Abstract
Background
Patients with Clostridioides difficile infection (CDI) with either eosinopenia or infected with a binary toxin strain have increased likelihood of mortality. However, the relationship between binary toxin and eosinopenia to synergistically increase mortality has not been studied in humans. We hypothesized that patients with CDI due to binary toxin strains and concomitant peripheral eosinopenia would have a higher likelihood of inpatient mortality.
Methods
This multicenter, retrospective cohort study included adult patients with CDI of known ribotypes stratified by eosinopenia, defined as an absence of eosinophils in the peripheral blood (Houston cohort). The primary outcome was inpatient mortality. Results were supported by a separate national cohort of veterans with CDI (Veterans’ cohort).
Results
In the Houston cohort, a total of 688 patients from 13 institutions in 6 cities were included. Of these, 132 (19%) had an eosinophil count of 0.0 cells/µL (0.0 cells*109/L) and 109 (16%) were infected with a binary toxin strain. After adjusting for covariates, the combination of eosinopenia and infection with a binary toxin strain was an independent predictor of inpatient mortality (odds ratio [OR], 7.8; 95% confidence interval [CI], 1.9–33.2; P = .005). In the separate Veterans’ cohort (n = 790), this combination was also a significant predictor of inpatient mortality (OR, 6.1; 95% CI, 1.5–23.9; P = .009).
Conclusions
In conclusion, the combination of eosinopenia and CDI due to a binary toxin strain was correlated with increased mortality in hospitalized patients from 2 independent cohorts. Prospective studies should further study this important subset of patients at the time of CDI diagnosis.
Keywords: anaerobe infections, health care–acquired infections, molecular epidemiology, multicenter study, outcomes assessment
Clostridioides difficile is an anaerobic, spore-forming, Gram-positive bacillus that was first isolated from the feces of healthy infants by Hall and O’Toole in 1935 [1]. The incidence of C. difficile infection (CDI) has increased over the past 2 decades, making C. difficile the most common pathogen causing health care–associated infections in the United States [2–6]. The ability of C. difficile to cause clinical disease in humans is dependent on toxin production. With few exceptions, all toxigenic strains of C. difficile identified in humans produce a cytotoxin, toxin B, and most coproduce an enterotoxin, toxin A [7]. In addition, some strains produce a binary toxin, Clostridium difficile transferase (CDT), encoded by genes cdtA and cdtB [8]. The recent epidemic ribotype 027 (RT 027) harbors binary toxin genes [9–13], as do other ribotypes (eg, RT 019, 075, and 078) [8, 11, 14–16]. Highly dependent on circulating strains, studies have demonstrated the prevalence of binary toxin strains infecting humans to range from 6% to 84% [10, 11, 14, 17–21]. Although controversial, the presence of binary toxin has been shown to increase the severity [17], nonhome discharge [22], mortality [12, 15, 17, 21], and recurrence of CDI [11].
A recent clinical trial demonstrated increased mortality rates in patients with CDI and eosinopenia but was limited by a lack of data on binary toxin strains [23]. In vitro and mouse models recently demonstrated that the binary toxin increases virulence by suppressing the protective effects of colonic eosinophilia, with an associated increase in the host inflammatory response [24]. These effects are thought to be due to the indirect apoptosis of systemic eosinophils via increased recognition of binary toxin by the toll-like receptor 2 located on eosinophils [24]. The relationship between binary toxin and eosinopenia to synergistically increase mortality has not been studied in humans.
Given this background, our research group has been conducting an ongoing clinical trial of patients with CDI for several years [22]. As part of this study, leftover stool samples from hospitalized patients are collected for C. difficile growth and polymerase chain reaction (PCR) ribotyping [25]. As we routinely collect clinical data on these patients, including results from hematologic blood draws and discharge status, we had the unique opportunity to study if patients with eosinopenia and binary toxin had increased likelihood of inpatient mortality compared with other groups. Our hypothesis was that CDI due to a binary toxin strain accompanied by peripheral eosinopenia would be associated with higher odds of inpatient mortality. To test our hypothesis, we conducted a large multicenter cohort study evaluating the association between inpatient mortality and CDI due to a binary toxin strain accompanied by eosinopenia. An ongoing collaboration with a co-investigator allowed us to confirm our results in an independent cohort of US veterans.
METHODS
Houston Cohort
Study Population
This multicenter, retrospective cohort study was conducted using data available from an ongoing clinical study of patients with CDI hospitalized in 2 large health systems (13 hospitals in total) in the Houston, Texas area (years: 2015–2018). The study included all patients ≥18 years of age with CDI who had specimen ribotype data available. Patients were excluded if they did not have clinical or laboratory data available, their test result represented a test for cure, receiving corticosteroids greater than prednisone 5 mg per day, or they were neutropenic. The study was approved by the University of Houston Committee for the Protection of Human Subjects with a waiver of informed consent (IRB study 00000128).
Definitions
Patients were tested for CDI at the discretion of the treating physician and medical team. The standard-of-care diagnostic in the included facilities was a C. difficile nucleic acid amplification test (NAAT) or enzyme-linked immunosorbent assay (EIA) in patients with unexplained and new-onset diarrhea (≥3 unformed stools in 24 hours). Recurrent C. difficile infection and health care facility–onset CDI (HO-CDI) cases were defined by the Centers for Disease Control and Prevention multidrug-resistant organism and CDI module [26]. Two episodes of CDI in the same patient were considered to be distinct events if they occurred >8 weeks apart, meaning the same patient could be included more than once. Clinical definitions of CDI severity were defined per the 2017 Infectious Diseases Society of America and Society for Healthcare Epidemiology of America C. difficile guidelines [27]. Admission and discharge location options included home, long-term care facility, another hospital, hospice, or death. The need for a higher level of care was determined by a trained abstractor and was defined as discharge to a facility offering higher-intensity medical and/or nursing services than residence before admission (eg, admitted from home and discharged to an SNF). Lastly, eosinopenia was defined as an eosinophil count of 0.0 cells/μL (0.0 cells*109/L), measured using hematologic blood draws obtained within 24 hours of stool collection.
Covariates and Outcomes
Medical records were reviewed retrospectively for demographic information, underlying comorbidities, laboratory data, and clinical outcomes. Notably, both health systems utilize a system-wide electronic health record (EHR) system, allowing medical data to be collected from the entire health system. Laboratory analytes measured at the time of CDI diagnosis (±24 hours) included white blood cell (WBC) count, eosinophil count, albumin level, and serum creatinine (SCr) level. The Charlson Comorbidity Index (CCI) score was calculated using comorbidities documented on or before the date of hospital admission [28]. The primary study outcome was inpatient mortality. Secondary outcomes included 30-day recurrence, 90-day recurrence, intensive care unit admission within 48 hours of CDI diagnosis, colectomy within 30 days of CDI diagnosis, and a need for a higher level of care upon discharge.
Sample Collection and Ribotyping
Leftover stool samples from patients diagnosed with CDI were collected and brought to a centralized research laboratory at the University of Houston. Stool samples were then plated onto C. difficile–selective cefoxitin-cycloserine-fructose agar plates and anaerobically incubated for 48–72 hours. Colonies were identified as C. difficile by PCR. Fluorescent PCR ribotyping was performed as previously described [25, 29, 30]. The library contains >100 known ribotypes (https://thewalklab.com/tools/) but does not distinguish between ribotypes 053 and 163, ribotypes 014 and 020, and ribotypes 078 and 126; therefore, these are reported as combined ribotypes (eg, 053-163, 014-020, and 078-126). Binary toxin is present in RT 019, 027, 075, and 078–126 [8–16].
Veterans’ Cohort
Study Population, Definitions, Covariates, and Outcomes
Data for the Veteran’s cohort were obtained from the Department of Veterans Affairs Informatics and Computing Infrastructure, which includes administrative, clinical, laboratory, and pharmacy data repositories that are linked using unique patient identifiers. Patients were included if they were 18–89 years old and had any inpatient or outpatient International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) code for CDI (008.45) plus a positive CDI stool test (eg, NAAT or EIA) during the visit or within 7 days of the visit from October 1, 2002, through September 30, 2014. Patients were excluded if they had an ICD-9-CM code for CDI in the year before cohort inclusion to select for primary cases. Patients were also excluded if they did not receive active CDI therapy or did not have complete laboratory data (eg, eosinophil count). The database was queried for hospitals that reported either RT 027 or binary toxin data in the EHR, most commonly due to use of the Expert C. difficile/Epi PCR diagnostic (Cepheid, Sunnyvale CA, USA), which reports binary toxin/ribotype 027 results. The University of Texas Health Science Center at San Antonio and the South Texas Veterans Health Care System Research and Development Committee institutional review boards, with a waiver of informed consent, approved the study (approval #HSC20130473H).
Statistical Analysis
An eosinophil count reported as 0.0 cells/μL (0.0 cells*109/L) was the cutoff used to stratify the cohort. For baseline characteristic comparison, binary and categorical variables were compared using the χ 2 or Fisher exact test, whereas continuous variables were compared using the Student t test or Wilcoxon rank-sum test, depending on the data distribution. A logistic regression model was developed modeling inpatient mortality as a function of the available patient covariates based on methods recommended by Hosmer and Lemeshow [31]. To prevent overfitting the model, covariates with a univariate Wald test P value <.20 were chosen as candidates for the multivariable model. Following the fit of this model, each covariate was assessed again using the P value of its Wald statistic. Variables with a P > .05 were removed, and the partial likelihood ratio test was used to compare the new, smaller model with the old model. Secondary outcomes were compared between cohorts by univariate comparison. Odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were calculated. The cumulative probability of survival during 90 days of follow-up was estimated using the Kaplan-Meier procedure, whereas the groups were compared using the log-rank test. The time to death was defined as the number of days between CDI diagnosis and death from any cause on or before day 90. All P values were from 2-sided tests, and results were deemed statistically significant at P < .05. All statistical analyses were performed using STATA, version 15.1 (StataCorp LLC, College Station, TX, USA), and RStudio (RStudio Inc., Boston, MA, USA).
RESULTS
Houston Cohort
Patient Characteristics
A total of 3118 patients met the inclusion criteria; 2309 patients were missing clinical outcomes data, 110 patients had missing laboratory values (eosinophil count or SCr), 10 patients’ tests represented a test for cure, and 1 patient’s sample was EIA-negative. The final study cohort consisted of 688 patients from 13 institutions in 6 cities (132 had a baseline eosinophil count of 0.0 cells/µL [0.0 cells*109/L], and 556 had a baseline eosinophil count >0.0 cells/µL [0.0 cells*109/L]) (Figure 1). Clinical outcome data were most commonly missing due to patient hospitalization before an EHR change-over by the health system that prevented access to the clinical data. Demographic and baseline characteristics of the patients are shown in Table 1. The mean age of the cohort was 64 ± 18 years, and 389 (56%) were female. The median CCI score was 2, 244 (35%) of the cases were defined as HO-CDI, 70 (10%) patients were experiencing a CDI recurrence, and 172 (25%) had been diagnosed with CDI at some point in their past. Most baseline characteristics were similar between groups, but patients with eosinopenia had a higher median WBC count at CDI diagnosis and race/ethnicity proportions differed between groups.
Figure 1.
Patient selection flow chart, Houston cohort. Abbreviations: EIA, enzyme-linked immunosorbent assay; SCr, serum creatinine.
Table 1.
Comparison of Patient Demographics, Comorbidities, and Laboratory Parameters Between Eosinophil Count Groups
| Houston Cohort | Veterans’ Cohort | |||||
|---|---|---|---|---|---|---|
| Covariate | Eosinopenia (n = 132) | Eosinophils Present (n = 556) | P Value | Eosinopenia (n = 67) | Eosinophils Present (n = 723) | P Value |
| Age, mean (±SD), y | 65.5 (18.7) | 63.7 (17.7) | .32 | 67 (12) | 66 (12) | .33 |
| Female, No. (%) | 71 (53.8) | 318 (57.2) | .48 | 3 (4.4) | 38 (5.3) | .78 |
| Race/ethnicity, No. (%) | .04 | |||||
| White, non-Hispanic | 94 (71.2) | 320 (57.6) | 43 (67.2) | 459 (65.9) | .84 | |
| Black, non-Hispanic | 18 (13.6) | 114 (20.5) | 19 (29.7) | 186 (26.7) | .61 | |
| Hispanic | 14 (10.6) | 98 (17.6) | 2 (3.1) | 25 (3.6) | .84 | |
| Asian | 2 (1.5) | 7 (1.3) | 0 (0.0) | 26 (3.7) | .03 | |
| Othera | 4 (3.0) | 17 (3.1) | ||||
| CCI, median (IQR) | 2 (1–3) | 2 (1–4) | .09 | 3 (2–5) | 3 (1–6) | .79 |
| SOT, No. (%) | 8 (6.1) | 57 (10.3) | .14 | 2 (3.0) | 6 (0.8) | .16 |
| HSCT, No. (%) | 0 (0.0) | 1 (0.2) | 1.00 | 0 (0.0) | 0 (0.0) | N/A |
| WBC, median (IQR), cells/μL | 12.3 (8.7–20.0) | 10.7 (7.3–15.8) | .001 | 15.4 (10.5–22.0) | 10.6 (7.4–15.1) | <.001 |
| SCr, median (IQR), mg/dL | 1.07 (0.78–1.70) | 1.06 (0.75–2.00) | .88 | 1.4 (1.0–2.3) | 1.2 (0.9–1.9) | .12 |
| Albumin, mean (±SD), g/dL | 3.1 (0.8) | 3.1 (0.7) | .98 | 2.7 (2.3–3.4) | 2.9 (2.4–3.5) | .31 |
| Severe CDI, No. (%) | 73 (55.3) | 289 (52.0) | .49 | 61 (91.0) | 512 (70.8) | <.001 |
| Testing method, No. (%) | .78 | N/A | ||||
| NAAT | 129 (97.7) | 538 (96.8) | 67 (100.0) | 723 (100.0) | ||
| EIA | 3 (2.3) | 18 (3.2) | 0 | 0 | ||
| HO-CDI, No. (%) | 40 (30.3) | 204 (36.7) | .17 | 56 (83.6) | 450 (62.2) | <.001 |
| Recurrent CDI, No. (%)b | 16 (12.1) | 54 (9.7) | .41 | 0 (0.0) | 0 (0.0) | N/A |
| History of CDI, No. (%)b | 36 (27.3) | 136 (24.5) | .50 | 0 (0.0) | 0 (0.0) | N/A |
| CDT + ribotype, No. (%) | 14 (10.6) | 95 (17.1) | .07 | 13 (19.4) | 214 (29.6) | .07 |
Abbreviations: CCI, Charlson Comorbidity Index; CDI, Clostridioides difficile infection; CDT+, binary toxin–positive; EIA, enzyme-linked immunosorbent assay; HO-CDI, health care facility–onset CDI; HSCT, hematopoietic stem cell transplantation; IQR, interquartile range; NAAT, nucleic acid amplification test; SCr, serum creatinine; SOT, solid organ transplantation; WBC, white blood cell count.
Conversion factors: WBC (109/L) = WBC (cells/μL)/1 000 000; albumin (g/L) = albumin (g/dL)/100; SCr (mg/dL) = SCr (μmol/L)/88.42.
aAmerican Indian, Alaska Native, Native Hawaiian, Pacific Islander, other race/ethnicity, or ≥2 races/ethnicities.
bNo patients with a history of CDI were included in the validation cohort.
C. difficile Strain Characteristics
The most commonly isolated ribotypes present in ≥10% of the sample included RT 014-020, 106, 027, and 002 (Supplementary Table 1). In total, 109 (16%) were binary toxin strains, of which RT 027 was the most common (n = 82). The proportion of CDI due to a binary toxin strain did not differ between those with eosinopenia and those with a measureable peripheral blood eosinophil count within 24 hours of CDI diagnosis (10.6% vs 17%; P = .07). The sensitivity and specificity of eosinopenia as a surrogate for binary toxin presence were 12.8% and 79.6%, respectively.
Clinical Outcomes
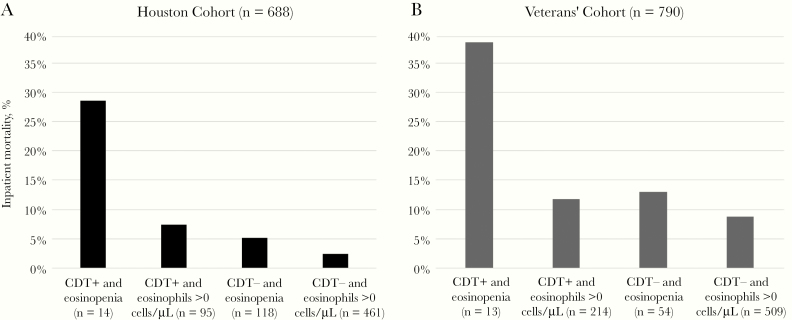
Fourteen patients both had eosinopenia and were infected with a binary toxin strain. In univariate analysis, both eosinopenia (OR, 2.5; 95% CI, 1.1–5.4; P = .03) and infection with a binary toxin strain (OR, 3.7; 95% CI, 1.7–8.2; P = .01) were associated with inpatient mortality. However, the combination of CDI due to a binary toxin strain and eosinopenia was associated with a higher odds of inpatient mortality (OR, 10.8; 95% CI, 3.2–37.0; P < .001). Patients were stratified into 4 groups: 1) binary toxin–positive with eosinopenia, 2) binary toxin–positive without eosinopenia, 3) binary toxin–negative with eosinopenia, and 4) binary toxin–negative without eosinopenia (Figure 2). Inpatient mortality was highest for group 1 compared with all other groups (28.6% vs 7.4%, 5.1%, and 2.4%, respectively; P < .001). Using Kaplan-Meier survival curves, time to death was also significantly shorter in patients with CDI due to a binary toxin strain and concomitant eosinopenia compared with all other groups (P = .03) (Supplementary Figure 1). Secondary outcomes did not differ significantly between groups (Table 2).
Figure 2.
Infection with a binary toxin strain and eosinopenia was associated with increased inpatient mortality in both cohorts (P < .001, each). Abbreviation: CDT, binary toxin.
Table 2.
Stratified Univariate Analysis of Primary and Secondary Outcomes
| Outcomes | Eosinopenia/CDT Present | Eosinopenia/CDT Present | Eosinopenia/CDT Present | Eosinopenia/CDT Present |
|---|---|---|---|---|
| Houston Cohort | Yes/Yes (n = 14) | No/Yes (n = 95) | Yes/No (n = 118) | No/No (n = 461) |
| Inpatient mortality, No. (%) | 4 (28.6) | 7 (7.4) | 6 (5.1) | 11 (2.4) |
| 30-d recurrence, No. (%) | 0 (0.0) | 5 (5.3) | 3 (2.5) | 22 (4.8) |
| 90-d recurrence, No. (%) | 3 (21.4) | 20 (21.1) | 5 (4.2) | 55 (11.9) |
| ICU admission, No. (%) | 4 (28.6) | 13 (13.7) | 24 (20.3) | 57 (12.4) |
| Higher LOC at discharge, No. (%) | 7 (50) | 23 (24.2) | 44 (37.3) | 128 (27.8) |
| Colectomy, No. (%) | 0 (0.0) | 0 (0.0) | 3 (2.5) | 4 (0.9) |
| Veteran’s Cohort | Yes/Yes (n = 13) | No/Yes (n = 214) | Yes/No (n = 54) | No/No (n = 509) |
| Inpatient mortality, No. (%) | 5 (38.5) | 25 (11.7) | 7 (13.0) | 45 (8.8) |
| 30-d recurrence, No. (%) | 1 (7.7) | 8 (3.7) | 0 (0.0) | 17 (3.3) |
| 90-d recurrence, No. (%) | 3 (23.1) | 13 (6.1) | 0 (0.0) | 26 (5.1) |
| ICU admission, No. (%) | 0 (0.0) | 9 (4.2) | 1 (1.9) | 7 (1.4) |
| Higher LOC at discharge, No. (%) | 8 (66.7) | 54 (29.7) | 16 (32.0) | 88 (19.5) |
| Colectomy, No. (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
Abbreviations: CDT, binary toxin; ICU, intensive care unit; LOC, level of care.
Using a multivariable model that adjusted for CCI score, WBC count, and serum albumin level, CDI due to a binary toxin strain with concomitant eosinopenia remained an independent predictor of inpatient mortality (OR, 7.8; 95% CI, 1.9–33.2; P = .005) (Table 3).
Table 3.
Univariate and Multivariable Analysis for Predictors of Inpatient Mortality Post–Clostridioides difficile Infection Diagnosis
| Houston Cohort | Validation Cohort | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariable Analysis | Univariate Analysis | Multivariable Analysis | |||||
| Covariate | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value |
| Eosinopenia + CDT | 10.8 (3.2–37.0) | <.001 | 7.8 (1.9–33.2) | .005 | 5.7 (1.8–17.8) | .003 | 6.1 (1.5–23.9) | .009 |
| Age, y | 1.02 (0.99–1.04) | .14 | 1.01 (0.99–1.03) | .19 | ||||
| CCI | 1.19 (1.02–1.39) | .02 | 1.23 (1.01–1.48) | .04 | 1.11 (1.04–1.19) | .002 | 1.09 (1.01–1.17) | .02 |
| SOT | 0.73 (0.17–3.14) | .67 | 9.0 (2.2–36.8) | .002 | ||||
| WBC, cells/μL | 1.05 (1.03–1.08) | <.001 | 1.03 (1.00–1.06) | .02 | 1.02 (1.01–1.04) | .005 | 1.01 (0.99–1.03) | .18 |
| SCr, mg/dL | 1.02 (0.88–1.19) | .76 | 0.96 (0.81–1.15) | .67 | ||||
| Albumin, g/dL | 0.15 (0.07–0.33) | <.001 | 0.18 (0.08–0.43) | <.001 | 0.48 (0.34–0.68) | <.001 | 0.53 (0.37–0.77) | <.001 |
| Severe CDI | 1.65 (0.75–3.64) | .21 | 17.4 (4.3–71.6) | <.001 | ||||
| HO-CDI | 2.95 (1.36–6.40) | .01 | 0.91 (0.57–1.47) | .71 | ||||
| History of CDIa | 0.49 (0.17–1.43) | .19 |
Abbreviations: CCI, Charlson Comorbidity Index; CDI, Clostridioides difficile infection; CDT, binary toxin; CI, confidence interval; HO-CDI, health care facility–onset CDI; OR, odds ratio; SCr, serum creatinine; SOT, solid organ transplantation; WBC, white blood cell count.
aNo patients with a history of CDI were included in the validation cohort.
Veterans’ Cohort
A total of 26 149 veterans with CDI were included, of whom 790 had eosinophil and RT 027 (n = 322) or binary toxin (n = 468) laboratory data available in the EHR (Table 1). The mean age of the cohort was 66 ± 12 years, and 749 (95%) were male. The median CCI score was 3, 506 (64%) of the cases were defined as HO-CDI, 0 (0%) patients were experiencing a CDI recurrence, and 0 (0%) had been diagnosed with CDI at some point in their past. Patients were once again stratified into 4 groups: 1) binary toxin–positive with eosinopenia, 2) binary toxin–positive without eosinopenia, 3) binary toxin–negative with eosinopenia, and 4) binary toxin–negative without eosinopenia (Figure 2). Thirteen patients both had eosinopenia and were infected with a binary toxin strain. Inpatient mortality was highest for group 1 compared with all other groups (38.5% vs 11.7%, 13.0%, and 8.8%, respectively). In univariate analysis, eosinopenia was associated with inpatient mortality (OR, 2.0; 95% CI, 1.0–3.9), whereas CDI due to a binary toxin strain was not. However, the combination of CDI due to a binary toxin strain and eosinopenia was a stronger predictor of mortality (OR, 5.7; 95% CI, 1.8–17.8). Secondary outcomes were similar to the results from the Houston cohort (Table 2).
In the multivariable model, the combination of CDI due to a binary toxin strain and eosinopenia remained an independent predictor of inpatient mortality after controlling for CCI score, WBC count, and serum albumin level (OR, 6.1; 95% CI, 1.5–23.9) (Table 3).
DISCUSSION
C. difficile transferase, also known as the binary toxin, is present in the major hypervirulent C. difficile strains including RT 027 and 078 [8, 9, 11–16]. Recent in vitro and animal models have shown that the binary toxin induces a pathogenic host inflammatory response characterized by the suppression of colonic eosinophils and increased apoptosis of systemic eosinophils [24]. However, outcomes associated with CDI due to a binary toxin strain and concomitant eosinopenia have not been studied in humans. We hypothesized that inpatient mortality would be higher in patients with CDI due to a binary toxin strain and concomitant eosinopenia compared with all other groups. Using 2 large, independent data sets, we demonstrated that the combination of CDI due to a binary toxin strain and eosinopenia led to a significantly greater odds of inpatient mortality. Patients infected with a binary toxin strain and concomitant eosinopenia were nearly 8 times more likely to die as an inpatient than patients who did not meet these criteria. The strengths of this study include its large, multicenter sample and C. difficile strain characterization by PCR ribotyping. We were also able to control for multiple patient characteristics known to confound CDI outcome by collecting a comprehensive list of relevant covariates. Furthermore, having 2 independent databases allowed us to confirm our results. Eosinopenia in the context of infection was first described in 1893 [32] and is a result of migration of eosinophils to the site of infection during the acute phase of inflammation [33]. Although the optimal cutoff to define a low eosinophil count is controversial, eosinopenia has been considered a criterion for systemic inflammatory response syndrome (SIRS) [34–36]. Patients with eosinopenia associated with infections are more likely to present with more severe disease and increased likelihood of poor outcomes, including mortality. Our results support this observation. However, the fact that mortality was significantly increased in patients infected with binary toxin strains and eosinopenia, compared with patients with eosinopenia and other strains, supports our hypothesis of a synergy between the 2 variables.
Causality should not be inferred from these studies but raises important hypotheses for future prospective cohort studies. For example, the binary toxin may simply be a surrogate marker of other virulence traits of C. difficile. As eosinophil counts are readily available from routine complete blood counts collected from hospitalized patients, it would be relatively straightforward to identify patients with eosinopenia and CDI in real time. Several commercially available CDI diagnostics use PCR to detect the specific genes that encode for the binary toxin, allowing for prospective validation of the results observed in this study. Our study has a number of limitations. First, we chose an absolute cutoff to define eosinopenia to directly compare our results with the only other human trial [23]. Our sample size was not large enough to explore an optimized cutoff, and this will need to be evaluated in future studies. Second, we chose to collect and analyze eosinopenia at a single time point within 24 hours of stool collection. Whether resolution of eosinopenia can be used as a prognostic marker of clinical response in CDI will require further study. Third, clinical data were obtained from the EHR as a secondary database analysis, and our results should be further validated using a prospective, multicenter study design. Although our primary objective was available for all patients, some of our secondary outcomes, such as 30- and 90-day recurrence, may have been underestimated. We did not evaluate binary toxin production or its biological activity, but rather assumed that all RT 019, 027, 075, and 078-126 isolates harbored genes capable of producing biologically active proteins, as previously demonstrated [14]. Eosinopenia is associated with other chronic medical conditions such as trauma, stroke, and others. Our study lacks the sample size to correctly assess for the confounding of other medical conditions to influence these results. Lastly, these results demonstrate an association between the variables, and causality should not be inferred.
In conclusion, the combination of eosinopenia and CDI due to a binary toxin strain was correlated with increased mortality in hospitalized patients from 2 independent cohorts. Prospective studies should be performed to further study this important subset of patients at the time of CDI diagnosis.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Author contributions. All authors made substantial contributions to the conception or design of the work; to the acquisition, analysis, or interpretation of data for the work; to drafting the work; or to revising it critically for important intellectual content. All authors approved of the final version. They agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Dr. Garey had full access to the data and is the guarantor for the data.
Financial support. This work was supported by the National Institutes of Health National Institute of Allergy and Infectious Diseases (U01AI124290-01).
Potential conflicts of interest. All authors declare no conflict of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hall IC, O’Toole E. Intestinal flora in new-born infants with a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child 1935; 49(2):390–402. [Google Scholar]
- 2. McDonald LC, Owings M, Jernigan DB. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg Infect Dis 2006; 12(3):409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khanna S, Pardi DS, Aronson SL, et al. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 2012; 107:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller BA, Chen LF, Sexton DJ, Anderson DJ. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol 2011; 32:387–90. [DOI] [PubMed] [Google Scholar]
- 5. Magill SS, O’Leary E, Janelle SJ, et al. ; Emerging Infections Program Hospital Prevalence Survey Team Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med 2018; 379:1732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magill SS, Edwards JR, Bamberg W, et al. ; Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team Multistate point-prevalence survey of health care-associated infections. N Engl J Med 2014; 370:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 2005; 18:247–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes 2014; 5:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 2005; 353:2433–41. [DOI] [PubMed] [Google Scholar]
- 10. Loo VG, Poirier L, Miller MA, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 2005; 353:2442–9. [DOI] [PubMed] [Google Scholar]
- 11. Stewart DB, Berg A, Hegarty J. Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg 2013; 17(1):118–24; discussion 24–5. [DOI] [PubMed] [Google Scholar]
- 12. Miller M, Gravel D, Mulvey M, et al. Health care-associated Clostridium difficile infection in Canada: patient age and infecting strain type are highly predictive of severe outcome and mortality. Clin Infect Dis 2010; 50:194–201. [DOI] [PubMed] [Google Scholar]
- 13. Warny M, Pepin J, Fang A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 2005; 366:1079–84. [DOI] [PubMed] [Google Scholar]
- 14. Stubbs S, Rupnik M, Gibert M, et al. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett 2000; 186:307–12. [DOI] [PubMed] [Google Scholar]
- 15. Bacci S, Mølbak K, Kjeldsen MK, Olsen KE. Binary toxin and death after Clostridium difficile infection. Emerg Infect Dis 2011; 17:976–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goorhuis A, Bakker D, Corver J, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis 2008; 47:1162–70. [DOI] [PubMed] [Google Scholar]
- 17. Barbut F, Gariazzo B, Bonne L, et al. Clinical features of Clostridium difficile-associated infections and molecular characterization of strains: results of a retrospective study, 2000–2004. Infect Control Hosp Epidemiol 2007; 28(2):131–9. [DOI] [PubMed] [Google Scholar]
- 18. Gonçalves C, Decré D, Barbut F, et al. Prevalence and characterization of a binary toxin (actin-specific ADP-ribosyltransferase) from Clostridium difficile. J Clin Microbiol 2004; 42:1933–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rupnik M, Kato N, Grabnar M, Kato H. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J Clin Microbiol 2003; 41:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bauer MP, Notermans DW, van Benthem BH, et al. ; ECDIS Study Group Clostridium difficile infection in Europe: a hospital-based survey. Lancet 2011; 377:63–73. [DOI] [PubMed] [Google Scholar]
- 21. Goldenberg SD, French GL. Lack of association of tcdC type and binary toxin status with disease severity and outcome in toxigenic Clostridium difficile. J Infect 2011; 62:355–62. [DOI] [PubMed] [Google Scholar]
- 22. Reveles KR, Dotson KM, Gonzales-Luna A, et al. Clostridioides (formerly Clostridium) difficile infection during hospitalization increases the likelihood of nonhome patient discharge. Clin Infect Dis 2019; 68:1887–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kulaylat AS, Buonomo EL, Scully KW, et al. Development and validation of a prediction model for mortality and adverse outcomes among patients with peripheral eosinopenia on admission for Clostridium difficile infection. JAMA Surg 2018; 153:1127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cowardin CA, Buonomo EL, Saleh MM, et al. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol 2016; 1:16108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alam MJ, Walk ST, Endres BT, et al. Community environmental contamination of toxigenic Clostridium difficile. Open Forum Infect Dis 2017; 4(1):ofx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention. Multidrug-Resistant Organism And Clostridioides difficile Infection (MDRO/CDI) Module. Atlanta: Centers for Disease Control and Prevention; 2019. [Google Scholar]
- 27. McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66(7):987–94. [DOI] [PubMed] [Google Scholar]
- 28. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 29. Aitken SL, Alam MJ, Khaleduzzaman M, et al. In the endemic setting, Clostridium difficile ribotype 027 is virulent but not hypervirulent. Infect Control Hosp Epidemiol 2015; 36:1318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Endres BT, Dotson KM, Poblete K, et al. Environmental transmission of Clostridioides difficile ribotype 027 at a long-term care facility; an outbreak investigation guided by whole genome sequencing. Infect Control Hosp Epidemiol 2018; 39:1322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hosmer DW, Lemeshow S, Sturdivant RX.. Applied Logistic Regression. Hoboken, NJ: John Wiley & Sons, Inc.; 2013. [Google Scholar]
- 32. Zappert J. Ueber das vorkommen der eosinophilen zellen in menschlichen blute. Z Klin Med 1893; 23:227–308. [Google Scholar]
- 33. Bass DA, Gonwa TA, Szejda P, Cousart MS, DeChatelet LR, McCall CE. Eosinopenia of acute infection: production of eosinopenia by chemotactic factors of acute inflammation. J Clin Invest 1980; 65(6):1265–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gil H, Magy N, Mauny F, Dupond JL. Value of eosinopenia in inflammatory disorders: an “old” marker revisited. Rev Med Interne 2003; 24:431–5. [DOI] [PubMed] [Google Scholar]
- 35. Abidi K, Khoudri I, Belayachi J, et al. Eosinopenia is a reliable marker of sepsis on admission to medical intensive care units. Crit Care 2008; 12:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lavoignet CE, Le Borgne P, Slimani H, et al. Relevance of eosinopenia as marker of sepsis in the emergency department. Rev Med Interne 2016; 37:730–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.