Abstract
Botanical dietary supplements are complex mixtures that can be highly variable in composition and quality, making safety evaluation difficult. A key challenge is determining how diverse products in the marketplace relate to chemically and toxicologically characterized reference samples (i.e., how similar must a product be in order to be well-represented by the tested reference sample?). Ginkgo biloba extract (GBE) was used as a case study to develop and evaluate approaches for determining sufficient similarity. Multiple GBE extracts were evaluated for chemical and biological-response similarity. Chemical similarity was assessed using untargeted and targeted chemistry approaches. Biological similarity was evaluated using in vitro liver models and short-term rodent studies. Statistical and data visualization methods were then used to make decisions about the similarity of products to the reference sample. A majority of the 26 GBE samples tested (62%) were consistently determined to be sufficiently similar to the reference sample, while 27% were different from the reference GBE, and 12% were either similar or different depending on the method used. This case study demonstrated that approaches to evaluate sufficient similarity allow for critical evaluation of complex mixtures so that safety data from the tested reference can be applied to untested materials.
Graphical abstract
Introduction
Botanical dietary supplements are used by approximately 18% of adults (Clarke et al., 2015), or over 40 million people, in the United States. Therefore, the safety of these products is an important public health concern. For each type of botanical dietary supplement, there are numerous products available in the marketplace. These products are complex mixtures that can vary significantly depending on the ingredients (e.g., plant part, growing conditions), formulation (e.g., presence of different excipients like stabilizing agents or fillers), processing procedures, and storage conditions. Adulteration, either economically-motivated (i.e., addition of less expensive plant material) or with pharmaceutical compounds, adds to the variability in botanical dietary supplements. In a companion paper, (Shipkowski et al., 2018) provide a detailed discussion of the many challenges associated with assessing the safety of botanical dietary supplements.
Studies to evaluate the toxicity (or efficacy) of a botanical dietary supplement are typically limited to a single sample (e.g., unfinished extract from a certain supplier or finished product from a specific manufacturer) based on the inherent assumption that the selected sample is representative of all products with the same name on the label. However, there has been little effort to systematically evaluate this assumption. Two major challenges in determining whether toxicological data from a tested sample can be used to evaluate the safety of an untested sample are 1) the development of methods for evaluating sufficient similarity (i.e., what are the best methods to compare complex mixtures?), and 2) the establishment of categorical or quantitative similarity criteria (i.e., what level of similarity is sufficient?).
The term sufficient similarity, also referred to as phytoequivalence in the parlance of botanical dietary supplements, applies to complex mixtures and indicates that two mixtures are similar enough that the toxicity or efficacy data from one of the mixtures (reference mixture) can be used as a surrogate for the mixture(s)-of-interest. A reference mixture can be selected based on a pre-determined quality standard or a high level of characterization (i.e., the known to which the unknown is being compared). To our knowledge, there has not been a concerted effort to evaluate sufficient similarity of botanicals using both chemical and biological-response measures. However, several organizations have made significant progress in establishing the authenticity of botanicals and identifying potential adulterants based on chemical analysis of samples, including the USDA (particularly the Food Composition and Methods Development Laboratory) (Harnly et al., 2012, Harnly et al., 2013, Harnly et al., 2016), the Association of Official Analytical Chemists (AOAC) International (2012), the United States Pharmacopeial Convention (USP) (2015), and the American Botanical Council (Gafner et al., 2015). These efforts have been instrumental in providing manufacturers with the tools needed to evaluate the quality of their products.
The approaches developed to evaluate authenticity and detect adulteration of botanical dietary supplements have relied on both untargeted and targeted chemistry approaches. Untargeted chemistry is defined here as any method that is used to detect the presence and relative abundance of constituents without prior knowledge of their identity. Untargeted chemistry approaches are intended to characterize as much of the sample composition as possible. These methods make no attempt to identify or quantify individual constituents within the mixture. However, they are not truly “untargeted” in that the methods used for sample preparation and the specific detection device used will influence the range of chemical structures included in the chromatographic profile. For example, the extraction process used to prepare samples for chemical analysis by chromatography will target components of a polarity range determined by the solvent. A true untargeted approach would employ multiple solvents with a range of polarities. In contrast to untargeted chemistry, targeted chemistry uses methods that have been qualified for each constituent-of-interest and requires authentic standards to quantify select marker constituents. Targeted chemistry can serve as a complement to untargeted chemistry by confirming the identity of specific analytes noted in chromatographic profiles generated using untargeted chemistry. Targeted chemistry is often used to quantify the purported active constituent(s) or marker constituents (if the active constituent is unknown) of a botanical product to confirm that the appropriate concentration is present in a standardized sample. For example, organizations such as the U.S. Pharmacopeia (http://www.usp.org/verification-services) and NSF International (http://www.nsf.org/services/by-industry/dietary-supplements) use targeted chemistry techniques to verify that the contents of a dietary supplement match label claims in their certification programs.
While there is not a history of sufficient similarity work involving botanical dietary supplements, there have been a limited number of examples evaluating similarity of complex environmental chemical mixtures such as water disinfection byproducts, petroleum substances, and pesticide mixtures (Rice et al., 2009, Marshall et al., 2013, Murray et al., 2013). These approaches have typically relied on quantification and comparison of select constituents in the mixture (Marshall et al., 2013) or structurally-defined classes of constituents (Murray et al., 2013), inferring that the toxicologically-active constituents are captured by the chemical analysis. However, it is important to note that there is usually a very large unidentified fraction in botanical products, and the active constituents are often unknown. Therefore, targeted analysis of select chemical constituents could be inadequate for determining similarity. In addition to examples that rely on chemical analysis alone, a few studies with environmental mixtures have also included in vitro biological data in the evaluation of similarity (Schenck et al., 2009, Grimm et al., 2016). In this manuscript, we explore sufficient similarity in terms of both chemical and biological-response similarity as applied to a complex botanical mixture: Ginkgo biloba extract (GBE).
Sufficient similarity framework
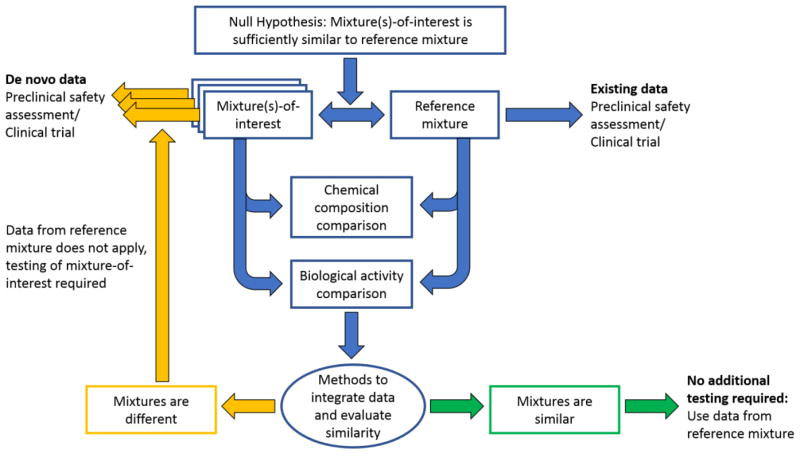
An overview of the framework for determining sufficient similarity is presented in Figure 1. The process begins with the hypothesis that an untested mixture or mixture-of-interest is “sufficiently similar” to a reference mixture (i.e., the mixture that has undergone comprehensive toxicological evaluation). Note, there can be multiple reference mixtures and/or multiple mixtures-of-interest. Next, the reference mixture and the mixture-of-interest undergo a chemical analysis to compare constituents and a biological activity assessment to compare responses generated from each mixture. The type(s) of chemical analysis (e.g., untargeted chemistry, targeted chemistry) and biological assessment (e.g., in vitro assays with specific biological targets, -omics) require careful consideration of existing information and should be tailored to the mixture under study. For example, are there known bioactive constituents that should be a focus of chemical characterization or are the active constituents unknown, requiring a more global chemical analysis? Are the biological targets of the reference mixture well-defined (e.g., nuclear receptor activation) or not, indicating a more comprehensive biological assessment is required (e.g., toxicogenomic evaluation)?
Figure 1.
Framework for determining sufficient similarity of mixture(s)-of-interest to a reference mixture that has been chemically and toxicologically characterized.
Following chemical and biological activity assessments, methods are required to integrate these divergent data streams and evaluate overall similarity of the mixture-of-interest to the reference mixture. Determination of sufficient similarity requires definition of the magnitude of acceptable difference. This critical step involves scientific judgment and requires focused attention from stakeholders and experts to develop standards that could be applied more broadly, as opposed to the current case-by-case approach. Mixtures-of-interest that are determined to be sufficiently similar to the reference mixture require no further testing, as toxicological data from the reference mixture can be used to estimate the hazard of the mixtures-of-interest. Mixtures-of-interest that are deemed to be different from the reference mixture cannot use data from the reference mixture and require additional toxicological characterization to complete a safety assessment.
This manuscript describes application of the general framework for determining sufficient similarity discussed above using GBE as a case study. Botanical dietary supplements, such as GBE, are ideally suited for exploring the issues surrounding sufficient similarity of complex mixtures because they involve both widespread, relatively high human exposure and pressing public health questions about quality and safety (Cohen, 2016). The components of the case study will be described first, followed by examples of the application of sufficient similarity methods using data generated at the National Toxicology Program (NTP). Detailed methods and results for each of the data streams used in the examples are beyond the scope of this summary and will be published separately. Instead, this paper addresses the process of evaluating sufficient similarity, which could potentially be applied to other complex mixtures using any number of chemical and biological-response data streams. The goals of this work are to develop and evaluate methods for determining chemical and biological-response similarity and identify challenges that require research attention.
Ginkgo biloba extract: A case study to explore sufficient similarity
GBE was selected for case study development by the NTP because it offers a relatively tractable starting place. GBE is derived from Ginkgo biloba leaves, and standardized GBEs have a well-characterized chemical profile compared to many other botanicals (van Beek and Montoro, 2009). Two major chemical classes in GBE, flavonol glycosides and terpene lactones, are considered to be responsible for its bioactivity. Flavonol glycosides have antioxidant potential (Ding et al., 2009), while terpene lactones (e.g., bilobalide, ginkgolides A, B, C, and J) are antagonists of platelet activating factor receptor (Braquet, 1987), glycine receptor (Heads et al., 2008), and γ aminobutyric acid (GABAA) receptor (Huang et al., 2004). Standardized GBE, including the unofficial industry “gold standard” EGb761® (Dr. Willmar Schwabe Pharmaceuticals), should contain 24% flavonol glycosides, 6% terpene lactones, and less than 5 ppm ginkgolic acids (van Beek and Montoro, 2009). Other “standardized” GBEs in the marketplace purportedly meet these established levels. However, efforts to chemically characterize GBEs from the market have found a great deal of inconsistency in constituent concentrations (Kressmann et al., 2002, Sloley et al., 2003, Fransen et al., 2010). The existence of a gold standard in the marketplace with EGb761® (DeFeudis, 2003) provides an important point of comparison for quality in the GBE case study.
One approach to quantifying the flavonol glycoside content in GBE samples is to convert the glycosides to the corresponding aglycones (i.e., quercetin, kaempferol, isorhamnetin) via hydrolysis, as analytical standards for the aglycones are commonly available. The aglycone values can then be used to estimate glycoside content using conversion factors. However, measurement of flavonol aglycones in hydrolyzed samples has been exploited by some manufacturers and has resulted in products that meet label claims for flavonol glycoside content but have a substandard flavonol profile because of either adulteration or degradation (Lin et al., 2008, Harnly et al., 2012). Comparing the pre- and post-hydrolysis data allows for detection of potential adulteration. Therefore, in both the untargeted and targeted chemical analyses of GBE in this case study, data were generated for both pre- and post-hydrolysis samples.
The in vivo toxicity of GBE has also been thoroughly characterized. In NTP toxicity and carcinogenicity studies in mice and rats, GBE displayed clear toxicological effects including hepatotoxicity (NTP, 2013, Rider et al., 2014). Furthermore, there is some information available on the mechanism of action (i.e., CAR- and PXR- mediated enzyme induction) associated with observed hepatotoxicity (Guo et al., 2010, Maeda et al., 2014, Maeda et al., 2015).
GBE samples
The first phase in case study development involved procuring GBEs for comparison (Table 1). Multiple unfinished GBEs (bulk GBE that serves as source material for finished GBE products) as well as a limited number of finished GBE products (commercially available GBE-containing tablets or capsules) were evaluated alongside the reference unfinished GBE. The reference sample (labeled as Sample 1 in tables and figures) was so designated because it underwent comprehensive chemical analysis both in 2003, at the time of procurement, and in the current program (2015–2016), as well as comprehensive toxicological evaluation in 3-month and 2-year toxicity and carcinogenicity studies in mice and rats (NTP, 2013, Rider et al., 2014). This reference GBE was subdivided into multiple aliquots that were kept under different storage conditions. Samples 1 and 1A were stored at ambient conditions from 2003 to 2014 and 2015, respectively, and then transferred to −20°C storage, while sample 1F was stored at −20°C since receipt in 2003.
Table 1.
Ginkgo biloba extract case study overview.
| Botanical | Ginkgo biloba extract | Sample identifiers |
|---|---|---|
|
| ||
| Materials | 3 National Toxicology Program reference extracts | 1, 1A, 1F |
| 20 Unfinished extractsa | A – T | |
| 1 Standard reference material (NIST), extract | U | |
| 1 Standard Reference Material (NIST), finished product | V | |
| 4 Finished products (containing EGb761®) | W, X, Y, Z | |
| 12 GBE constituentsb | ||
|
| ||
| Data Streams | Samples Tested | |
|
| ||
| Chemistry Biology | Targeted and untargeted chemical analyses | 1, 1A, 1F, A-Z |
| In vitro | Primary human hepatocyte assays | 1, 1A, 1F, A-Z, Constituents |
| Attagene HepG2 reporter gene assays | 1, 1A, 1F, A-Z, constituents | |
| In vivo | 5-day rat studies | 1, G, N, P, T |
|
| ||
| Comparison methods | ||
|
| ||
| Strength-of-evidence | ||
| Empirical equivalence testing | ||
| Visual interval evaluation | ||
Unfinished GBEs were obtained from several suppliers: Supplier 1 (samples A-C), Supplier 2 (sample D), Supplier 3 (sample E), Supplier 4 (sample F), Supplier 5 (sample G-I), Supplier 6 (sample J), Supplier 7 (sample K), Supplier 8 (sample L), Supplier 9 (samples M-N), Supplier 10 (sample O), Supplier 11 (sample P), Supplier 12 (sample Q), Supplier 13 (sample R), Supplier 14 (sample S), Supplier 15 (sample T).
Bilobalide, ginkgolide A, ginkgolide B, ginkgolide C, ginkgolide J, ginkgolic acid 15, ginkgolic acid 17, ginkgotoxin, rutin trihydrate, isorhamnetin, kaempferol, quercetin.
The unfinished GBE samples consisted of 20 GBEs purchased from multiple suppliers in 2014 (samples A-T; Table 1). The goal of sample procurement was to include multiple GBEs from a variety of suppliers. Two standard reference materials (SRMs) from the National Institutes of Standards and Technology (NIST) were also included: GBE - SRM 3247 (sample U) and GBE-containing tablets - SRM 3248 (sample V). Sample U represents the same type of material as the unfinished GBE samples and should be directly comparable, whereas sample V was included to illustrate the differences between unfinished GBE and GBE-containing finished products. Four EGB761®-containing finished products were also included for comparison (samples W, X, Y, and Z). A correction was applied to samples W-Z to allow for comparison of the finished products to the unfinished GBE samples. In effect, the amount of samples W, X, Y, and Z assessed was increased to render equivalent amounts of GBE across tested samples. This adjustment was not possible with sample V because the label did not specify the amount of GBE in the finished product. It is important to note that sample W is a combination botanical product, containing both GBE and another “active” botanical (Gotu kola) in the finished product, according to the label. Additionally, 12 GBE constituents (bilobalide, ginkgolide A, ginkgolide B, ginkgolide C, ginkgolide J, ginkgolic acid 15, ginkgolic acid 17, ginkgotoxin, rutin, isorhamnetin, kaempferol, and quercetin) were included for comparison. All of the samples purchased for the case study (samples A-Z and GBE constituents) were stored at −20°C upon receipt.
Overview of data streams, integration approaches, and determining similarity
The intent of the current work is to present the framework for integrating data streams and evaluating sufficient similarity of nominally-related samples. The case study for GBE included multiple chemical and biological data streams: untargeted and targeted chemistry, in vitro assays, and a short-term in vivo assay (Table 1). As noted previously, comprehensive descriptions of the materials and methods, raw data, and data processing for each data stream are not included in the current manuscript. A brief overview describing the nature of each data stream used in the current case study is presented below. Despite the diversity of data types included in the case study (e.g., untargeted chemistry versus gene expression data from human hepatocytes), there were commonalities in data interpretation. First, each data stream was considered individually and any data processing steps required for interpretation were performed. For example, untargeted chemistry data required peak alignment, while gene expression data required calculation of fold increase from control values. Next, multidimensional analysis tools were used to assess the relationships among all tested GBE samples for each data stream. Principal component analysis (PCA) was used for untargeted chemistry, while hierarchical clustering was used for all other data streams. Finally, similarity of each tested sample (samples A-Z) was determined in relation to the reference sample (sample 1).
Following analysis of the individual data streams, methods were used to combine chemical and biological data to draw conclusions about overall similarity of the mixtures-of-interest (samples A-Z) to the reference mixture (sample 1). The methods used to integrate data and evaluate similarity were the strength-of-evidence approach, empirical equivalence testing, and visual interval evaluation. It is important to emphasize that all these methods involve distillation and simplification of complex, nuanced data. The goal of this work was to provide pragmatic approaches for making decisions about sufficient similarity of complex mixtures in a safety assessment scenario. As such, the proposed approaches were intended to be simple, easy to apply, and adaptable to diverse mixture types and data streams. Application of these methods to distinct types of complex mixtures and data streams will be required to assess the limitations of their use. In addition, these simple approaches should be compared to more sophisticated analyses to evaluate their performance and inform future iterations.
In the strength-of-evidence approach, each data stream was first evaluated in isolation. Then, scientific judgment was used to categorize tested GBE samples as ‘similar’ to, ‘maybe similar’ to, or ‘different’ from the reference GBE for each dataset. Categorization was based on the following simple rules for all datasets: samples in the same statistical group or cluster as the reference sample were considered to be ‘similar’, samples in the most different group or cluster from the reference were considered to be ‘different’, and samples that were neither in the same nor in the most different groups were considered ‘maybe similar’. See an illustration of these rules in Figure 2. An overall determination of similarity for each GBE sample was then based on a strength-of-evidence evaluation that considered the similarity calls for each data source. The second method for evaluating sufficient similarity was empirical equivalence testing, which is based on a statistical test for equivalence (Marshall et al., 2013). Finally, the third method, referred to as visual interval evaluation, was developed to visualize similarity of samples using both chemical and biological response data. The second and third methods for determining sufficient similarity share some common features, including conversion of chemical data to distance measures relative to the reference sample, integration of chemical and biological data prior to determination of sufficient similarity, and separate analyses required for each endpoint-of-interest.
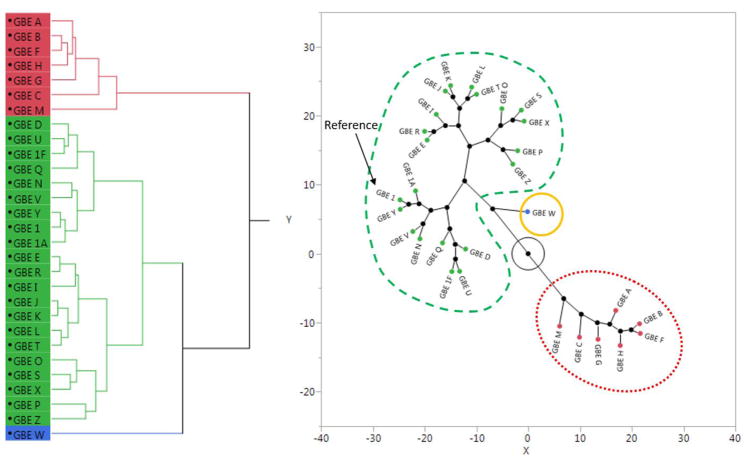
Figure 2.
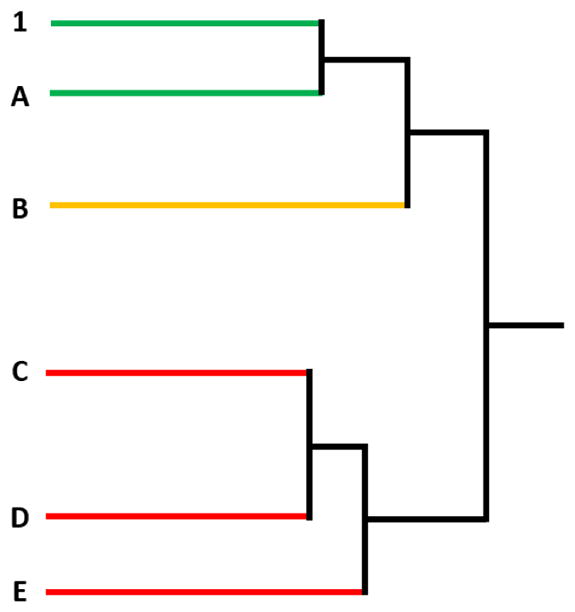
Illustration of hierarchical clustering-based categorization of samples as “similar” to, “maybe similar” to, and “different” from the GBE reference (sample 1). According to rule 1, sample A is considered to be “similar” to sample 1 because they are in the “most similar” cluster. According to rule 2, samples C, D, and E are “different” from sample 1 because they are in the “most different” clusters. According to rule 3, sample B is “maybe similar” to sample 1 because it does not belong to either of the two categories described above.
Untargeted chemistry
For the untargeted chemical analysis, chromatographic profiles were generated using a universal detector, where detector response is proportional to the analyte amount independent of the chemical structure. Specifically, analysis was done using high performance liquid chromatography with evaporative light scattering detection (HPLC-ELSD). For each of the GBE samples in Table 1, a rich dataset of signal intensity versus time was generated using corresponding HPLC-ELSD chromatograms (unpublished data). An initial untargeted approach was implemented by treating each chromatogram as an image and comparing the patterns. The solvent front was eliminated from analysis to preclude early eluting non-GBE-specific peaks from confounding the chemometric analysis. Preprocessing steps included taking the first derivative to remove baseline shifts, normalization of the total intensity of each chromatogram, and aligning retention times based on the largest peaks. A PCA was performed to visualize the relationship between samples (unpublished data). Visual inspection of the chromatographic profiles revealed that samples A, B, C, F, G, and H were notably different from all other GBE samples. These samples were removed from further analysis to achieve better granularity in determining the relationships among the remaining samples. In general, samples U-Z were in the same cluster as the reference sample, and were therefore determined to be “similar’ to the reference. All other samples were determined to be “maybe similar” or “different” from the reference depending on the distance of the cluster from the reference sample-containing cluster (Table 3).
Table 3.
Strength-of-evidence summary. For each measured endpoint in each data stream samples were classified as “similar” to the reference sample - GBE 1 (green color, value of 1), “different” from the reference (red color, value of −1) or “maybe similar” to the reference (yellow color, value of 0).
| Data stream Endpoint | A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Untargeted chemistry Unhydrolyzed samples | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 0 | 0 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Untargeted chemistry Hydrolyzed samples | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Untargeted chemistry score | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −0.5 | −0.5 | 0 | 0 | −0.5 | −0.5 | −0.5 | −1 | −0.5 | −0.5 | −0.5 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Targeted chemistry Unhydrolyzed | −1 | −1 | −1 | 0 | 0 | −1 | −1 | −1 | 0 | 0 | 0 | 0 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −1 | 1 | 1 | 1 | 1 |
| 1 | ||||||||||||||||||||||||||
| Targeted chemistry Hydrolyzed | −1 | −1 | −1 | 1 | 1 | −1 | −1 | −1 | 0 | 0 | 1 | 0 | −1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | ||||||||||||||||||||||||||
| Targeted chemistry score | −1 | −1 | −1 | 0.5 | 0.5 | −1 | −1 | −1 | 0 | 0 | 0.5 | 0 | −1 | 0.5 | 0 | 0.5 | 0.5 | 0.5 | 0 | 0.5 | 0.5 | −0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| SC-PHH gene expression | −1 | −1 | −1 | 1 | 1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| SC-PHH score | −1 | −1 | −1 | 1 | 1 | −1 | −1 | −1 | 1 | 1 | 1 | 1 | −1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| Attagene Cis-factorial assay | −1 | −1 | −1 | 1 | 1 | −1 | −1 | −1 | 1 | 0 | 0 | 0 | −1 | −1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | −1 | 1 | 1 | 1 | 1 |
| Attagene Trans-factorial assay | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −1 | 0 | 1 | 1 |
| Attagene GPCR assay | 0 | 0 | 0 | −1 | −1 | 0 | 0 | 0 | −1 | 1 | 1 | 1 | 1 | 1 | −1 | −1 | −1 | −1 | −1 | 1 | 1 | 0 | −1 | 0 | 0 | 0 |
| Attagene score | −0.3 | −0.3 | −0.3 | 0.0 | 0.0 | −0.3 | −0.3 | −0.3 | 0.0 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | −0.3 | −0.3 | 0.0 | 0.0 | −0.3 | 0.3 | 0.3 | −0.3 | −0.3 | 0.3 | 0.7 | 0.7 |
| In vivo 5-day studies | −1 | 1 | −1 | 1 | ||||||||||||||||||||||
| In vivo score | −1 | 1 | −1 | 1 | ||||||||||||||||||||||
| Overall similarity score | −0.8 | −0.8 | −0.8 | 0.1 | 0.1 | −0.8 | −0.9 | −0.8 | 0.1 | 0.2 | 0.5 | 0.3 | −0.5 | 0.5 | 0.0 | −0.2 | 0.3 | 0.3 | 0.0 | 0.6 | 0.7 | 0.3 | 0.3 | 0.7 | 0.8 | 0.8 |
Abbreviations: SC-PHH (Sandwich cultures of primary human hepatocytes); GPCR (G-protein coupled receptor)
Samples containing a single peak or no discernable peaks were excluded from multifactorial analysis of samples and were categorized as “different” from the reference GBE.
Sample U was included in two separate targeted chemistry analyses, resulting in two data sets for the sample. These runs resulted in a determination of “maybe similar” and “similar” to the reference. Other samples were run twice in various assays, but did not result in any differences in categorization, so a single value was included for simplicity.
Targeted chemistry
In the targeted chemical analysis, GBE constituent concentrations were quantified using analytical standards. The NTP quantified 12 GBE marker constituents (bilobalide, ginkgolide A, ginkgolide B, ginkgolide C, ginkgolide J, ginkgolic acid 15, ginkgolic acid 17, ginkgotoxin, rutin trihydrate, isorhamnetin, kaempferol, and quercetin) and compared constituent weight percentages across samples (unpublished data). Constituent concentration data was evaluated using hierarchical clustering with JMP software (version 12; SAS; Cary, NC). Interestingly, the samples that were determined to be “similar” and “maybe similar” to the reference sample changed depending on whether the samples were hydrolyzed versus unhydrolyzed. However, the samples that were determined to be “different” (samples A, B, C, F, G, H, and M) were consistent with the exception of sample V (Table 3).
Selection of assays to measure biological activity
The goal in selection of in vitro and in vivo assays for inclusion in the GBE case study was to cover a broad range of biological space while focusing on the target tissue (i.e., liver) identified in subchronic and chronic GBE studies. Many systemic diseases lead to changes in the liver, making it a good indicator organ for general toxicity (Shimizu, 2008, Edwards and Wanless, 2013). As noted previously, the liver was identified as an important target site based on the findings from the 3-month and 2-year toxicity and carcinogenicity studies with GBE (NTP, 2013, Rider et al., 2014). It was anticipated that the selected liver-based assays would also provide information on the bioactivity of a wide range of mixtures (not only GBE) and would, therefore, be useful in future assessments of sufficient similarity with different types of complex mixtures (e.g., commercial mixtures, environmental samples). The selected in vitro assays included primary human hepatocyte-based assays and a series of multiplexed gene-reporter assays in HepG2 cells (Attagene assays). In addition, a 5-day in vivo rat study was used to assess a subset of GBE samples and provide a link to the 3-month and 2-year studies conducted with the reference GBE.
Primary human hepatocyte-based assays
All GBE samples were evaluated in sandwich cultures of primary human hepatocytes (SC-PHH), comprised of collagen (I) basement matrix layered with a confluent monolayer of primary hepatocytes overlaid with Matrigel (Hewitt et al., 2007, Swift et al., 2010). In this report, the endpoints-of-interest included gene expression changes for pharmacologically-important drug metabolizing enzymes as sentinels to toxicologically-important hepatic signaling pathways: cytochrome P450 1A2 (CYP1A2) for aryl hydrocarbon receptor (AhR), CYP2B6 for constitutive androstane receptor (CAR), CYP3A4 for pregnane X receptor (PXR), ABCB11 for farnesoid X receptor (FXR), and HMGCS2 for peroxisome proliferator-activated receptor alpha (PPARα). Concentrations of the GBE samples were selected based on preliminary cytotoxicity data, which differed between samples. Concentration-response data were analyzed using a weighted area under the curve (AUC) method. Unexamined concentrations across the broad range of exposure levels were imputed and the AUC was calculated in Graphpad Prism (version 7; La Jolla, CA) with both negative and positive peaks calculated and the net area serving as the AUC value (unpublished data). Hierarchical clustering of the AUC values resulted in three groupings of the GBE samples (Figure 3). The reference sample (including all storage conditions – 1, 1A, and 1F) was in the largest group together with the NIST SRMs (U and V), the EGb761®-containing formulations (X, Y, and Z), and samples D, E, I, J, K, L, N, O, P, Q, R, S, and T, designating these samples as “similar” to the reference GBE. Samples that were identified as “different” from the reference GBE included A, B, C, F, G, H, and M, while sample W was identified as “maybe similar” to the reference GBE (Figure 3, Table 3).
Figure 3.
Example of the process for determining similarity within a data stream (i.e., primary human hepatocyte gene expression data). The Ginkgo biloba extract samples separated into three clusters, which are represented in the dendrogram (left) and constellation plots (right) by red, green, and blue. Similarity groupings are identified by color and line style in the constellation plot: “similar” to the reference sample (green - dashed), “maybe similar” (yellow - solid), and “different” (red - dotted).
Attagene assays
A series of assays developed by Attagene were also used to assess biological activity of the samples (Romanov et al., 2008, Martin et al., 2010). Briefly, the system allows for simultaneous quantitative evaluation of activity signals from various reporter constructs containing promoters responsive to transcription factors (Cis-Factorial assay), nuclear receptors (Trans-Factorial assay), or GPCRs (GPCR-Factorial assay) in HepG2 cells. A series of dilutions for each sample was assessed in each assay. Quality control and data processing were completed by Attagene, and resulting data were input into JMP for hierarchical clustering (unpublished data). Concentrations that resulted in significant indeterminate values due to cytotoxicity or detection issues (the lowest and highest concentrations in the Cis-Factorial assay, and the highest concentration in the Tran-Factorial assay) were excluded from further analysis. Consistent with both chemical and hepatocyte analyses, all samples from the reference GBE (samples 1, 1A, 1F) were in the same group, indicating a lack of difference among samples stored in different conditions. However, in comparing all other samples to the reference GBE (sample 1), there was little consistency across assays (Table 3). Notably, samples D, E, I, Q, and R were always in the same category, but were classified as “similar” to the reference GBE in the Cis Factorial assay, “maybe similar” to the reference GBE in the Trans-Factorial assay, and “different” from the reference GBE in the GPCR assay (Table 3).
Short-term in vivo rat studies
Finally, a 5-day in vivo study with male rats was used to evaluate liver weight and global gene expression changes in liver tissue for a subset of five chemically-distinct GBEs (samples 1, G, N, P, and T). Based on preliminary data from the targeted chemical analysis, five chemically-diverse samples (1, G, N, P, and T) were chosen for 5-day oral gavage studies in male F344 rats (Charles River Kingston, Stone Ridge, NY) with a wide dose range (0, 3, 30, 100, 300, and 1000 mg/kg/day in corn oil) and a sample size of 6 animals per group. The maximum dose of 1000 mg/kg was chosen because this dose was associated with significant liver lesions in both 3-month and 2-year studies performed by the NTP (NTP, 2013, Rider et al., 2014). Animal care and use were in accordance with the Public Health Service Policy on Humane Care and Use of Animals. All animal studies were conducted in an Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) International accredited animal facility and approved by the testing laboratory’s Animal Care and Use Committee. Overall, the genomic and non-genomic findings from the 5-day studies suggest there is a high degree of similarity between GBE 1, N, and T, albeit with N and T showing slightly less potency as follows: GBE 1 > GBE T > GBE N (unpublished data). GBE G and P did not exhibit similar responses to GBE 1. The similarity grouping largely tracks with both the targeted and untargeted chemistry findings where samples N and T are more closely related to sample 1, and samples G and P are quite different compared to sample 1 (Table 3).
Integrating the chemistry and biological-response data
Strength-of-evidence
In the first method used to integrate chemistry and biological-response data, referred to as the strength-of-evidence approach, numerical values were assigned to similarity determinations and averaged across data streams. The similarity determinations from the untargeted and targeted chemistry and in vitro and in vivo biological assays are summarized in Table 3. For each measured endpoint in each data stream (untargeted chemistry, targeted chemistry, SC-PHH, Attagene, in vivo), samples were classified as “similar” to the reference sample, “different” from the reference or “maybe similar” to the reference. An average score was then calculated for each data stream. Finally, an overall similarity score for each sample was calculated by taking the average of all data stream scores for each sample. Note that for most samples, the chemistry data and biological-response data are weighted equally. However, for samples that also have in vivo data (samples G, N, P, and T) the biological-response data has more weight, comprising 3 out of the 5 data streams contributing to the overall average score.
Using this approach, the samples that are most like GBE 1 (overall similarity score ≥0.5) include samples X, Y, and Z (finished GBE products containing EGb761®), sample U (the NIST GBE standard reference material), as well as unfinished GBE samples K and T. Samples D, E, I, J, L, N, Q, R, V, and W, also were judged to be similar, albeit less similar (overall similarity score > 0 and < 0.5). The samples that were most different from the reference (overall similarity score ≤−0.5) included samples A, B, and C (3 samples with slightly different label specifications from a single supplier) and samples F, G, and H. Sample P also fell in the different category (similarity score of −0.2), while samples O and S were found to be in the category of “maybe similar” to the reference (similarity score = 0).
Empirical equivalence testing
Equivalence testing of potency values is another method that can be used to determine sufficient similarity. In this approach, the distance between benchmark dose estimates for mixtures-of-interest and a reference mixture are compared for a specific endpoint (Marshall et al., 2013). First, benchmark doses are estimated for the reference mixture and the mixtures-of-interest based on the quantification of constituent chemicals within the mixtures. Next, expert judgment is used to determine a meaningful biological response deviation and ascribe a similarity space around the reference sample benchmark dose. Finally, a statistical test is used to determine whether the estimated benchmark doses of the mixtures-of-interest fall within or outside of the similarity space around the benchmark dose of the reference. In this case, liver weight change was selected as the critical endpoint for generating benchmark doses. The rationale for focusing on liver weight increase was that it was measured in the 5-day rat studies, served as a link to the hepatotoxicity observed in previous work (NTP, 2013, Rider et al., 2014), and provided robust dose-response data.
Determination of the change in response that represents a biologically-meaningful deviation is a key challenge in applying this approach. It is important to note that a biologically-meaningful change is different from a toxicologically-significant change. This is particularly true in the case of liver weight, which can fluctuate significantly without reflecting overt toxicity. The process for defining the similarity region around the reference sample benchmark dose for liver weight change was as follows:
Calculation of a benchmark response (BMR) of one standard deviation above the control mean response. Relative liver weight of 45.9 mg/g body weight (bw) plus standard deviation of 2.2 equals a BMR for relative liver weight of 48.1 mg/g bw.
Determination of the corresponding benchmark dose (BMD). A BMD of 10.9 mg/kg GBE corresponds to the BMR of 48.1 mg/g bw.
Judgment of biologically-meaningful change in response from the reference sample. A significant increase in liver weight was observed with 30 mg/kg of sample 1, which was the lowest observed effect level (LOEL). Therefore, we selected a change in response that fell below the response observed at the 30 mg/kg dose. In order to develop an approach that could potentially be applied broadly, we selected one standard error below the response at the LOEL: 49.2 mg/g bw (response observed at 30 mg/kg dose) – 0.59 standard error = 48.6 mg/g bw.
Calculation of the dose associated with a biologically-meaningful change in response. The estimated dose associated with a response of 48.6 mg/g bw is 14.7 mg/kg GBE.
Subtraction of the BMD from the biologically-meaningful dose to get the similarity boundary. The difference between 14.7 and 10.9 mg/kg GBE is the similarity bound of 3.77 mg/kg GBE.
To determine similarity, the distance calculated above is compared to the distance between the reference and each of the mixtures-of-interest (Table 4).
Table 4.
Equivalence testing method for determining sufficient similarity. Targeted chemistry data from unhydrolyzed samples was used to estimate the distance from the reference. Italicized values fall outside of the similarity region (> 3.77, the upper 95% confidence limit).
| SAMPLE | Distance Estimatea | Upper 95% confidence limita | Comparison with positive and negative controlsb |
|---|---|---|---|
| GBE 1 (REF) | 0 | 0 | + |
| GBE 1F | 0.05 | 0.09 | + |
| GBE 1A | 0.11 | 0.19 | + |
| GBE W | 0.32 | 0.56 | + |
| GBE 1A* | 0.35 | 0.63 | + |
| GBE X | 0.42 | 0.74 | + |
| GBE Y | 0.49 | 0.86 | + |
| GBE Z | 0.55 | 0.97 | + |
| GBE U* | 0.63 | 1.12 | + |
| GBE K | 0.72 | 1.28 | + |
| GBE L | 0.77 | 1.37 | + |
| GBE Q | 0.82 | 1.45 | + |
| GBE O | 0.83 | 1.47 | + |
| GBE N | 0.84 | 1.50 | + |
| GBE T | 0.85 | 1.52 | + |
| GBE S | 0.88 | 1.56 | −/+ |
| GBE J | 0.89 | 1.58 | −/+ |
| GBE E | 0.97 | 1.72 | −/+ |
| GBE D | 0.99 | 1.75 | −/+ |
| GBE U | 0.99 | 1.76 | −/+ |
| GBE I | 1.01 | 1.79 | −/+ |
| GBE R | 1.03 | 1.82 | −/+ |
| GBE P | 1.53 | 2.72 | − |
| GBE V | 2.05 | 3.64 | − |
| GBE A* | 2.17 | 3.85 | − |
| GBE G | 2.20 | 3.90 | − |
| GBE B* | 2.25 | 3.99 | − |
| GBE M* | 2.28 | 4.05 | − |
| GBE F* | 2.28 | 4.06 | − |
| GBE C* | 2.32 | 4.12 | − |
| GBE H* | 2.54 | 4.52 | − |
Samples were evaluated in a separate targeted chemical analysis. Sample 1A and U were evaluated twice to account for run-to-run variability.
Distance measures and upper 95% confidence limit on the distance between the BMDs for the reference mixture and the sample mixtures.
The dark green and dark red boxes indicate samples that were tested in vivo and had responses that were “similar” to or “different” from the reference response, respectively. The light green boxes represent an assumption of similarity and the light red boxes an assumption of difference. The yellow boxes represent an area of uncertainty between the tested samples. The plus symbols indicate similarity, the minus symbols indicate differences, and the plus/minus symbols indicate uncertainty.
This method provides a well-defined and transparent approach for determining the similarity region that can be applied to other complex mixtures. However, the size of the similarity region may require adjustment based on actual dose-response data from additional samples. For example, using the chemical and biological-response data for GBE samples, the statistically-defined similarity region included sample P, which was found to be biologically-dissimilar to the reference in the 5-day rat study. Therefore, the similarity region could be adjusted to account for the known biological difference. The details of the statistical approach behind this method are outside the scope of this manuscript; however, the methods are described by Marshall et al. (2013). It is also important to note that each endpoint would require a separate evaluation. For example, the evaluation presented in Table 4 includes targeted chemical data from unhydrolyzed samples and liver weight change. This is just one possible combination that can be evaluated. In order to build confidence in the results, multiple combinations should be evaluated using different chemical and biological response data.
Visual interval evaluation
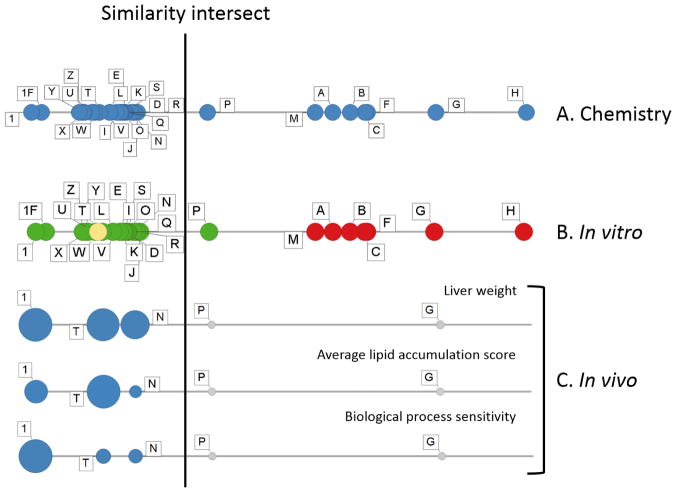
The chemistry and biology data were also evaluated using a line plot of chemical data combined with biological-response data, in an approach we are calling visual interval evaluation (Figure 4). These line plots were generated by calculating the distance (Pearson’s r calculated in Partek Genomics Suite version 6.6; St. Louis, MO) from the reference GBE (sample 1) to each of the samples (A-Z) using an average of distances generated from each chemical evaluation (i.e., hydrolyzed and unhydrolyzed samples in targeted and untargeted analyses) (Figure 4A). Note that the line begins at the reference GBE (sample 1) and subsequent samples moving from left to right are increasingly chemically different from sample 1, with sample H being the most chemically divergent. Following generation of the chemistry-based line plot, the biological data from the in vitro assay (Figure 4B) or in vivo data (Figure 4C) were superimposed on the chemistry evaluation to identify a natural break in the data, which was designated as the similarity cut-off. For the in vitro primary human hepatocyte data (Figure 4B), the color corresponds to the similarity call from the hierarchical clustering analysis described earlier (also presented in Table 3), with green indicating similar, yellow indicating possibly similar, and red indicating different biological activity as compared to the reference GBE. In the in vivo liver weight and gene expression examples (Figure 4C), the size of the circle corresponds to the magnitude of liver weight increase with the blue color corresponding to significant change and the grey indicating no change from control. This visualization allows us to compare the correlation of the chemistry and biology data. For the most part, the superimposed biological data are consistent with the results from the chemical analysis.
Figure 4.
Line plot comparison of Ginkgo biloba samples across chemical (A) and biological-response data from in vitro primary human hepatocytes (PHH; B) and in vivo data from the 5-day rat study (C). A similarity intersect was drawn based on consideration of chemical and biological-response data. In (A), (B), and (C), each circle represents the data for one of the GBE samples, and the position of the circles on the line represents the chemical similarity of the samples to the reference (sample 1) with increasing chemical difference indicated by increasing distance from sample 1 (e.g., sample H is the most chemically different from sample 1). In (B), the circles represent the chemistry data and the green, yellow, and red colors represent the similarity determination (similar, maybe similar, not similar, respectively). A subset of in vivo endpoints are included in (C) including liver weight and gene expression measures indicating the degree of change in genes associated with lipid accumulation (average lipid accumulation score) and the most sensitive gene response associated with any set of genes associated with a biological process or pathway (biological process sensitivity). In (C), the magnitude of biological effect is reflected by the size of the dot, blue represents a significant change from control and gray represents no significant difference from control.
Conclusions and the path forward
A critical challenge in preclinical safety assessments and clinical efficacy studies of botanical dietary supplements is the inherent complexity and variability in these products, which complicates comparison of data generated with one sample to untested samples in the marketplace (Pferschy-Wenzig and Bauer, 2015, Shipkowski et al., 2018). Methods for comparing across these complex mixtures are required to understand how broadly data generated from a single sample can be applied to other samples. In the current work, we first describe a framework for evaluating sufficient similarity of complex mixtures, then work through application of methods using GBE as a case study.
In the case study, multiple chemical and biological-response measures were used to evaluate GBE samples and determine whether they were “sufficiently similar” to a reference GBE. Sample 1 was selected as the reference because it was comprehensively evaluated for toxicity and carcinogenicity (NTP, 2013, Rider et al., 2014). A summary of similarity conclusions from each of the three methods described above is presented in Table 5. Of the 26 unique samples evaluated for similarity, 16 samples (62%) were consistently classified as similar to the reference sample, and 7 samples (27%) were consistently classified as different, while 3 samples (12%) displayed variable classification depending on the method used.
Table 5.
Comparison of conclusions from each method to determine sufficient similarity of GBE samples to the reference GBE. Bolded letters indicate samples that were classified consistently across all methods and italicized letters indicate samples that were not.
| Method | Call | Samples (most to least similar/different) |
|---|---|---|
| Weight-of- evidence | Similar | Y = Z > X = U > T > K = N > L = Q = R = V = W > J > D = E = I |
| Maybe similar | O = S | |
| Different | G > A = B = C = F = H > M > P | |
| Equivalence testing | Similar | W > X > Y > Z > U* > K > L > Q > O > N > T > S > J > E > D > U* > I > R > P > V |
| Different | H > C > F > M > B > G > A | |
| Visual interval evaluation | Similar | Z > Y > X > U > W > T > V > L > I > K > O > D > Q > E > S > N > R > J |
| Different | H > G > F > C > B > A > M > P |
Sample U was run twice in targeted chemical evaluation and exhibited slightly different constituent concentrations between runs.
Notably, the samples that were consistently found to be similar to the reference included the two GBE NIST standard reference materials (U and V) and three formulated samples containing the standardized GBE EGb761® (X, Y, and Z). These findings support the initial selection of the reference (sample 1) as a high-quality GBE for comprehensive evaluation and indicate that the toxicity and carcinogenicity data generated with sample 1 (NTP, 2013, Rider et al., 2014) is representative of standardized GBE in general. Of the twenty unfinished GBE samples, exactly half were consistently identified as similar to the reference (D, E, I, J, K, L, N, Q, R, and T). In contrast, 7 GBE samples (A, B, C, F, G, H, and M) appeared to be consistently less biologically-active than the reference GBE across multiple assays. These samples were notably different based on chemical analysis alone, regardless of whether targeted or untargeted approaches were used. These samples do not appear to contain GBE in any measurable quantity and, it is not clear what they do contain. A majority of dissimilar samples (A, B, C, and F) had a single peak in the chromatogram around the area where flavonol glycosides such as rutin elute (unpublished data), which could indicate the presence of a pure adulterant to mimic the expected flavonol aglycone content of standardized GBE. Three of the four samples (A, B, and C) were procured from a single supplier (Table 1). Further chemical analysis to identify constituents would be required to elucidate the content of dissimilar samples. The remaining samples that are somewhere between similar and dissimilar likely represent lower-quality samples that could contain GBE, along with other unidentified ingredients. Again, further chemical analysis is required to identify potential adulterants in these samples.
In designing the GBE case study, several considerations went into biological assay selection. First, we wanted to incorporate in vitro and short term in vivo models that would be relevant to the hepatotoxicity observed in the previous 3-month and 2-year rodent studies (NTP, 2013, Rider et al., 2014). Hence, liver-centric models were the focus of the biological assays. Second, we wanted to build a bridge between previous and current animal studies and more human-relevant models. This was particularly important due to the species-dependent activity of the CAR pathway (Lau et al., 2012, Cherian et al., 2015), which has been implicated in GBE hepatotoxicity (Maeda et al., 2014, Maeda et al., 2015). In vitro methods to compare various samples of the same botanical can be useful because they provide quick and human-relevant information (with the use of human cell lines), and they can be tailored to specific biological activities (e.g., CAR and PXR activity). The short-term in vivo assays were used with a subset of chemically-divergent samples to provide a more direct link to longer-term animal studies.
All three methods used to integrate chemical and biological data in this case study (strength-of-evidence, equivalence testing, and visual interval evaluation) require reduction of high-dimensional data and involve some form of expert judgment. While simplification of complex data sets is useful in facilitating our judgment of similarity, some information is lost in the process. For example, untargeted chemistry provides information on the specific peaks that differ among samples, which is not used in the current similarity evaluation. In addition to the distillation of data from the individual data streams, the approaches for combining the data streams are simple, in that there is no weighting of the data to account for differences in reliability or relevance. Developing more sophisticated approaches for combining chemical and biological-response data is an area that requires research attention. Decisions on how to weight different data types will necessarily rely on how data are valued in determining similarity. For example, going from most confidence to least confidence in a judgment of similarity, could include mixtures that display:
The same pharmacokinetic profile and biological responses measured in human clinical trials.
The same toxicity and potency across a broad range of endpoints in an in vivo bioassay.
The same apical effect observed in an in vivo bioassay and comparable estimates of potency.
The same outcome and similar dose-response relationships observed in an in vitro assay that examines an outcome relevant to the apical effect.
Similar outcome and similar dose-response relationships observed in an in vitro assay that examines a general toxicity outcome.
Similar chemical composition using untargeted chemical analyses.
Similar marker constituent concentrations.
Note that evaluation of similarity for authentication purposes would have a completely different hierarchy of considerations.
Interestingly, in the current case study with GBE, the most widely-used measure of similarity, which is listed above as providing the weakest confidence in a similarity judgment (i.e., marker constituent concentrations) would have provided a reasonable basis for determining sufficient similarity. However, we contend that this is due to several unique features of GBE. Notably, the marker constituents which correlate to hepatotoxicity, terpene lactones, are also relatively abundant in standardized GBE (~6%), and vary widely in concentration across GBE samples in the marketplace. We would not expect measurement of marker constituent concentrations to correlate with biological responses in cases where the marker constituents are not associated with biological activity. This could include cases where a low-abundance or unknown constituent(s), or an adulterant is responsible for biological activity. Future case studies will include botanicals with low-abundance marker constituents and unknown active constituents.
While all three methods resulted in relatively consistent conclusions, there were differences between the approaches that could influence their application. The strength-of-evidence approach collapsed the greatest amount of data for a bigger picture view across data streams. While this afforded a complete representation of the data, it required the most condensation of data and loss of granularity within the data sets. An advantage of the strength-of-evidence method is that the bird’s eye view allows us to see which data are driving the similarity decisions in any given case. For example, we can see that although sample W was found to be sufficiently similar, the chemical similarity was driving this conclusion, while the biological pattern of activity displayed some differences from the reference. Sample W poses an interesting challenge because it is a formulation containing both EGb761® and Gotu kola. Therefore, it is plausible that while the EGb761® content of sample W is similar to the reference GBE, additional chemical constituents from Gotu kola could be mediating the observed differences in biological response.
Both the equivalence testing method and the visual interval evaluation method rely on chemical data to rank samples from most to least similar, and then use biological data to make decisions about when the chemical differences pass a threshold necessary to elicit differences in biological responses. As such, these methods could miss subtle differences in chemistry that drive disproportionately large differences in biological responses. Sample W, as described above, is the best example of this scenario. An advantage of the equivalence testing method is that it is a quantitative and transparent approach with minimal data required. While the example presented in Table 4 only used targeted chemistry data from unhydrolyzed samples and liver weight, the calculations could be automated to run through any number of iterations using a variety of chemistry and biological-response data combinations. Finally, the visual interval approach is useful for visualization of the relationship between chemical and biological-response data. The correlation of chemical and biological data is clearly evident in this approach. However, the visual interval approach requires additional methods development to impart a clear, reproducible structure for decision-making.
This case study demonstrated that methods for assessing similarity can work very well for botanicals, like GBE, with known marker constituents and known biological activity. Additionally, the case study provided samples with extremely divergent chemical and biological-response profiles. Further comparison of these methods is needed for more challenging cases, where less is known about the chemistry and biological activity of the botanical dietary supplement and differences among samples in the class are subtler. Toward this goal, the NTP is developing additional sufficient similarity case studies with Echinacea purpurea extract and black cohosh extract. Relative to GBE, black cohosh has a larger chemically-unidentified fraction and less is known about the link between the constituents and observed genetic toxicity in vivo and in vitro (Mercado-Feliciano et al., 2012, Smith-Roe et al., 2018). Finally, Echinacea purpurea extract is equivalent to black cohosh in terms of the percent of chemically-identified versus unidentified fractions, but has a less well-defined toxicity profile (NTP, unpublished data). Working through additional case studies with other botanical dietary supplements will provide opportunities for refining methods and identifying issues that require further research attention. While results from the GBE case study are promising, further work is required understand the applicability and limitations of the approaches described here.
Highlights.
Botanical dietary supplements with similar labels can vary widely in content
Approaches to determine sufficient similarity of complex mixtures are presented
Chemical and biological activity data are integrated for similarity evaluation
Ginkgo biloba extract is used as a case study to illustrate methods
While a majority of Ginkgo samples were similar, some were notably different
Acknowledgments
We were grateful for funding from the Office of Dietary Supplements to support case study development. We would also like to thank Ikhlas Khan, Kerri LeVanseler, and James MacGregor for their participation in the 2016 NTP Workshop “Addressing Challenges in the Assessment of Botanical Dietary Supplement Safety” panel on sufficient similarity. Finally, we would like to thank Kembra Howdeshell, Nigel Walker, and Jeffrey Swartout for their review of this manuscript.
References
- 1.AOAC INTERNATIONAL. Guidelines for Validation of Botanical Identification Methods. Journal of Aoac International. 95:268–272. doi: 10.5740/jaoacint.11-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.BRAQUET P. The Ginkgolides: Potent Platelet-Activating Factor Antagonists Isolated from Ginkgo biloba L.: Chemistry, Pharmacology and Clinical Application. Drugs of the Future. 1987;12:643–699. [Google Scholar]
- 3.CHERIAN MT, CHAI SC, CHEN TS. Small-molecule modulators of the constitutive androstane receptor. Expert Opinion on Drug Metabolism & Toxicology. 2015;11:1099–1114. doi: 10.1517/17425255.2015.1043887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CLARKE T, BLACK L, STUSSMAN B, BARNES P, NAHIN R. National Health Statistics Reports. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics; 2015. Trends in the Use of Complementary Health Approaches Among Adults: United States, 2002–2012. [Google Scholar]
- 5.COHEN PA. The Supplement Paradox: Negligible Benefits, Robust Consumption. JAMA. 2016;316:1453–1454. doi: 10.1001/jama.2016.14252. [DOI] [PubMed] [Google Scholar]
- 6.DEFEUDIS FV. A brief history of EGb 761 and its therapeutic uses. Pharmacopsychiatry. 2003;36(Suppl 1):S2–7. doi: 10.1055/s-2003-40450. [DOI] [PubMed] [Google Scholar]
- 7.DING XP, QI J, CHANG YX, MU LL, ZHU DN, YU BY. Quality control of flavonoids in Ginkgo biloba leaves by high-performance liquid chromatography with diode array detection and on-line radical scavenging activity detection. Journal of Chromatography A. 2009;1216:2204–2210. doi: 10.1016/j.chroma.2008.08.115. [DOI] [PubMed] [Google Scholar]
- 8.EDWARDS L, WANLESS IR. Mechanisms of liver involvement in systemic disease. Best Practice & Research in Clinical Gastroenterology. 2013;27:471–483. doi: 10.1016/j.bpg.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 9.FRANSEN HP, PELGROM SMGJ, STEWART-KNOX B, DE KASTE D, VERHAGEN H. Assessment of health claims, content, and safety of herbal supplements containing Ginkgo biloba. Food & Nutrition Research. 2010:54. doi: 10.3402/fnr.v54i0.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GAFNER S, BLUMENTHAL M, FOSTER S, CARDELLINA J, KHAN IA, UPTON R. The ABC-AHP-NCNPR botanical adulterants program’s laboratory guidance on analytical methods to detect adulterants in botanical materials. Planta Medica. 2015;81:888–888. [Google Scholar]
- 11.GRIMM FA, IWATA Y, SIRENKO O, CHAPPELL GA, WRIGHT FA, REIF DM, BRAISTED J, GERHOLD DL, YEAKLEY JM, SHEPARD P, SELIGMANN B, ROY T, BOOGAARD PJ, KETELSLEGERS HB, ROHDE AM, RUSYN I. A chemical-biological similarity-based grouping of complex substances as a prototype approach for evaluating chemical alternatives. Green Chemistry. 2016;18:4407–4419. doi: 10.1039/c6gc01147k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GUO L, MEI N, LIAO W, CHAN PC, FU PP. Ginkgo biloba extract induces gene expression changes in xenobiotics metabolism and the Myc-centered network. OMICS. 2010;14:75–90. doi: 10.1089/omi.2009.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.HARNLY J, CHEN P, HARRINGTON PD. Probability of Identification: Adulteration of American Ginseng with Asian Ginseng. Journal of Aoac International. 2013;96:1258–1265. doi: 10.5740/jaoacint.13-290. [DOI] [PubMed] [Google Scholar]
- 14.HARNLY J, CHEN P, SUN JH, HUANG HL, COLSON KL, YUK J, MCCOY JAH, REYNAUD DTH, HARRINGTON PB, FLETCHER EJ. Comparison of Flow Injection MS, NMR, and DNA Sequencing: Methods for Identification and Authentication of Black Cohosh (Actaea racemosa) Planta Medica. 2016;82:250–262. doi: 10.1055/s-0035-1558113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.HARNLY JM, LUTHRIA D, CHEN P. Detection of Adulterated Ginkgo biloba Supplements Using Chromatographic and Spectral Fingerprints. Journal of Aoac International. 2012;95:1579–1587. doi: 10.5740/jaoacint.12-096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.HEADS JA, HAWTHORNE RL, LYNAGH T, LYNCH JW. Structure-activity analysis of ginkgolide binding in the glycine receptor pore. Journal of Neurochemistry. 2008;105:1418–1427. doi: 10.1111/j.1471-4159.2008.05244.x. [DOI] [PubMed] [Google Scholar]
- 17.HEWITT NJ, LECLUYSE EL, FERGUSON SS. Induction of hepatic cytochrome P450 enzymes: methods, mechanisms, recommendations, and in vitro-in vivo correlations. Xenobiotica. 2007;37:1196–1224. doi: 10.1080/00498250701534893. [DOI] [PubMed] [Google Scholar]
- 18.HUANG SH, DUKE RK, CHEBIB M, SASAKI K, WADA K, JOHNSTON GAR. Ginkgolides, diterpene trilactones of Ginkgo biloba, as antagonists at recombinant alpha(1)beta(2)gamma(2L) GABA(A) receptors. European Journal of Pharmacology. 2004;494:131–138. doi: 10.1016/j.ejphar.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 19.KRESSMANN S, MULLER WE, BLUME HH. Pharmaceutical quality of different Ginkgo biloba brands. Journal of Pharmacy and Pharmacology. 2002;54:661–669. doi: 10.1211/0022357021778970. [DOI] [PubMed] [Google Scholar]
- 20.LAU AJ, YANG GX, RAJARAMAN G, BAUCOM CC, CHANG TKH. Species-Dependent and Receptor-Selective Action of Bilobalide on the Function of Constitutive Androstane Receptor and Pregnane X Receptor. Drug Metabolism and Disposition. 2012;40:178–186. doi: 10.1124/dmd.111.042879. [DOI] [PubMed] [Google Scholar]
- 21.LIN LZ, CHEN P, OZCAN M, HARNLY JM. Chromatographic profiles and identification of new phenolic components of Ginkgo biloba leaves and selected products. J Agric Food Chem. 2008;56:6671–9. doi: 10.1021/jf800488x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MAEDA J, INOUE K, ICHIMURA R, TAKAHASHI M, KODAMA Y, SAITO N, YOSHIDA M. Essential role of constitutive androstane receptor in Ginkgo biloba extract induced liver hypertrophy and hepatocarcinogenesis. Food Chem Toxicol. 2015;83:201–9. doi: 10.1016/j.fct.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 23.MAEDA J, KIJIMA A, INOUE K, ISHII Y, ICHIMURA R, TAKASU S, KURODA K, MATSUSHITA K, KODAMA Y, SAITO N, UMEMURA T, YOSHIDA M. In Vivo Genotoxicity of Ginkgo Biloba Extract in gpt Delta Mice and Constitutive Androstane Receptor Knockout Mice. Toxicological Sciences. 2014;140:298–306. doi: 10.1093/toxsci/kfu090. [DOI] [PubMed] [Google Scholar]
- 24.MARSHALL S, GENNINGS C, TEUSCHLER LK, STORK LG, TORNERO-VELEZ R, CROFTON KM, RICE GE. An empirical approach to sufficient similarity: combining exposure data and mixtures toxicology data. Risk Anal. 2013;33:1582–95. doi: 10.1111/risa.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MARTIN MT, DIX DJ, JUDSON RS, KAVLOCK RJ, REIF DM, RICHARD AM, ROTROFF DM, ROMANOV S, MEDVEDEV A, POLTORATSKAYA N, GAMBARIAN M, MOESER M, MAKAROV SS, HOUCK KA. Impact of Environmental Chemicals on Key Transcription Regulators and Correlation to Toxicity End Points within EPA’s ToxCast Program. Chemical Research in Toxicology. 2010;23:578–590. doi: 10.1021/tx900325g. [DOI] [PubMed] [Google Scholar]
- 26.MERCADO-FELICIANO M, CORA MC, WITT KL, GRANVILLE CA, HEJTMANCIK MR, FOMBY L, KNOSTMAN KA, RYAN MJ, NEWBOLD R, SMITH C, FOSTER PM, VALLANT MK, STOUT MD. An ethanolic extract of black cohosh causes hematological changes but not estrogenic effects in female rodents. Toxicology and Applied Pharmacology. 2012;263:138–147. doi: 10.1016/j.taap.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MURRAY F, ROTH R, NICOLICH M, GRAY T, SIMPSON B. The relationship between developmental toxicity and aromatic-ring class profile of high-boiling petroleum substances. Regulatory Toxicology and Pharmacology. 2013;67:S46–S59. doi: 10.1016/j.yrtph.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 28.NTP; NTP, editor. Technical Report Series. Research Triangle Park, NC: NIEHS/NTP; 2013. NTP Technical Report on the Toxicology and Carcinogenesis Studies of Ginkgo biloba Extract (CAS No. 90045-36-6) in F344/N Rats and B6C3F1/N Mice (Gavage Studies) [PubMed] [Google Scholar]
- 29.PFERSCHY-WENZIG EM, BAUER R. The relevance of pharmacognosy in pharmacological research on herbal medicinal products. Epilepsy & Behavior. 2015;52:344–362. doi: 10.1016/j.yebeh.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 30.RICE GE, TEUSCHLER LK, BULL RJ, SIMMONS JE, FEDER PI. Evaluating the similarity of complex drinking-water disinfection by-product mixtures: overview of the issues. J Toxicol Environ Health A. 2009;72:429–36. doi: 10.1080/15287390802608890. [DOI] [PubMed] [Google Scholar]
- 31.RIDER CV, NYSKA A, CORA MC, KISSLING GE, SMITH C, TRAVLOS GS, HEJTMANCIK MR, FOMBY LM, COLLETON CA, RYAN MJ, KOOISTRA L, MORRISON JP, CHAN PC. Toxicity and carcinogenicity studies of Ginkgo biloba extract in rat and mouse: liver, thyroid, and nose are targets. Toxicol Pathol. 2014;42:830–43. doi: 10.1177/0192623313501235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ROMANOV S, MEDVEDEV A, GAMBARIAN M, POLTORATSKAYA N, MOESER M, MEDVEDEVA L, GAMBARIAN M, DIATCHENKO L, MAKAROV S. Homogeneous reporter system enables quantitative functional assessment of multiple transcription factors. Nature Methods. 2008;5:253–260. doi: 10.1038/nmeth.1186. [DOI] [PubMed] [Google Scholar]
- 33.SCHENCK KM, SIVAGANESAN M, RICE GE. Correlations of water quality parameters with mutagenicity of chlorinated drinking water samples. J Toxicol Environ Health A. 2009;72:461–7. doi: 10.1080/15287390802608940. [DOI] [PubMed] [Google Scholar]
- 34.SHIMIZU Y. Liver in systemic disease. World Journal of Gastroenterology. 2008;14:4111–4119. doi: 10.3748/wjg.14.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.SHIPKOWSKI KA, BETZ JM, BIRNBAUM LS, BUCHER JR, COATES PM, HOPP DC, MACKAY D, OKETCH-RABAH H, WALKER NJ, WELCH C, RIDER CV. Naturally Complex: Perspectives and Challenges Associated with Botanical Dietary Supplement Safety Assessment. Food and Chemical Toxicology. 2018 doi: 10.1016/j.fct.2018.04.007. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.SLOLEY BD, TAWFIK SR, SCHERBAN KA, TAM YK. Quality control analyses for ginkgo extracts require analysis of intact flavonol glycosides. Journal of Food and Drug Analysis. 2003;11:102–107. [Google Scholar]
- 37.SMITH-ROE SL, SWARTZ CD, SHEPARD KG, BRYCE SM, DERTINGER SD, WAIDYANATHA S, KISSLING GE, AUERBACH SS, WITT KL. Black cohosh extracts and powders induce micronuclei, a biomarker of genetic damage, in human cells. Environ Mol Mutagen. 2018 doi: 10.1002/em.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.SWIFT B, PFEIFER ND, BROUWER KLR. Sandwich-cultured hepatocytes: an in vitro model to evaluate hepatobiliary transporter-based drug interactions and hepatotoxicity. Drug Metabolism Reviews. 2010;42:446–471. doi: 10.3109/03602530903491881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.THE UNITED STATES PHARMACOPEIAL CONVENTION. Dietary Supplements Compendium. Rockville, MD: The United States Pharmacopeial Convention; 2015. [Google Scholar]
- 40.VAN BEEK TA, MONTORO P. Chemical analysis and quality control of Ginkgo biloba leaves, extracts, and phytopharmaceuticals. Journal of Chromatography A. 2009;1216:2002–2032. doi: 10.1016/j.chroma.2009.01.013. [DOI] [PubMed] [Google Scholar]