The formation of a supramolecular core–shell like assembly upon interaction of the receptor with potassium sulfate enables its selective extraction.
The formation of a supramolecular core–shell like assembly upon interaction of the receptor with potassium sulfate enables its selective extraction.
Abstract
Selective extraction of sulfates in the form of alkali metal salts using charge-neutral molecular receptors is one of the holy grails of supramolecular chemistry. Herein we describe, for the first time, a squaramide-based ion pair receptor equipped with a crown ether site that is able to extract potassium sulfate from the aqueous to the organic phase (an analogous monotopic anion receptor lacking the crown ether unit lacks this ability). 1H NMR, UV-vis, DOSY-NMR, DLS, and MS experiments and the solid-state single crystal structure provided evidence of the formation of a supramolecular core–shell like assembly upon interaction of the receptor with potassium sulfate. The presence of monovalent potassium salts, in contrast, promoted the formation of simple 1 : 1 complexes. Unlike the 4 : 1 assembly, the 1 : 1 complexes are poorly soluble in organic media. This feature was utilized to overcome the Hofmeister bias and allow for selective extraction of extremely hydrophilic sulfates over lipophilic nitrate anions, which was unambiguously proved by quantitative AES and ion chromatography measurements. A simple modification of the receptor structure led to a “naked eye” optical sensor able to selectively detect sulfates under both SLE and LLE conditions.
Introduction
Due to the numerous roles played by sulfates in the environment and biological systems, the design of artificial receptors able to selectively recognize sulfates is of great interest in supramolecular chemistry.1 Sulfates have a very high hydration energy (–1090 kJ mol–1),2 making strong and selective binding of this anion by neutral receptors a very challenging task, especially under aqueous or interfacial conditions. On the other hand, selective binding of extremely hydrophilic sulfate anions over lipophilic anions, such as nitrate anions (hydration energy –306 kJ mol–1),2 is highly desired and requires the Hofmeister bias to be overcome.3 One field of potential application is environment protection, for instance in the disposal of sulfate- and nitrate-containing high-level liquid waste (HLLW) stored at the Hanford site.4 The presence of sulfates in HLLW and their low solubility limit in borosilicate glass is a serious issue and is problematic upon vitrification of HLLW. Sulfates cause corrosion of glass melters and the constituent electrodes and are responsible for the decrease of durability of glass logs, directly posing a safety hazard during vitrification and storage. One of the ways of sequestering sulfates from their aqueous solutions involves anion or ion pair receptor facilitated liquid–liquid extraction (LLE). In this context it has been demonstrated that properly preorganized macrocyclic, tripodal or charged anion receptors can be used as sulfate extractants.5 However, in such cases a bulky counterion needs to be present and the process has the nature of an exchange rather than extraction. To the best of our knowledge, simple extraction of sulfates associated with alkali metals from the aqueous to the organic phase has not yet been reported in the literature. We envisioned that utilizing the advantage of ion pair receptors, as compounds which can bind ion pairs cooperatively and are able to interact with salts under interfacial conditions, may fill this gap in methodology.6 Nature solved the problem of sulfate binding by means of the sulfate binding protein (SBP), where four sulfate oxygen atoms are bound through seven hydrogen bonding interactions donated by specific residues of the protein.7 Thus we addressed the question of whether several relatively simple and rigid non-multimacrocyclic ion pair receptors can form hydrogen bonding inorganic–organic core–shell like assemblies and enable selective extraction of sulfates from the aqueous to the organic phase. The basis for distinguishing nitrate and sulfate anions should be the formation of complexes of various stoichiometry depending on the valency and geometry of the anions tested, which should affect the solubility of the complexes in the organic phase. To verify our hypothesis we designed and synthesized squaramide based ion pair receptors in which the anion binding site is directly linked with an electron deficient phenyl ring and with a benzo 18-crown-6 unit (Scheme 1).
Scheme 1. Squaramide-based receptor 1 and sensor 2.
Results and discussion
The synthesis of receptors 1 and 2 is outlined in Scheme 2. Briefly, receptor 1 was obtained by sequential amidation of dimethyl squarate with 3,5-trifluoromethylaniline followed by 4-aminobenzo-18-crown-6. In the case of receptor 2, which is designed as a simple optical sensor, 4-nitroaniline was used in the first step of synthesis. All intermediates and receptors can be simply purified by crystallization and are thus easily accessible on a large scale.
Scheme 2. Synthesis of receptors 1 and 2. Reagents and conditions: (i) H2, Pd/C, MeOH–THF, 12 h, r.t., and quantitative; (ii) methanol, 72 h, r.t., 95% for S1 and 84% for S2; (iii) methanol, 48 h, r.t., 90% for 1 and 75% for 2.
Initial evidence that squaramide 1 might act as an ion pair receptor and is able to interact with anions in the presence of cations in an enhanced manner came from UV-vis spectroscopy in acetonitrile. The addition of incremental amounts of TBA salts of various anions to a solution of 1 caused bathochromic shifts in the absorption maximum of the receptor, enabling the determination of apparent stability constants. We found that receptor 1 binds these anions with moderate to very high strength, in the order I– < NO3– < Br– < NO2– < Cl– (Table 1). Due to the very strong interaction of receptor 1 with chlorides and the variability of data due to water capture during titrations, we were able to obtain reliable stability constants when we standardized conditions and carried out experiments in the presence of 0.5% water. In the case of a basic anion, such as acetate, dihydrogen phosphate or hydrogen phosphate deprotonation was observed. Interestingly, when receptor 1 was titrated in the presence of one equivalent of sodium or potassium cations (added as NaClO4 or KPF6), an enhancement in anion binding was observed in all cases. This can be attributed to an increased acidity of squaramide protons upon interaction of the benzocrown unit with cations, which reinforce interactions with anions.8
Table 1. Association constants (Ka) for interactions between receptor 1 and selected anions and apparent association constants for interaction of 1 with anions in the presence of one equivalent of sodium perchlorate or potassium hexafluorophosphate a .
| 1 | 1 + 1 equiv. Na+ | 1 + 1 equiv. K+ | |
| Cl– | 176 000 b | 492 500 b | 766 000 b |
| NO2– | 100 900 | 363 200 | 509 600 |
| Br– | 49 800 | 142 000 | 217 000 |
| NO3– | 2500 | 5900 | 7100 |
| I– | 1940 | 3850 | 4500 |
| HSO4– | K11 = 15 000 | ||
| K21 = 108 900 | — c | — c | |
| SO42– | K11 = 58 600 | ||
| K21 = 63 300 | — c | — c |
aUV-vis, solvent CH3CN, temperature 293 K, [1] = 3.14 × 10–5 M, anions added as TBA salts [TBAX] ∼3 × 10–3 M; M–1, and errors < 10%.
bTitrations performed in 0.5% water in acetonitrile.
cThe data obtained could not be fitted to a 1 : 1 or 2 : 1 binding mode; 4 : 1 fitting produced no reliable association constants (see the ESI).
As expected, for receptor 1 possessing a benzo-18-crown-6 unit, the highest enhancement in anion binding was observed for titrations performed in the presence of potassium rather than sodium cations. This correlates well with the association constants obtained for complexes of receptor 1 with sodium (added as NaClO4) and potassium (added as KPF6) cations (KNa+ = 48 700 M–1 and KK+ = 133 300 M–1). This is also supported with titrations performed in the presence of 5% water in acetonitrile using NaCl and KCl instead of salts generated in situ. We found that in the presence of water, receptor 1 is still able to recognize these salts with stability constants of 2300 and 4300 M–1 for NaCl and KCl, respectively.
Strikingly, when a divalent sulfate anion was tested as a tetrabutylammonium salt in acetonitrile the 2 : 1 binding mode was better suited while the 1 : 1 fitting produced a high error rate. Furthermore, an analogous titration experiment conducted in the presence of potassium cations produced an isotherm with a two-step binding profile, indicating the occurrence of a more complex binding equilibrium depending on the concentration of in situ generated K2SO4.9 This also demonstrates the differing ability of free receptor 1 and its complex with potassium cations to recognize anions. To gain more insight into the ion pair binding mechanism and prove disparity in monovalent and divalent anions, in particular in nitrate and sulfate binding by receptor 1, 1H NMR experiments were conducted in CD3CN. The analysis of the binding isotherms thus obtained supported the data collected from UV-vis titrations. Specifically, the data collected from nitrate titration in the absence and presence of potassium cation can be simply fitted to the 1 : 1 binding mode, suggesting the formation of a [1 × KNO3] complex. On the other hand, titrations carried out with both TBA2SO4 or in situ generated K2SO4 resulted in inconsistent perturbation in the titration profile. In particular, upon incremental addition of sulfate anions into an acetonitrile solution of receptor 1 containing one equivalent of potassium cations, the signals corresponding to the aromatic and crown ether protons shifted inconsistently. These signals initially shifted downfield, but after exceeding approx. 0.25 equivalents of sulfate anions they moved upfield, suggesting that in equilibrium a 4 : 1 complex (receptor : anion) is present (Fig. 1).10
Fig. 1. Titration curves obtained by following crown ether and aromatic protons upon incremental addition of in situ generated K2SO4 into solution of 1 in CD3CN.
Further confirmation that an inorganic/organic assembly is formed in solution, after addition of SO42– and K+ to the solution of 1 in acetonitrile, came from DOSY-NMR and dynamic light scattering (DLS) studies. In order to prove the formation of a large complex, diffusion NMR experiments were carried out in CD3CN. The formation of such an assembly should cause a significant decrease in the diffusion coefficient (D) values. The collected data clearly indicate the formation of supramolecular structures when TBA2SO4 and KPF6 were added to a solution of receptor 1 (c = 3.04 × 10–3 M) in CD3CN. The presence of in situ generated K2SO4 promoted a significant decrease in the diffusion coefficient of the ligand from D = 10.53 × 10–10 to 7.56 × 10–10 m2 s–1, with ΔD = –28%. On the other hand, the addition of TBAPF6 to the solution of 1 did not affect its diffusion coefficient. Furthermore, DLS measurements were performed to determine the size of the supramolecular assembly in CH3CN. In the case of the addition of TBA2SO4 and KPF6 to the solution of 1, the Z-average provided confirmation for the formation of a large supramolecular assembly. The value of the hydrodynamic diameter was found to be dH = 28 nm while receptor 1 alone was not detectable. Similarly, receptor 1 pretreated with in situ generated KNO3 was also not detectable by DLS, suggesting that KNO3 does not promote the formation of a large assembly in solution.
Final evidence of the formation of 4 : 1 and 1 : 1 complexes for sulfate and nitrate promoted assemblies came from X-ray analysis.11 Despite making a number of attempts to crystallize complexes of receptor 1 with K2SO4, we were unable to obtain crystals. However, slow diffusion of diethyl ether into an MeCN/MeOH solution of 1 pretreated with an excess of Na2SO4 enabled us to obtain crystals suitable for X-ray diffraction analysis. The resulting structure revealed the formation of a complex with 4 : 1 stoichiometry, entirely embedding the sulfate into a shell formed by four ligands (Fig. 2a). The sulfate anion is surrounded by disordered receptor moieties oriented so as to form eight hydrogen bonds between the oxygen atoms of the tetrahedron-shaped sulfate and the amide hydrogen atoms. The electroneutrality of the [1 × Na2SO4] complex is ensured by two sodium cations trapped in benzocrown cavities by two out of four ligands. A different single-crystal structure was formed after slow diffusion of diethyl ether into an MeCN/MeOH solution of 1 obtained after solid–liquid extraction with KNO3. In the [1 × KNO3] crystal structure the asymmetric part of the unit cell consists of two ligands and two KNO3 ion pairs. Like in the case of the previous structure, the potassium cations reside in the benzocrown cavities and are in addition coordinated from both sides by nitrate anions, giving an infinite 1-D polymeric assembly (Fig. 2b). Strong hydrogen bond interactions between squaramide protons and NO3– anions result in the formation of supramolecular layers of moieties. Interestingly, in a salt-free environment, ligand 1 crystallizes as a monohydrate [1 × H2O] with water molecules located in the benzocrown part and additionally coordinated by H atoms of the squaramide moiety, giving an altogether dimeric arrangement (Fig. 2c).
Fig. 2. Molecular assemblies in the single crystal structures of (a) [1 × Na2SO4], (b) [1 × KNO3] and (c) [1 × H2O].
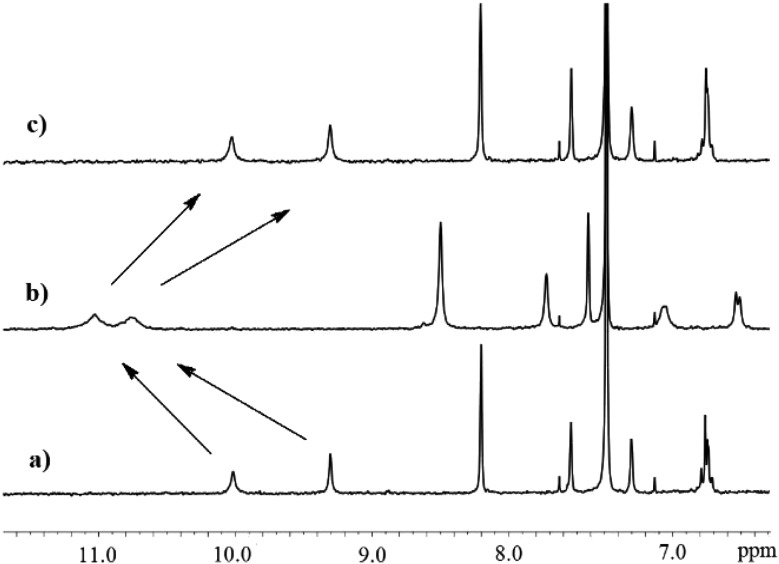
Taking into consideration the different properties of nitrate and sulfate complexes with 1 formed in solution and in the solid state, we envisioned that using a squaramide-based ion pair receptor would be able to differentiate between these salts under interfacial conditions and preferentially extract sulfates into an organic phase. As a first test, LLE experiments were conducted under 1H NMR control using a 2 mM solution of 1 in CDCl3 and various aqueous 50 mM salt solutions (KCl, KBr, KNO3, KNO2, Na2SO4, KH2PO4 and K2SO4). With the exception of K2SO4 extraction, in all cases the precipitation of solids was noted, showing the insolubility of the supramolecular species formed in CDCl3. This opens up a unique opportunity to distinguish these salts under interfacial conditions by preferentially increasing the solubility of sulfate complexes with receptor 1 rather than complexes of 1 with other salts. Indeed, the comparison of the 1H NMR spectrum of free 1 in water saturated CDCl3 revealed substantial changes in the resonance signals after contact with an aqueous solution of K2SO4 (Fig. 3b).
Fig. 3. Partial 1HNMR spectra of receptor 1: (a) 2.0 mM in wet CDCl3, (b) after extraction of 50 mM K2SO4 and (c) after back extraction to distilled water.
This clearly indicates that receptor 1 is able to extract extremely hydrophilic sulfate anions from the aqueous to the organic phase and form complexes in the organic layer.12 This is also supported by electron spray ionization mass spectrometric measurements of the extracted organic solution, which clearly shows characteristic peaks (m/z) appearing at 2750.7 [4 × 1 + K2SO4 + K+] and 1316.3 [4 × 1 + SO42–] (see the ESI†). Control experiments with an analogous urea based ion pair receptor or squaramide based ion pair receptor equipped with a benzo-15-crown-5 ether or anion receptor lacking a crown ether unit revealed an inability to extract either K2SO4 or Na2SO4 under such conditions (the receptors denoted in the ESI as S4, S5 and S3,† respectively). Furthermore, the ability to release potassium sulfate by receptor 1 was confirmed by a 1H NMR back extraction experiment, which resulted in the signals returning to the initial position after contacting with water (Fig. 3c).
In order to establish the extraction efficiency in the experiments described above, we used atomic emission spectroscopy (AES) and quantified the potassium content in the organic phase. The fraction of receptor 1 molecules occupied by a potassium cation after extraction of 2 mM solution of 1 in chloroform with 50 mM K2SO4 aqueous solution was determined to be 36%. After back extraction the potassium content was found to be negligible. This suggests that receptor 1 may act as a suitable candidate as a sulfate transporter. By contrast, neither a binary mixture of a squaramide based anion receptor (denoted as receptor S3 in the ESI†) with 4-nitrobenzo-18-crown-6-ether nor a urea based ion pair receptor (denoted as receptor S4 in the ESI†) is an effective ionophore (see the ESI†). Efforts were then made to prove the selectivity of receptor 1 towards sulfates and its ability to overcome the Hofmeister bias. Thus we carried out competitive extraction experiments under ion chromatography control. Specifically, an aqueous mixture of KNO3 and K2SO4 (5 mM each) was extracted with a 5 mM or 20 mM solution of 1 in chloroform, respectively. In both cases a significant decrease in the concentration of sulfates was detected while nitrates were affected only slightly. The drop in concentration was calculated to be 25 and 74% for sulfates and 4.9 and 7.2% for nitrates, using 5 and 20 mM 1 in chloroform, respectively (Fig. 4). Importantly, the selectivity of receptor 1 towards sulfates was retained even when the spectrum of potassium salts in the aqueous mixture was extended to chlorides, bromides, nitrites and dihydrogen phosphates. On the other hand, the use of a solution containing basic salts such as divalent or trivalent phosphates causes no phase separation most likely due to the deprotonation of the receptor. This indicates the limitations of this system operating at neutral and acidic pH (see the ESI†).
Fig. 4. Chromatograms obtained during extraction experiments after tenfold dilution: (a) source phase, (b) after extraction with 5 mM 1 in CHCl3 and (c) after extraction with 20 mM 1 in CHCl3.
In order to prove the high performance and very high selectivity of receptor 1 towards sulfates we performed more competitive extraction experiments using an aqueous solution containing one order of magnitude higher concentration of potassium nitrate (50 mM) than potassium sulfate. For this reason, the concentration of anions was monitored in the aqueous phase after back extraction. We found that under such conditions receptor 1 is still able to preferentially extract extremely hydrophilic sulfates rather than nitrates (Fig. 5). Taking into consideration the assumed stoichiometry of complexes formed with receptor 1 in the organic phase (1 : 1 for KNO3 and 4 : 1 for K2SO4) the extraction efficiency was calculated to be 6 and 64% for potassium nitrate and potassium sulfate, respectively. From a practical point of view, this applies to approx. 0.32/0.12 molar ratio of sulfate/nitrate ions in the extract.
Fig. 5. Chromatogram obtained during back extraction experiments after tenfold dilution. Organic phase: 20 mM receptor 1 in chloroform.
Finally, we modified the receptor structure to act as a simple optical sensor (receptor 2) capable of “naked eye” recognition of sulfates. The addition of potassium sulfate to 2.1 mM solution of 2 in DMSO did indeed result in a drastic color change from orange to purple, while the addition of potassium nitrate did not affect the color change. Similarly, only an aqueous solution of K2SO4 induced a color change from yellow to red after contacting with 0.5 mM solution of 2 in nitrobenzene (Fig. 6). To gain more insight into the binding ability of receptor 2 and to establish its mechanism of action, we added tetrabutylammonium nitrate and tetrabutylammonium sulfate to the solution of 2 in deuterated DMSO. We found that the nitrate anion does not influence the chemical shifts of signals corresponding to the squaramide protons of receptor 2. This clearly demonstrates that in such competitive media receptor 2 is not able to form complexes with this anion. On the other hand, the addition of tetrabutylammonium sulfate to the solution of 2 in deuterated DMSO, apart from the color change, resulted in the disappearance of squaramide signals in the 1H NMR spectrum, suggesting deprotonation rather than formation of complexes. This was supported by UV-vis measurements. Particularly, the addition of TBANO3 to the solution of receptor 2 in DMSO does not affect its UV-vis spectrum. Contrarily, upon addition of sulfate anions or basic anions such as acetate or hydroxide, a new bathochromic band in the UV-vis spectra was observed. Based on these findings we concluded that the color change upon addition of sulfates to the solution of 2 originates more likely from the deprotonation event, which is unusual for these anions, making possible the optical differentiation of nitrates and sulfates. This shows its sensing limitations because basic anions also cause a change in the color of the receptor 2 (see the ESI†). Nevertheless, this system can serve as a facile supportive sensor for monitoring competitive NO3–/SO42– extraction processes using receptor 1.
Fig. 6. SLE and LLE experiments. (A) (From left) solution of 2 in DMSO, after addition of solid KNO3, and after addition of solid K2SO4. (B) (From left) solution of 2 in nitrobenzene and water, solution of 2 in nitrobenzene and aqueous KNO3, and solution of 2 in nitrobenzene and aqueous K2SO4.
Conclusions
In summary, a new ion pair receptor was synthesized and characterized by standard spectroscopic methods as well as by DOSY, DLS and single X-ray crystal diffraction analyses. Based on titration experiments we evidenced the high ability of receptor 1 to form complexes with ion pairs. In contrast to potassium salts of the monovalent anions tested, which form 1 : 1 stoichiometry complexes with receptor 1, in the case of potassium sulfate a 4 : 1 complex was evidenced. This feature was utilized to differentiate the solubility of complexes in organic media in favor of the 4 : 1 assembly and enable selective extraction of potassium sulfate from aqueous solution to chloroform. To the best of our knowledge, compound 1 is the first ion pair receptor capable of extracting the extremely hydrophilic sulfate anion in the form of an alkali metal salt from the aqueous to the organic phase. We have demonstrated that receptor 1 extracts potassium sulfate selectively even in the presence of lipophilic anions such as nitrates, as shown independently by qualitative ion chromatography analysis. A simple modification of the receptor structure allowed us to obtain a “naked eye” optical sensor 2 capable of detecting sulfates under both SLE and LLE conditions and to do so with selectivity relative to potassium nitrate.
Conflicts of interest
The authors declare no conflicts of interest.
Supplementary Material
Acknowledgments
This work was supported by Grant no. 2018/30/E/ST5/00841 from the National Science Centre, Poland.
Footnotes
†Electronic supplementary information (ESI) available: General information, synthetic details, 1H-NMR and UV-vis titration data, extraction experimental details, X-ray crystal data for 1 × Na2SO4 (CCDC 1877168), 1 × KNO3 (CCDC 1877169) and 1 × H2O (CCDC 1877170). CCDC 1877168–1877170. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9sc02923k
References
- (a) Ravikumarw I., Ghosh P. Chem. Soc. Rev. 2012;42:3077–3098. doi: 10.1039/c2cs15293b. [DOI] [PubMed] [Google Scholar]; (b) Olomu A. B., Vickers C. R., Waring R. H., Clements D., Babbs C., Warnes T. W., Elias E. N. Engl. J. Med. 1988;318:1089–1092. doi: 10.1056/NEJM198804283181703. [DOI] [PubMed] [Google Scholar]; (c) Amerongen A. V. N., Bolscher J. G. M., Bloemena E., Veerman E. C. I. Biol. Chem. 1998;379:1–26. doi: 10.1515/bchm.1998.379.1.1. [DOI] [PubMed] [Google Scholar]; (d) Murch S. H., MacDonald T. T., Walker-Smith J. A., Levin M., Lionetti P., Klein N. J. Lancet. 1993;341:711–714. doi: 10.1016/0140-6736(93)90485-y. [DOI] [PubMed] [Google Scholar]; (e) Fritz K. M., Fulton S., Johnson B. R., Barton C. D., Jack J. D., Word D. A., Burke R. A. J. North Am. Benthol. Soc. 2010;29:673–689. [Google Scholar]; (f) Dawson P. A., Beck L., Markovich D. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13704–13709. doi: 10.1073/pnas.2231298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Y., in Ion Properties, ed. M. Dekker, New York, 1997. [Google Scholar]
- (a) Custelcean R., Moyer B. A. Eur. J. Inorg. Chem. 2007;10:1321–1340. [Google Scholar]; (b) Moyer B. A. and Singh R. P., in Fundamentals and Applications of Anion Separation, ed. A. B. Bruce and R. P. Raj, Kluwert Academic/Plenum, New York, 2004. [Google Scholar]
- (a) Moyer B. A., Custelcean R., Hay B. P., Sessler J. L., Bowman-James K., Day V. W., Kang S. O. Inorg. Chem. 2013;52:3473–3490. doi: 10.1021/ic3016832. [DOI] [PubMed] [Google Scholar]; (b) Katayev E. A., Ustynyuk A., Sessler J. L. Coord. Chem. Rev. 2006;250:3004–3037. [Google Scholar]
- (a) Jia C., Wu B., Li S., Huang X., Zhao Q., Li Q. S., Yang X. J. Angew. Chem., Int. Ed. 2011;50:486–490. doi: 10.1002/anie.201004461. [DOI] [PubMed] [Google Scholar]; (b) Fowler C. J., Haverlock T. J., Moyer B. A., Shriver J. A., Gross D. E., Marquez M., Sessler J. L., Hossain M. A., Bowman-James K. J. Am. Chem. Soc. 2008;130:14386–14387. doi: 10.1021/ja806511b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Borman C. J., Custelcean R., Hay B. P., Bill N. L., Sessler J. L., Moyer B. A. Chem. Commun. 2011;47:7611–7613. doi: 10.1039/c1cc12060c. [DOI] [PubMed] [Google Scholar]; (d) Williams C. A., Seipp N. J., Bryantsev V. S., Moyer B. A. Sep. Sci. Technol. 2018;53:1864–1873. [Google Scholar]; (e) Kim S. K., Lee J., Williams N. J., Lynch V. M., Hay B. P., Moyer B. A., Sessler J. L. J. Am. Chem. Soc. 2014;136:15079–15085. doi: 10.1021/ja5086996. [DOI] [PubMed] [Google Scholar]; (f) Williams N. J., Seipp C. A., Garrabrant K. A., Custelcean R., Holguin E., Keum J. K., Ellias J. R., Moyer B. A. Chem. Commun. 2018;54:10048–10051. doi: 10.1039/c8cc05115a. [DOI] [PubMed] [Google Scholar]; (g) Akhuli B., Ra-vikumar I., Ghosh P. Chem. Sci. 2012;3:1522–1530. [Google Scholar]
- (a) Wintererst M. P., Levitskaia T. G., Moyer B. A., Sessler J. L., Delmau L. H. J. Am. Chem. Soc. 2008;130:4129–4139. doi: 10.1021/ja7102179. [DOI] [PubMed] [Google Scholar]; (b) White D. J., Laing N., Miller H., Parsons S., Tasker P. A., Coles S. Chem. Commun. 1999;20:2077–2078. [Google Scholar]; (c) Kim S. K., Lynch V. M., Young N. J., Hay B. P., Lee C.-H., Kim J. S., Moyer B. A., Sessler J. L. J. Am. Chem. Soc. 2012;134:2087–20843. doi: 10.1021/ja310673p. [DOI] [PubMed] [Google Scholar]; (d) He Q., Zhang Z., Brewster J. T., Lynch V. M., Kim S. K., Sessler J. L. J. Am. Chem. Soc. 2016;138:9779–9782. doi: 10.1021/jacs.6b05713. [DOI] [PubMed] [Google Scholar]; (e) Romański J., Piątek P. J. Org. Chem. 2013;78:4341–4347. doi: 10.1021/jo4003322. [DOI] [PubMed] [Google Scholar]; (f) He Q., Williams N. J., Oh J. H., Lynch V. M., Kim S. K., Moyer B. A., Sessler J. L. Angew. Chem., Int. Ed. 2018;57:1–6. doi: 10.1002/anie.201805127. [DOI] [PubMed] [Google Scholar]; (g) Mahoney J. M., Beatty A. M., Duggan P. J., Smith B. D. Inorg. Chem. 2004;43:5902–5907. doi: 10.1021/ic0494859. [DOI] [PubMed] [Google Scholar]; (h) Jagleniec D., Siennicka S., Dobrzycki Ł., Karbarz M., Romański J. Inorg. Chem. 2018;57:12941–12952. doi: 10.1021/acs.inorgchem.8b02163. [DOI] [PubMed] [Google Scholar]; (i) He Q., Vargas-Zúñiga G. I., Kim S. H., Kim S. K., Sessler J. L. Chem. Rev. 2019 doi: 10.1021/acs.chemrev.8b00734. [DOI] [Google Scholar]
- Pflugrath J. W., Quiocho F. A. Nature. 1985;314:257–260. doi: 10.1038/314257a0. [DOI] [PubMed] [Google Scholar]
- (a) Mäkelä T., Kiesilä A., Kalenius E., Rissanen K. Chem.–Eur. J. 2016;22:14264–14272. doi: 10.1002/chem.201602362. [DOI] [PubMed] [Google Scholar]; (b) Mäkelä T., Rissanen K. Dalton Trans. 2016;45:6481–6490. doi: 10.1039/c6dt00414h. [DOI] [PubMed] [Google Scholar]; (c) Karbarz M., Romański J. Inorg. Chem. 2016;55:3616–3623. doi: 10.1021/acs.inorgchem.6b00132. [DOI] [PubMed] [Google Scholar]; (d) Mäkelä T., Kalenius E., Rissanen K. Inorg. Chem. 2015;54:9154–9156. doi: 10.1021/acs.inorgchem.5b01577. [DOI] [PubMed] [Google Scholar]
- (a) Elmes R. B. P., Yuen K. K. Y., Jolliffe K. A. Chem.–Eur. J. 2014;20:7373–7380. doi: 10.1002/chem.201400292. [DOI] [PubMed] [Google Scholar]; (b) Young P. G., Jolliffe K. A. Org. Biomol. Chem. 2012;10:2664–2672. doi: 10.1039/c2ob06964d. [DOI] [PubMed] [Google Scholar]; (c) Dungan V. J., Ngo H. T., Young P. G., Jolliffe K. A. Chem. Commun. 2013;49:264–266. doi: 10.1039/c2cc37686e. [DOI] [PubMed] [Google Scholar]; (d) Gale P. A., Hiscock J. R., Jie C. Z., Hursthouse M. B., Light M. E. Chem. Sci. 2010;1:215–220. [Google Scholar]
- Bąk K., Chmielewski M. Chem. Commun. 2014;50:1305–1308. doi: 10.1039/c3cc48463g. [DOI] [PubMed] [Google Scholar]
- (a) APEX3, Bruker AXS Inc., Madison, Wisconsin, USA, 2017.; (b) SAINT, Bruker AXS Inc., Madison, Wisconsin, USA, 2017.; (c) SADABS, Bruker AXS Inc., Madison, Wisconsin, USA, 2016.; (d) TWINABS, Bruker AXS Inc., Madison, Wisconsin, USA, 2012.; (e) Sheldrick G. M. Acta Crystallogr., Sect. A: Found. Adv. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Sheldrick G. M. Acta Crystallogr., Sect. C: Struct. Chem. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Parsons S., Flack H. D., Wagner T. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2013;69:249–259. doi: 10.1107/S2052519213010014. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Cowley J. M., in International Tables for Crystallography, ed. J. C. A. Wilson, Kluwer, Dordrecht, The Netherlands, 1992,vol. Cpp. 223–245. [Google Scholar]; (i) Macrae C. F., Bruno I. J., Chisholm J. A., Edgington P. R., McCabe P., Pidcock E., Rodriguez-Monge L., Taylor R., van de Streek J., Wood P. A. J. Appl. Crystallogr. 2008;41:466–470. [Google Scholar]; (j) Putz H. and Brandenburg GbR K., Diamond – Crystal and Molecular Structure Visualization, Crystal Impact, Kreuzherrenstr, 102, 53227 Bonn, Germany.
- Jagleniec D., Karbarz M., Romański J., Polish Patent Application No. P.429164, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.