median overall survival; PFS, median progression-free survival; MGMTm, MGMT promoter methylated status; MGMTu, MGMT promoter unmethylated status; NG, not given; n.r., not reached; HR, hazard ratio; 95%CI, 95 percent confidence interval.
, included other treatments: BCNU alone or supportive care; ※, standard RT and hypofractioned RT is grouped together; GBM, glioblastoma; LGG, low-grade gliomas; AA, anaplastic astrocytoma; AOD, anaplastic oligodendroglia tumors; AOA, anaplastic oligoastrocytoma; RT, radiotherapy; TMZ, temozolomide; BCNU, carmustine; PCV, procarbazine, lomustine, and vincristine; OS, median overall survival; PFS, median progression-free survival; MGMTm, MGMT promoter methylated status; MGMTu, MGMT promoter unmethylated status; NG, not given; n.r., not reached; HR, hazard ratio; 95%CI, 95 percent confidence interval.
Epigenetic Regulation of MGMT CpG Island
The methylation sites and frequencies of CpG islands vary among MGMT-deficient tumor cell lines, xenografts of glioblastoma and in situ glioblastoma. Pieper et al. used Linker-mediated PCR (LMPCR) to detect the methylation status of MGMT promoter. It was found that the changes in methylation level of MGMT promoter mainly occurred at four CpG loci in cell lines expressing MGMT and those lacking MGMT, rather than being distributed uniformly throughout the CpG island. Two of them are located at about 130 nucleotides (+130) downstream of the transcription initiation site (TSS), including the sites recently studied using MPS. Two other nucleotides (−200) are located upstream of the transcription initiation site, and the transcription factor binding sites in both cell lines are not methylated. The transcription initiation site is defined as 0 (43). Watts et al. performed bisulfite sequencing PCR (BSP) on 108 CpG loci of 8226/s and 8226/v promoter CpG islands, respectively, and found that 8226/v has three methylation-rich regions which differs from those of 8226/s: −446 to −353, −265 to −162 and +112 to +212 (11). Costello et al. analyzed the methylation status of CpG loci in MGMT promoter −252 to −155 and −90 to +65 regions of glioma cell line by LM-PCR, and found that 21 of 25 loci were negatively correlated with MGMT gene expression (12). Because the authors detected the methylation level in high, low and non-expressing cell lines in these two regions, a quantitative relationship could not be established. Qian et al. used BSP method to detect the methylation level of CpG loci from −249 to +259 in MGMT CpG island region. It was found that HT29, a cell line expressing MGMT, was almost not methylated in this region, whereas BE, a cell line not expressing MGMT, was heavily methylated in each clone in this region. The most frequently methylated regions ranged from −249 to −103, +107 to +196 (44). Malley et al. used pyrophosphatic acid sequencing to detect the methylation of CpG islands in the entire MGMT promoter of glioblastoma cell lines, xenografts and normal brain tissues (41 samples). It was found that the +152 to +214 were the key regions promoting the transcription of MGMT (45). Subsequently, methylation of MGMT promoter was studied in human glioma samples. It was found that the methylation of CpG loci at −186 to −172 and +93 to +153 regions was most correlated with MGMT gene expression, but previous MSP loci were not found in this region, although the methylation level of MSP loci was similar to that reported previously (46). Bady et al. used human methylation 450 gene chip (HM-450K) to detect 14 CpG loci of MGMT promoter in 63 glioblastoma samples. It was found that the methylation of −193 and +173 CpG loci was negatively correlated with gene expression and had a good predictive accuracy for prognosis (47). Similarly, Mur et al. obtained genome-wide methylation profiles of 247 glioma samples from HM-450K platform, including 25 CpG loci in CpG island of MGMT promoter region. The methylation of + 173 CpG loci was significantly associated with overall survival (48). These researchers also found that MGMT promoter CpG islands are not suitable for methylation and this do not regulate expression or predict the prognosis of patients. Everhard et al. found that MGMT promoter regions −452 to −399 were highly methylated in both tumors and normal brain tissues. The region −90 to +69 is the first CpG region of small promoter, TSS and non-coding exon, which is equivalent to the methylation-free region (46) in both normal brain tissue and tumors. Thus, transcriptional silencing does not require methylation of the entire CpG island, but only methylation of several gene-specific core CpG sites. Therefore, methylation at some regions of promoter CpG islands is particularly associated with gene expression.
The Influence of Chemotherapy and Radiotherapy on the Methylation Status, Activity and Protein Expression of MGMT
Methylation of the promoter region of the MGMT gene is known to predict the response to alkylating agent's treatment in glioma patients. However, knowledge about the change in the methylation status of the MGMT promoter after chemotherapy, radiotherapy or both is still incomplete. Wiewrodt et al. analyzed MGMT activity in 40 paired primary and recurrent glioblastomas, 16 patients after RT only, 24 patients with RT combined with chemotherapy (TMZ and/or CCNU or ACNU). In both recurrent groups, the MGMT activity was higher than in primary tumors. In contrast, for patients who received RT only, there was no significant difference between primary glioblastomas and recurrences. The MGMT activity was significant, however, in patients with primary glioblastomas and recurrences that received RT plus alkylating agent therapy (49). Brandes et al. analyzed MGMT promoter methylation status of 38 paired primary and recurrent glioblastomas treated with adjuvant radiotherapy and chemotherapy. They found that MGMT methylation status was changed in 14 patients (37%) who had recurrent tumors and more frequently in those with methylated MGMT than in unmethylated patients (50). Christmann et al. compared MGMT activity and MGMT promoter methylation in 46 primary glioblastoma samples and 19 recurrent glioblastoma samples. They found that MGMT activity increased after treatment, and methylation of MGMT promoter was detected in 39% primary tumors, while only 5.3% recurrent glioblastomas displayed MGMT promoter methylation (17). Elsewhere, Felsberg et al. analyzed the methylation status of MGMT promoter in 80 paired primary and recurrent glioblastomas, of which 16 patients received radiotherapy alone and 64 patients received radiotherapy and TMZ chemotherapy. They found the MGMT methylation status of 89% patients was not altered (51). It is worth noting, besides, that the response of the human MGMT promoter to genotoxic stress may be weak. Although Fritz et al. and Chan et al. had reported MGMT mRNA transcription can be induced by DNA-damaging treatments, both of their experiments were limited in rat H4IIE hepatoma cells (52, 53). As for human MGMT promoter, Grombacher et al. found that it could be induced by dexamethasone when transfected into rat H4IIE and human HeLa S3 (Mex+) cells, but methylating agents and ionizing radiation only worked in H4IIE cells (54). Boldogh et al. analyzed the mechanism of human MGMT expression induction, they found that protein kinase C-mediated signaling played an important role, involving activation of AP-1 sites on MGMT promoter by TPA (55). Aasland et al. further identified that human MGMT promoter can be induced by glucocorticoids, but not by genotoxic stress, in human malignant glioblastoma cells (56). They put forward that a cluster of SP1 sites in human MGMT promoter prevented transcriptional up-graduation and overshadowed activation signals from other weaker transcriptional factors. The transcription factor SP1 was sequestrated by p53, which was induced following radiochemotherapy (57). Coincidentally, an earlier retrospective clinical data from Pitter at al. showed longer survival of no glucocorticoid usage GBM patient cohorts, alongside corresponding data in animal models (58). Thus, radiation and chemotherapy may have minor influence on transient transcriptional activation of human MGMT. The finding of a protection of tumor cells by dexamethasone and other steroids suggests that a controlled use of glucocoricoids in GBM therapy is desirable. In conclusion, these studies revealed that chemotherapy may provoke an up-regulation of MGMT expression in gliomas through selection of high MGMT expressing cells during chemotherapy. Selective survival of glioma cells with high MGMT expression during alkylating agent therapy may change MGMT status when recurrence.
MGMT in Glioblastoma Stem Cells
Cancer stem cells have been implicated in the progression and recurrence of GBMs. It has been recognized that even after effective treatment of tumors, minimal residual stem cells may be activated to enter a new stage of differentiation and proliferation. In this way, cancer stem cells promote the recurrence of tumors. Thus, we postulate that glioblastoma stem cells may cause resistance to TMZ, which enables them survive during chemotherapy. Liu et al. and Pistollato et al. revealed that glioblastoma (GBM) stem cells, identified with the stem cell marker CD133, express high level of MGMT and displayed strong tumor resistance to TMZ (59, 60). Beier et al. reported that there are distinct stem cell populations that, despite having similar MGMT promoter methylation status, differ in MGMT protein expression. And they also found that TMZ preferentially kills cancer stem cells in glioblastoma in MGMT-negative cell lines (61, 62). Mantwill et al. stated that MGMT is not expressed in all stem cell lines, which indicates that these cells have different grades of TMZ resistance (63). Happold et al. observed that differentiation of glioma stem cells resulted in a gradual loss of MGMT expression and increased TMZ sensitivity (64, 65). Although MGMT is highly expressed in stem cells, it is not clear why the alkylating agents are not effective in recurrent GBMs. Do the differentiated cells retain the TMZ resistance features of stem cells? These challenges necessitate the search for the mechanisms that regulate the expression of MGMT in different cell stages.
Targeting MGMT Protein
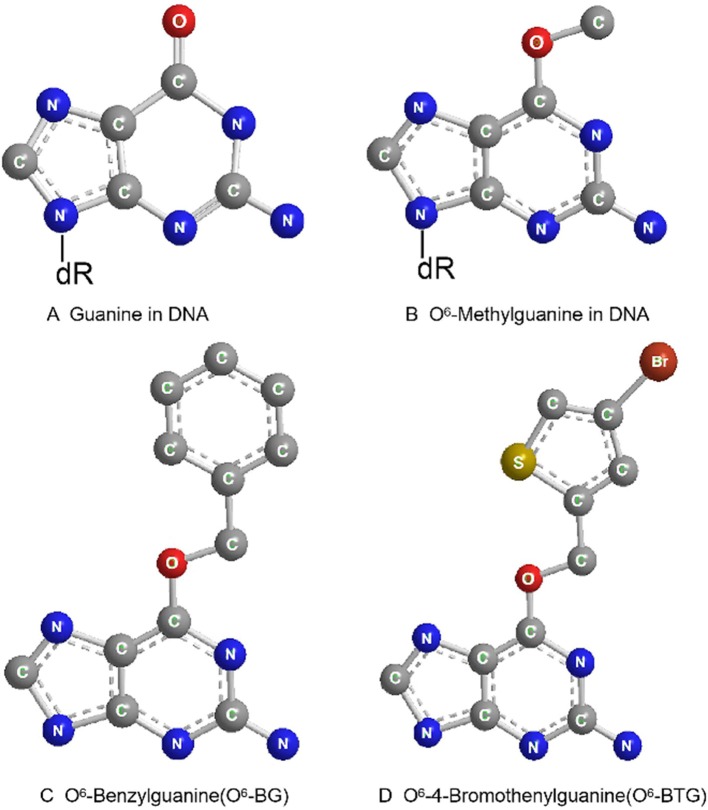
O6-benzylguanine (O6-BG) is the analog of O6-meG which is a low molecular weight pseudosubstrate for MGMT. It inactivates MGMT through alkyl group transfer (Figure 2). It can pass the blood brain barrier and has, therefore, the potential to be a treatment for gliomas. It has been widely used as an MGMT inhibitor and as a sensitizer of glioma cells to alkylating agent TMZ (66, 67). Koch et al. found that local intracranial interaction of O6-BG with TMZ after intraoperative removal of brain tumors might delay tumor recurrence without any side effect (68). Phase I, II and III clinical trials of O6-BG combined with TMZ have revealed that this combination successfully aberrates other tumors, such as brain tumor, melanoma, lymphoma and colon cancer (69–72). A later phase II clinical research by Quinn et al. found that 06-BG combined with gliadel wafer prolonged the survival time of patients. However, it also increased the risk of hydrocephalus, cerebrospinal fluid (CSF) leak, and CSF/brain infection (73).
Figure 2.
Molecular structure of guanine in DNA (A), O6-Methylguanine in DNA (B), O6-Benzylguanine (C), and O6-4-Bromothenylguanine (D).
Another pseudosubstrate, O6-(4-bromothenyl) guanine (O6-BTG), has 10-fold higher potency than O6-BG in inactivating MGMT protein and is orally bioavailable without inherently toxic (Figure 2). It has been reported that O6-BTG efficiently and rapidly inactivates MGMT in various tumors in vivo and in v vitro and significantly increases tumor sensitivity to TMZ (74–77). A phase I trial of O6-BTG in combination with TMZ in advanced solid tumors established an oral ATase-depleting dose of lomeguatrib (Trade name for O6-BTG) and developed a combination regimen with TMZ that was 75% of the maximum tolerable dose of the single agent. The dose-limiting toxicity of O6-BTG was myelosuppression (78). Papachristodoulou et al. reported that a liposomal O6BTG can efficiently target MGMT, thereby sensitizing murine and human glioma cells to TMZ in vitro and magnetic resonance image-guided microbubble-enhanced low-intensity pulsed focused ultrasound mediates the delivery of the stable liposomal MGMT inactivator into the tumor region resulting in complete MGMT depletion in vivo (79).
Although the developed MGMT inhibitors, O6-BG and O6-BTG, are effective, their systemic toxicity due to non-specific targeting to normal cells cannot be ignored. Going forward, the high glucose consumption hallmark of tumor cells presents a new avenue that can be exploited for development of selective inhibitors by conjugating agents to glucose. It has been reported that both O6BG-Glu and O6BTG-Glu are highly effective at inhibiting MGMT in several cancer cell lines, including T98G glioblastoma. These agents also enhance the cell-killing effect of temozolomide (80–82). Besides, Tomaszowski et al. found that glucose conjugates are subject of transport out of the cell by the ATP-binding cassette (ABC) transporter mediated efflux, which impacts the efficiency of MGMT inhibition. In this study, the importance of proper linker selection for a successful ligand-based drug delivery strategy was underscored (83). Similarly, conjugating pseudosubstrates to folate esters is another promising strategy to target tumor cells (84). So far, few studies have investigated the cellular effects of glucose or folate esters conjugated inhibitors. Further detailed studies should unravel the mechanisms of these inhibitors to provide better treatment agents.
Watson et al. conducted phase II clinical trials to evaluate the efficacy of lomeguatrib in patients with melanoma. They found that lomeguatrib plus TMZ had a greater MGMT inactivation than did TMZ treatment alone (85). Another study by the same group also reported that lomeguatrib can be applied in prostate, primary CNS, and colorectal cancers to inactivate MGMT (86). However, lomeguatrib increases myelosuppression, and other studies revealed that it did not improve the response rate to TMZ (69, 70, 74, 87–89).
RNA interference is another promising therapy targeting MGMT. Kato et al. reported that when combined with TMZ, the MGMT-siRNA/liposome complex exerted a strong synergistic antitumor effect (90). Zhang et al. found that miR-181d downregulated MGMT by directly interacting with MGMT 3'UTR, and this potentiated the TMZ sensitizer as an MGMT targeting therapy (91). Nie et al. found that miR-198 directly targeted MGMT by binding to the 3′-UTR of MGMT, thereby inhibiting the MGMT mRNA translation in GBM cells. MiR-198 restored the tumor sensitivity to TMZ in glioblastomas overexpressing MGMT (92).
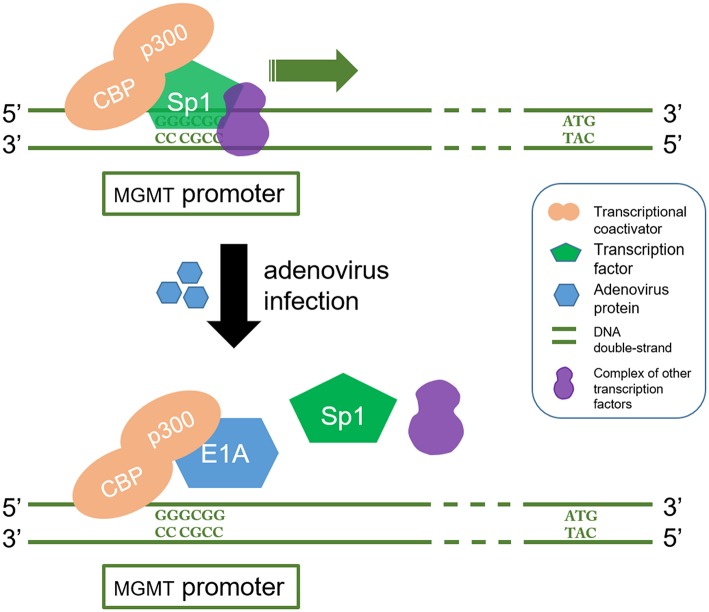
Oncolytic viruses which inactivate or leverage the cellular DNA-repair machinery to achieve productive replication have also been exploited to design agents targeting MGMT. Adenoviruses express proteins which can downregulate MGMT expression. It has been reported that overexpression of adenovirus E1A, which binds p300, efficiently inhibits both basal and TSA inducible promoter activity of MGMT and may thus reduce chemoresistance (93, 94). CBP/p300 is a transcriptional coactivator which interacts with multiple transcription factors including those involved in MGMT gene. It plays an important role in many cellular processes, and the structural and functional versatility of CBP/p300 are yet to be fully elucidated. For this reason, the utilization of adenovirus targeting MGMT is far from being clinically implemented (95) (Figure 3).
Figure 3.
E1A interferes with CBP/p300 in which regulate the transcription process of MGMT gene. CBP/p300 is a transcription coactivator and Sp1 is a transcription factor. CBP/p300 recruits Sp1 protein to recognize and bind to the GC region (5'-GGGCGG-3') in the MGMT promoter, which in turn acts as a transcriptional activator. On the other hand, E1A is a product of oncolytic adenovirus. It stops the Sp1 from being recruited by binding to p300. This blocks the MGMT gene transcription signal.
Jiang et al. reported that a combination of TMZ and viral therapy may overcome the chemoresistance of gliomas to TMZ (96). Further, it has been shown that oncolytic virus-mediated manipulation of DNA damage responses can also be applied to kill GSCs (97). The discovery of this oncolytic viral therapy opens a new era in cancer therapy. However, there are safety concerns regarding the use of virus-based therapy.
Other drugs that target MGMT protein such as disulfiram (DSF) have also been studied. For instance, Paranjpe reported that DSF directly suppressed MGMT protein expression through sole site Cys145 (98).
Conclusion and Perspectives
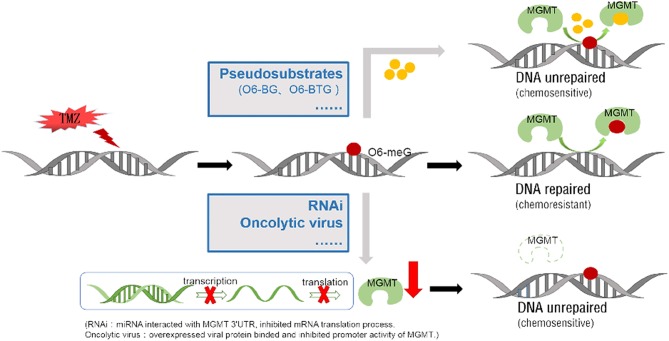
MGMT is a DNA methyltransferase which repairs damaged DNA thus avoiding cell death caused by alkylating agents. The expression of MGMT gene is mainly regulated by epigenetic modification. Several methods have been developed for MGMT detection including immunohistochemistry, methylation-specific PCR, pyrophosphate sequencing, MGMT activity test, real-time quantitative PCR among others. Methylation of MGMT promoter can predict whether alkylating agents are effective for glioblastoma and low-grade gliomas. The prognostic value of MGMT methylation is still controversial and calls for further clarification. Epigenetic regulation of specific sites of MGMT CpG island influences MGMT transcription. Chemotherapy and radiotherapy may modulate MGMT methylation status, activity and protein expression. TMZ is a promising chemotherapeutic agent for glioma, but the rapid development of drug resistance poses a huge challenge. Overexpression of MGMT is an important mechanism of TMZ resistance. Several strategies have been pursued to improve the anti-tumor effects of TMZ. These include development of pseudosubstrates, RNAi, viral proteins and many others agents (Figure 4). Given on-going research advancements in this field, the current poor prognosis of glioma patients is expected to improve.
Figure 4.
Two main approaches to block methylated DNA repair by targeting the MGMT protein. TMZ can add methyl to the guanine on the DNA molecule (form as 06-meG), which then induce DNA cross-linking. MGMT protein can repair DNA damage by binding and transferring the methyl on it. Low molecular weight O6-meG analogs, like O6-BG and O6-BTG, are used as pseudosubstrates to bind MGMT protein and reduce the methyl transfer activity. Specific miRNA/ liposome complexes which interact with MGMT 3'UTR then inhibit the MGMT mRNA translation process (RNAi). The proliferated oncolytic virus in a host can inhibit the host's MGMT promoter's activation, by means of the E1A binding to the p300.
Author Contributions
AS designed the study and determined the final version. QW revised the manuscript and polished the language. WY drafted the manuscript. LZ made the figures and tables. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- MGMT
O6-methylguanine-DNA methyltransferase
- MSP
methylation-specific PCR
- IHC
immunohistochemistry
- PCV
methylbenzylhydrazine, cyclohexanisolone and vincristine
- OS
overall survival
- PFS
progression-free survival
- IDH1
isocitrate dehydrogenase
- LMPCR
Linker-mediated PCR
- TSS
transcription initiation site
- BSP
Bisulfite Sequencing PCR
- O6-BG
O6-benzylguanine
- TMZ
temozolomide
- CSF
cerebrospinal fluid
- O6-BTG
O6-(4-bromothenyl) guanine
- DSF
disulfiram.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (81701144) and China Postdoctoral Science Foundation (2017M612010).
References
- 1.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. (1987) 196:261–82. 10.1016/0022-2836(87)90689-9 [DOI] [PubMed] [Google Scholar]
- 2.Harris LC, Potter PM, Tano K, Shiota S, Mitra S, Brent TP. Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene. Nucleic Acids Res. (1991) 19:6163–7. 10.1093/nar/19.22.6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. (2002) 12:656–64. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. (2005) 33:D501–4. 10.1093/nar/gki025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day RS, III, Ziolkowski CH, Scudiero DA, Meyer SA, Lubiniecki AS, Girardi AJ, et al. Defective repair of alkylated DNA by human tumour and SV40-transformed human cell strains. Nature. (1980) 288:724–7. 10.1038/288724a0 [DOI] [PubMed] [Google Scholar]
- 6.Fornace AJ, Jr, Papathanasiou MA, Hollander MC, Yarosh DB. Expression of the O6-methylguanine-DNA methyltransferase gene MGMT in MER+ and MER- human tumor cells. Cancer Res. (1990) 50:7908–11. [PubMed] [Google Scholar]
- 7.Pieper RO, Futscher BW, Dong Q, Ellis TM, Erickson LC. Comparison of O-6-methylguanine DNA methyltransferase (MGMT) mRNA levels in Mer+ and Mer- human tumor cell lines containing the MGMT gene by the polymerase chain reaction technique. Cancer Commun. (1990) 2:13–20. 10.3727/095535490820874812 [DOI] [PubMed] [Google Scholar]
- 8.Kroes RA, Erickson LC. The role of mRNA stability and transcription in O6-methylguanine DNA methyltransferase (MGMT) expression in Mer+ human tumor cells. Carcinogenesis. (1995) 16:2255–7. 10.1093/carcin/16.9.2255 [DOI] [PubMed] [Google Scholar]
- 9.Bouras E, Karakioulaki M, Bougioukas KI, Aivaliotis M, Tzimagiorgis G, Chourdakis M. Gene promoter methylation and cancer: an umbrella review. Gene. (2019) 710:333–40. 10.1016/j.gene.2019.06.023 [DOI] [PubMed] [Google Scholar]
- 10.Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. (1994) 269:17228–37. [PubMed] [Google Scholar]
- 11.Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. (1997) 17:5612–9. 10.1128/MCB.17.9.5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. (1994) 14:6515–21. 10.1128/MCB.14.10.6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micko ASG, Hoftberger R, Wohrer A, Millesi M, Knosp E, Wolfsberger S. MGMT assessment in pituitary adenomas: comparison of different immunohistochemistry fixation chemicals. Pituitary. (2018) 21:266–73. 10.1007/s11102-018-0862-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Reardon DA, Desjardins A, Vredenburgh JJ, Quinn JA, Austin AD, et al. O6-methylguanine-DNA methyltransferase (MGMT) immunohistochemistry as a predictor of resistance to temozolomide in primary CNS lymphoma. J Neurooncol. (2013) 114:135–40. 10.1007/s11060-013-1162-y [DOI] [PubMed] [Google Scholar]
- 15.Mason S, McDonald K. MGMT testing for glioma in clinical laboratories: discordance with methylation analyses prevents the implementation of routine immunohistochemistry. J Cancer Res Clin Oncol. (2012) 138:1789–97. 10.1007/s00432-012-1312-1 [DOI] [PubMed] [Google Scholar]
- 16.Preusser M, Charles Janzer R, Felsberg J, Reifenberger G, Hamou MF, Diserens AC, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. (2008) 18:520–32. 10.1111/j.1750-3639.2008.00153.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christmann M, Nagel G, Horn S, Krahn U, Wiewrodt D, Sommer C, et al. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. Int J Cancer. (2010) 127:2106–18. 10.1002/ijc.25229 [DOI] [PubMed] [Google Scholar]
- 18.Yachi K, Watanabe T, Ohta T, Fukushima T, Yoshino A, Ogino A, et al. Relevance of MSP assay for the detection of MGMT promoter hypermethylation in glioblastomas. Int J Oncol. (2008) 33:469–75. 10.3892/ijo_00000029 [DOI] [PubMed] [Google Scholar]
- 19.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. (1996) 93:9821–6. 10.1073/pnas.93.18.9821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Switzeny OJ, Christmann M, Renovanz M, Giese A, Sommer C, Kaina B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin Epigenetics. (2016) 8:49. 10.1186/s13148-016-0204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tost J, El abdalaoui H, Gut IG. Serial pyrosequencing for quantitative DNA methylation analysis. Biotechniques. (2006) 40:721–2. 10.2144/000112190 [DOI] [PubMed] [Google Scholar]
- 22.Schnell O, Albrecht V, Pfirrmann D, Eigenbrod S, Krebs B, Romagna A, et al. MGMT promoter methylation is not correlated with integrin expression in malignant gliomas: clarifying recent clinical trial results. Med Oncol. (2018) 35:103 10.1007/s12032-018-1162-z [DOI] [PubMed] [Google Scholar]
- 23.Toffolatti L, Scquizzato E, Cavallin S, Canal F, Scarpa M, Stefani PM, et al. MGMT promoter methylation and correlation with protein expression in primary central nervous system lymphoma. Virchows Arch. (2014) 465:579–86. 10.1007/s00428-014-1622-6 [DOI] [PubMed] [Google Scholar]
- 24.Melguizo C, Prados J, Gonzalez B, Ortiz R, Concha A, Alvarez PJ, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med. (2012) 10:250. 10.1186/1479-5876-10-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uno M, Oba-Shinjo SM, Camargo AA, Moura RP, Aguiar PH, Cabrera HN, et al. Correlation of MGMT promoter methylation status with gene and protein expression levels in glioblastoma. Clinics. (2011) 66:1747–55. 10.1590/S1807-59322011001000013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Li Z, Liu C, Chen L, Liu L, Hu Z, et al. Comparative assessment of three methods to analyze MGMT methylation status in a series of 350 gliomas and gangliogliomas. Pathol Res Pract. (2017) 213:1489–93. 10.1016/j.prp.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 27.Kaina B, Christmann M. DNA repair in personalized brain cancer therapy with temozolomide and nitrosoureas. DNA Repair. (2019) 78:128–41. 10.1016/j.dnarep.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 28.Criniere E, Kaloshi G, Laigle-Donadey F, Lejeune J, Auger N, Benouaich-Amiel A, et al. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. (2007) 83:173–9. 10.1007/s11060-006-9320-0 [DOI] [PubMed] [Google Scholar]
- 29.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. (2005) 352:997–1003. 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 30.Reifenberger G, Hentschel B, Felsberg J, Schackert G, Simon M, Schnell O, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. (2012) 131:1342–50. 10.1002/ijc.27385 [DOI] [PubMed] [Google Scholar]
- 31.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. (2000) 343:1350–4. 10.1056/NEJM200011093431901 [DOI] [PubMed] [Google Scholar]
- 32.Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. (2004) 10:1871–4. 10.1158/1078-0432.CCR-03-0384 [DOI] [PubMed] [Google Scholar]
- 33.Everhard S, Kaloshi G, Criniere E, Benouaich-Amiel A, Lejeune J, Marie Y, et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. (2006) 60:740–3. 10.1002/ana.21044 [DOI] [PubMed] [Google Scholar]
- 34.Aoki K, Natsume A. Overview of DNA methylation in adult diffuse gliomas. Brain Tumor Pathol. (2019) 36:84–91. 10.1007/s10014-019-00339-w [DOI] [PubMed] [Google Scholar]
- 35.Qi F, Yin Z, Wang G, Zeng S. Clinical and prognostic significance of O(6)-methylguanine-DNA methyltransferase promoter methylation in patients with melanoma: a systematic meta-analysis. Ann Dermatol. (2018) 30:129–35. 10.5021/ad.2018.30.2.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahlrot RH, Larsen P, Boldt HB, Kreutzfeldt MS, Hansen S, Hjelmborg JB, et al. Posttreatment effect of MGMT methylation level on glioblastoma survival. J Neuropathol Exp Neurol. (2019) 78:633–40. 10.1093/jnen/nlz032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandith AA, Qasim I, Zahoor W, Shah P, Bhat AR, Sanadhya D, et al. Concordant association validates MGMT methylation and protein expression as favorable prognostic factors in glioma patients on alkylating chemotherapy (Temozolomide). Sci Rep. (2018) 8:6704. 10.1038/s41598-018-25169-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malmstrom A, Gronberg BH, Marosi C, Stupp R, Frappaz D, Schultz H, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. (2012) 13:916–26. 10.1016/S1470-2045(12)70265-6 [DOI] [PubMed] [Google Scholar]
- 39.Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. (2012) 13:707–15. 10.1016/S1470-2045(12)70164-X [DOI] [PubMed] [Google Scholar]
- 40.van den Bent MJ, Dubbink HJ, Sanson M, van der Lee-Haarloo CR, Hegi M, Jeuken JW, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. (2009) 27:5881–6. 10.1200/JCO.2009.24.1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wick W, Hartmann C, Engel C, Stoffels M, Felsberg J, Stockhammer F, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. (2009) 27:5874–80. 10.1200/JCO.2009.23.6497 [DOI] [PubMed] [Google Scholar]
- 42.Gilbert MR, Wang M, Aldape KD, Stupp R, Hegi M, Jaeckle KA, et al. RTOG 0525: a randomized phase III trial comparing standard adjuvant temozolomide (TMZ) with a dose-dense (dd) schedule in newly diagnosed glioblastoma (GBM). J Clin Oncol. (2011) 29:2006 10.1200/jco.2011.29.15_suppl.2006 [DOI] [Google Scholar]
- 43.Pieper RO, Patel S, Ting SA, Futscher BW, Costello JF. Methylation of CpG island transcription factor binding sites is unnecessary for aberrant silencing of the human MGMT gene. J Biol Chem. (1996) 271:13916–24. 10.1074/jbc.271.23.13916 [DOI] [PubMed] [Google Scholar]
- 44.Qian XC, Brent TP. Methylation hot spots in the 5' flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. (1997) 57:3672–7. [PubMed] [Google Scholar]
- 45.Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. (2011) 121:651–61. 10.1007/s00401-011-0803-5 [DOI] [PubMed] [Google Scholar]
- 46.Everhard S, Tost J, El Abdalaoui H, Criniere E, Busato F, Marie Y, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. (2009) 11:348–56. 10.1215/15228517-2009-001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. (2012) 124:547–60. 10.1007/s00401-012-1016-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mur P, Rodriguez de Lope A, Diaz-Crespo FJ, Hernandez-Iglesias T, Ribalta T, Fiano C, et al. Impact on prognosis of the regional distribution of MGMT methylation with respect to the CpG island methylator phenotype and age in glioma patients. J Neurooncol. (2015) 122:441–50. 10.1007/s11060-015-1738-9 [DOI] [PubMed] [Google Scholar]
- 49.Wiewrodt D, Nagel G, Dreimuller N, Hundsberger T, Perneczky A, Kaina B. MGMT in primary and recurrent human glioblastomas after radiation and chemotherapy and comparison with p53 status and clinical outcome. Int J Cancer. (2008) 122:1391–9. 10.1002/ijc.23219 [DOI] [PubMed] [Google Scholar]
- 50.Brandes AA, Franceschi E, Tosoni A, Bartolini S, Bacci A, Agati R, et al. O(6)-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. (2010) 12:283–8. 10.1093/neuonc/nop050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. (2011) 129:659–70. 10.1002/ijc.26083 [DOI] [PubMed] [Google Scholar]
- 52.Fritz G, Tano K, Mitra S, Kaina B. Inducibility of the DNA repair gene encoding O6-methylguanine-DNA methyltransferase in mammalian cells by DNA-damaging treatments. Mol Cell Biol. (1991) 11:4660–8. 10.1128/MCB.11.9.4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan CL, Wu Z, Eastman A, Bresnick E. Irradiation-induced expression of O6-methylguanine-DNA methyltransferase in mammalian cells. Cancer Res. (1992) 52:1804–9. [PubMed] [Google Scholar]
- 54.Grombacher T, Mitra S, Kaina B. Induction of the alkyltransferase (MGMT) gene by DNA damaging agents and the glucocorticoid dexamethasone and comparison with the response of base excision repair genes. Carcinogenesis. (1996) 17:2329–36. 10.1093/carcin/17.11.2329 [DOI] [PubMed] [Google Scholar]
- 55.Boldogh I, Ramana CV, Chen Z, Biswas T, Hazra TK, Grosch S, et al. Regulation of expression of the DNA repair gene O6-methylguanine-DNA methyltransferase via protein kinase C-mediated signaling. Cancer Res. (1998) 58:3950–6. [PubMed] [Google Scholar]
- 56.Aasland D, Reich TR, Tomicic MT, Switzeny OJ, Kaina B, Christmann M. Repair gene O(6) -methylguanine-DNA methyltransferase is controlled by SP1 and up-regulated by glucocorticoids, but not by temozolomide and radiation. J Neurochem. (2018) 144:139–51. 10.1111/jnc.14262 [DOI] [PubMed] [Google Scholar]
- 57.Tomicic MT, Meise R, Aasland D, Berte N, Kitzinger R, Kramer OH, et al. Apoptosis induced by temozolomide and nimustine in glioblastoma cells is supported by JNK/c-Jun-mediated induction of the BH3-only protein BIM. Oncotarget. (2015) 6:33755–68. 10.18632/oncotarget.5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pitter KL, Tamagno I, Alikhanyan K, Hosni-Ahmed A, Pattwell SS, Donnola S, et al. Corticosteroids compromise survival in glioblastoma. Brain. (2016) 139(Pt 5):1458–71. 10.1093/brain/aww046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pistollato F, Abbadi S, Rampazzo E, Persano L, Della Puppa A, Frasson C, et al. Intratumoral hypoxic gradient drives stem cells distribution and MGMT expression in glioblastoma. Stem Cells. (2010) 28:851–62. 10.1002/stem.415 [DOI] [PubMed] [Google Scholar]
- 60.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. (2006) 5:67. 10.1186/1476-4598-5-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. (2007) 67:4010–5. 10.1158/0008-5472.CAN-06-4180 [DOI] [PubMed] [Google Scholar]
- 62.Beier D, Rohrl S, Pillai DR, Schwarz S, Kunz-Schughart LA, Leukel P, et al. Temozolomide preferentially depletes cancer stem cells in glioblastoma. Cancer Res. (2008) 68:5706–15. 10.1158/0008-5472.CAN-07-6878 [DOI] [PubMed] [Google Scholar]
- 63.Mantwill K, Naumann U, Seznec J, Girbinger V, Lage H, Surowiak P, et al. YB-1 dependent oncolytic adenovirus efficiently inhibits tumor growth of glioma cancer stem like cells. J Transl Med. (2013) 11:216. 10.1186/1479-5876-11-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Happold C, Stojcheva N, Silginer M, Weiss T, Roth P, Reifenberger G, et al. Transcriptional control of O(6) -methylguanine DNA methyltransferase expression and temozolomide resistance in glioblastoma. J Neurochem. (2018) 144:780–90. 10.1111/jnc.14326 [DOI] [PubMed] [Google Scholar]
- 65.Chumakova A, Lathia JD. Outlining involvement of stem cell program in regulation of O6-methylguanine DNA methyltransferase and development of temozolomide resistance in glioblastoma: an editorial highlight for 'Transcriptional control of O(6) -methylguanine DNA methyltransferase expression and temozolomide resistance in glioblastoma' on page 780. J Neurochem. (2018) 144:688–90. 10.1111/jnc.14280 [DOI] [PubMed] [Google Scholar]
- 66.Middleton MR, Margison GP. Improvement of chemotherapy efficacy by inactivation of a DNA-repair pathway. Lancet Oncol. (2003) 4:37–44. 10.1016/S1470-2045(03)00959-8 [DOI] [PubMed] [Google Scholar]
- 67.Bobola MS, Silber JR, Ellenbogen RG, Geyer JR, Blank A, Goff RD. O6-methylguanine-DNA methyltransferase, O6-benzylguanine, and resistance to clinical alkylators in pediatric primary brain tumor cell lines. Clin Cancer Res. (2005) 11:2747–55. 10.1158/1078-0432.CCR-04-2045 [DOI] [PubMed] [Google Scholar]
- 68.Koch D, Hundsberger T, Boor S, Kaina B. Local intracerebral administration of O(6)-benzylguanine combined with systemic chemotherapy with temozolomide of a patient suffering from a recurrent glioblastoma. J Neurooncol. (2007) 82:85–9. 10.1007/s11060-006-9244-8 [DOI] [PubMed] [Google Scholar]
- 69.Quinn JA, Desjardins A, Weingart J, Brem H, Dolan ME, Delaney SM, et al. Phase I trial of temozolomide plus O6-benzylguanine for patients with recurrent or progressive malignant glioma. J Clin Oncol. (2005) 23:7178–87. 10.1200/JCO.2005.06.502 [DOI] [PubMed] [Google Scholar]
- 70.Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, et al. Phase I trial of temozolomide plus O6-benzylguanine 5-day regimen with recurrent malignant glioma. Neuro Oncol. (2009) 11:556–61. 10.1215/15228517-2009-007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-methylguanine-DNA methyltransferase inactivation and chemotherapy. Br Med Bull. (2008) 85:17–33. 10.1093/bmb/ldm036 [DOI] [PubMed] [Google Scholar]
- 72.Broniscer A, Gururangan S, MacDonald TJ, Goldman S, Packer RJ, Stewart CF, et al. Phase I trial of single-dose temozolomide and continuous administration of O6-benzylguanine in children with brain tumors: a pediatric brain tumor consortium report. Clin Cancer Res. (2007) 13(22 Pt 1): 6712–8. 10.1158/1078-0432.CCR-07-1016 [DOI] [PubMed] [Google Scholar]
- 73.Quinn JA, Jiang SX, Carter J, Reardon DA, Desjardins A, Vredenburgh JJ, et al. Phase II trial of gliadel plus O6-benzylguanine in adults with recurrent glioblastoma multiforme. Clin Cancer Res. (2009) 15:1064–8. 10.1158/1078-0432.CCR-08-2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barvaux VA, Lorigan P, Ranson M, Gillum AM, McElhinney RS, McMurry TB, et al. Sensitization of a human ovarian cancer cell line to temozolomide by simultaneous attenuation of the Bcl-2 antiapoptotic protein and DNA repair by O6-alkylguanine-DNA alkyltransferase. Mol Cancer Ther. (2004) 3:1215–20. [PubMed] [Google Scholar]
- 75.Turriziani M, Caporaso P, Bonmassar L, Buccisano F, Amadori S, Venditti A, et al. O6-(4-bromothenyl)guanine (PaTrin-2), a novel inhibitor of O6-alkylguanine DNA alkyl-transferase, increases the inhibitory activity of temozolomide against human acute leukaemia cells in vitro. Pharmacol Res. (2006) 53:317–23. 10.1016/j.phrs.2005.12.001 [DOI] [PubMed] [Google Scholar]
- 76.Middleton MR, Kelly J, Thatcher N, Donnelly DJ, McElhinney RS, McMurry TB, et al. O(6)-(4-bromothenyl)guanine improves the therapeutic index of temozolomide against A375M melanoma xenografts. Int J Cancer. (2000) 85:248–52. [DOI] [PubMed] [Google Scholar]
- 77.Clemons M, Kelly J, Watson AJ, Howell A, McElhinney RS, McMurry TB, et al. O6-(4-bromothenyl)guanine reverses temozolomide resistance in human breast tumour MCF-7 cells and xenografts. Br J Cancer. (2005) 93:1152–6. 10.1038/sj.bjc.6602833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ranson M, Middleton MR, Bridgewater J, Lee SM, Dawson M, Jowle D, et al. Lomeguatrib, a potent inhibitor of O6-alkylguanine-DNA-alkyltransferase: phase I safety, pharmacodynamic, and pharmacokinetic trial and evaluation in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res. (2006) 12:1577–84. 10.1158/1078-0432.CCR-05-2198 [DOI] [PubMed] [Google Scholar]
- 79.Papachristodoulou A, Signorell RD, Werner B, Brambilla D, Luciani P, Cavusoglu M, et al. Chemotherapy sensitization of glioblastoma by focused ultrasound-mediated delivery of therapeutic liposomes. J Control Release. (2019) 295:130–9. 10.1016/j.jconrel.2018.12.009 [DOI] [PubMed] [Google Scholar]
- 80.Reinhard J, Eichhorn U, Wiessler M, Kaina B. Inactivation of O(6)-methylguanine-DNA methyltransferase by glucose-conjugated inhibitors. Int J Cancer. (2001) 93:373–9. 10.1002/ijc.1336 [DOI] [PubMed] [Google Scholar]
- 81.Kaina B, Muhlhausen U, Piee-Staffa A, Christmann M, Garcia Boy R, Rosch F, et al. Inhibition of O6-methylguanine-DNA methyltransferase by glucose-conjugated inhibitors: comparison with nonconjugated inhibitors and effect on fotemustine and temozolomide-induced cell death. J Pharmacol Exp Ther. (2004) 311:585–93. 10.1124/jpet.104.071316 [DOI] [PubMed] [Google Scholar]
- 82.Tomaszowski KH, Schirrmacher R, Kaina B. Multidrug efflux pumps attenuate the effect of MGMT inhibitors. Mol Pharm. (2015) 12:3924–34. 10.1021/acs.molpharmaceut.5b00341 [DOI] [PubMed] [Google Scholar]
- 83.Tomaszowski KH, Hellmann N, Ponath V, Takatsu H, Shin HW, Kaina B. Uptake of glucose-conjugated MGMT inhibitors in cancer cells: role of flippases and type IV P-type ATPases. Sci Rep. (2017) 7:13925. 10.1038/s41598-017-14129-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Javanmard S, Loktionova NA, Fang Q, Pauly GT, Pegg AE, Moschel RC. Inactivation of O(6)-alkylguanine-DNA alkyltransferase by folate esters of O(6)-benzyl-2'-deoxyguanosine and of O(6)-[4-(hydroxymethyl)benzyl]guanine. J Med Chem. (2007) 50:5193–201. 10.1021/jm0705859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watson AJ, Middleton MR, McGown G, Thorncroft M, Ranson M, Hersey P, et al. O(6)-methylguanine-DNA methyltransferase depletion and DNA damage in patients with melanoma treated with temozolomide alone or with lomeguatrib. Br J Cancer. (2009) 100:1250–6. 10.1038/sj.bjc.6605015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Watson AJ, Sabharwal A, Thorncroft M, McGown G, Kerr R, Bojanic S, et al. Tumor O(6)-methylguanine-DNA methyltransferase inactivation by oral lomeguatrib. Clin Cancer Res. (2010) 16:743–9. 10.1158/1078-0432.CCR-09-1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, et al. Phase II trial of temozolomide plus O6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma. J Clin Oncol. (2009) 27:1262–7. 10.1200/JCO.2008.18.8417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tawbi HA, Villaruz L, Tarhini A, Moschos S, Sulecki M, Viverette F, et al. Inhibition of DNA repair with MGMT pseudosubstrates: phase I study of lomeguatrib in combination with dacarbazine in patients with advanced melanoma and other solid tumours. Br J Cancer. (2011) 105:773–7. 10.1038/bjc.2011.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Romani M, Pistillo MP, Banelli B. Epigenetic targeting of glioblastoma. Front Oncol. (2018) 8:448. 10.3389/fonc.2018.00448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kato T, Natsume A, Toda H, Iwamizu H, Sugita T, Hachisu R, et al. Efficient delivery of liposome-mediated MGMT-siRNA reinforces the cytotoxity of temozolomide in GBM-initiating cells. Gene Ther. (2010) 17:1363–71. 10.1038/gt.2010.88 [DOI] [PubMed] [Google Scholar]
- 91.Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, et al. miR-181d: a predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. (2012) 14:712–9. 10.1093/neuonc/nos089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nie E, Jin X, Wu W, Yu T, Zhou X, Shi Z, et al. MiR-198 enhances temozolomide sensitivity in glioblastoma by targeting MGMT. J Neurooncol. (2017) 133:59–68. 10.1007/s11060-017-2425-9 [DOI] [PubMed] [Google Scholar]
- 93.Bhakat KK, Mitra S. Regulation of the human O(6)-methylguanine-DNA methyltransferase gene by transcriptional coactivators cAMP response element-binding protein-binding protein and p300. J Biol Chem. (2000) 275:34197–204. 10.1074/jbc.M005447200 [DOI] [PubMed] [Google Scholar]
- 94.Alonso MM, Gomez-Manzano C, Bekele BN, Yung WK, Fueyo J. Adenovirus-based strategies overcome temozolomide resistance by silencing the O6-methylguanine-DNA methyltransferase promoter. Cancer Res. (2007) 67:11499–504. 10.1158/0008-5472.CAN-07-5312 [DOI] [PubMed] [Google Scholar]
- 95.Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci. (2013) 70:3989–4008. 10.1007/s00018-012-1254-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang H, Alonso MM, Gomez-Manzano C, Piao Y, Fueyo J. Oncolytic viruses and DNA-repair machinery: overcoming chemoresistance of gliomas. Expert Rev Anticancer Ther. (2006) 6:1585–92. 10.1586/14737140.6.11.1585 [DOI] [PubMed] [Google Scholar]
- 97.Kanai R, Rabkin SD, Yip S, Sgubin D, Zaupa CM, Hirose Y, et al. Oncolytic virus-mediated manipulation of DNA damage responses: synergy with chemotherapy in killing glioblastoma stem cells. JNCI J Natl Cancer Inst. (2012) 104:42–55. 10.1093/jnci/djr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Paranjpe A, Zhang R, Ali-Osman F, Bobustuc GC, Srivenugopal KS. Disulfiram is a direct and potent inhibitor of human O6-methylguanine-DNA methyltransferase (MGMT) in brain tumor cells and mouse brain and markedly increases the alkylating DNA damage. Carcinogenesis. (2014) 35:692–702. 10.1093/carcin/bgt366 [DOI] [PMC free article] [PubMed] [Google Scholar]