Abstract
Background
Circular RNAs (circRNAs) which are shown as a class of RNAs exhibit the importance in the regulation of gene expression and the development of biological process. However, the expression profile and molecular mechanism of circRNA ATXN7 (circATXN7) is still mostly uncertain in gastric cancer (GC).
Methods
qRT-PCR analysis was performed to detect the expression of circATXN7, miR-4319 and ENTPD4 in GC tissues and cells. CCK-8, colony formation, EdU, flow cytometry, TUNEL and transwell assays were conducted to assess the effect of circATXN7 or miR-4319 on cell proliferation, apoptosis and invasion. In vivo assays were utilized to further analyze the function of circATXN7 on the tumorigenesis and progression of GC. The interaction between miR-4319 and circATXN7 (or ENTPD4) was verified using luciferase reporter and RNA pull-down assays.
Results
The results showed an upregulated circATXN7 expression in GC tissues and cell lines. Besides, silenced circATXN7 hampered the proliferation and invasion as well as promoted the apoptosis in GC cells. Moreover, low expression of miR-4319 was found in GC. It was determined that circATXN7 acted as a sponge for miR-4319 and had a negative association with miR-4319. We also found that miR-4319 upregulation restrained GC cell proliferation and migration whereas enhanced apoptosis. Subsequently, ENTPD4, the target gene of miR-4319, was found overexpressed in GC. Additionally, it was negatively correlated with miR-4319 whereas positively associated with circATXN7. In vivo experiments, circATXN7 silence was confirmed to inhibit GC tumor growth.
Conclusions
CircATXN7 promoted GC development through sponging miR-4319 and regulating ENTPD4, which identified circATXN7 as a new biomarker in GC.
Keywords: circATXN7, miR-4319, ENTPD4, Gastric cancer
Background
Gastric cancer (GC) is a common type of malignancies in gastrointestinal part of human body, and is the leading cause of death that related to cancers [1]. Due to the improvement of medical level in recent years, a stable decrease was showed in the occurrence and mortality of GC. However, this cancer still poses a great threat to human health with an unsatisfactory long-term survival rate [2]. Consequently, discovering the novel biomarkers and critical molecular mechanisms is indispensable to develop potent therapy for the patients with GC.
Circular RNAs (circRNAs), characterized as a covalently closed loop, is a group of endogenous RNAs that has no ability of coding proteins, and generated from back-splicing [3]. Besides, circRNAs expression are more stable than their linear counterparts on account of their loop feature, and the sponge effect of circRNAs was more potent than that of linear RNAs [4]. Furthermore, it was revealed that circRNAs are involved in regulating the tumorigenesis and development of malignancies [5]. For example, circPVT1 was identified as a factor for proliferation and a biomarker for prognosis in GC [6]. Meanwhile, circSMARCA5 which responded by androgen is overexpressed in prostate cancer and enhances the proliferation [7]. Furthermore, circMTO1 suppresses the progression of hepatocellular carcinoma by acting as microRNA-9 sponge [8]. In the past years, circRNAs were hypothesized to be the competing endogenous RNA (ceRNA), competitively combining with miRNAs and regulating mRNAs, including GC [9]. For instance, circMYLK, as a ceRNA, promotes the tumor growth and metastasis of bladder cancer via regulating VEGFA/VEGFR2 signaling [10]. CircDOCK1 restrains the apoptosis of oral squamous cell carcinoma via inhibiting miR‑196a‑5p and targeting BIRC3 [11]. In addition, circLARP4 was reported to inhibit GC cell proliferation and invasion by regulating miR-424-5p/LATS1 axis [12]. Although circRNA circATXN7 has been unveiled to be dramatically upregulated in tissues and cells of non-small cell lung cancer and facilitates the progression of this cancer [13], its specific performance in GC remains unknown. Thus, exploring the biological function and molecular mechanism of circATXN7 in GC is of great meaning for developing a novel biomarker for GC treatment.
This study was devoted to investigating the specific role of circATXN7 in GC. According to the results of this research, we found that the circATXN7/miR-4319/ENTPD4 axis effectively affected the proliferation, apoptosis and invasion of GC, which offered an effective diagnostic and therapeutic method for GC.
Human tissue samples
30 GC samples and matched non-tumor tissues were collected from patients who received treatment at the Second Hospital of Shandong University from May 2013 to June 2018. Fresh GC samples were frozen in liquid nitrogen and stored at − 80 °C. No treatments were performed on patients before this study. Written informed consent was signed by every patient, the study protocol was accepted by the Ethics Committee of the Second Hospital of Shandong University.
Cell culture
The gastric cancer cells (MGC-803, SGC-7901, MKN-45, AGS, BGC-823) and gastric epithelial cell (GSE-1) were obtained from Chinese Academy of Sciences (Beijing, China). These cells were cultured in DMEM medium (Thermo Fisher Scientific, Waltham, USA) plus 10% fetal bovine serum (FBS; Thermo Fisher Scientific), 100 U/mL penicillin (Sigma-Aldrich, Milan, Italy) and streptomycin (Sigma-Aldrich). Cells were cultivated under conditions (37 °C, 5% CO2). Medium was changed every 3 days.
Cell transfection
When cells were passaged at 70–80% confluence, cells were placed into 6-well plates. AGS and MGC-803 cells were transfected with shRNAs against circATXN7 (sh-circATXN7#1/2) and their negative controls (sh-NCs). The pcDNA3.1/ENTPD4 and the empty pcDNA3.1 (+) circRNA Mini Vector were bought from Invitrogen (Carlsbad, California, USA). The miR-4319 mimics, NC mimics, miR-4319 inhibitor were purchased from Invitrogen. After transfection, cells were gathered.
Actinomycin D (actD) and RNase R treatment assay
In order to block transcription, Actinomycin D or dimethylsulfoxide (Sigma-Aldrich) as negative control was added. For RNase R treatment assay, total RNA was cultivated for 30 min with or without 3 U/μg of RNase R (Epicentre Technologies, Madison, USA). After treatment, qRT-PCR analysis was employed to evaluate the expression levels of circATXN7 and ATXN7mRNA.
Quantitative real-time PCR
The assay was conducted as previously [14].
Cell proliferation assay
Cells were plated in 96-well plates under a density of 1 × 103 cells each well in RPMI 1640 culture and cultivated over 24, 48, 72 or 96 h. Ten microliters CCK-8 (Sigma-Aldrich) were added and cultured for further 4 h. The absorbance was recorded at 450 nm using microplate reader (Nikon, Tokyo, Japan).
Transwell invasion assay
After treatment, MGC-803 and AGS cells in the medium of serum-free were placed in the top chamber (Corning, Steuben County, New York, USA), which was pre-coated with Matrigel (Franklin Lakes, NJ, USA). 10% FBS was added into the basolateral chamber. Remaining cells were fixed with paraformaldehyde (Solarbio) and stained with crystal violet (Solarbio). The invaded cells were counted in five random fields by light microscope (Nikon).
Flow cytometry analysis
The apoptosis of AGS and MGC-803 cells was detected by using Annexin V-FITC Apoptosis kit (MultiSciences, Shanghai, China). Briefly, the apoptosis cells were cleaned with PBS (Solarbio) and suspended. After, cells were fixed with 70% ethanol which was cooled by ice. Finally, the rate of flow cytometry was evaluated.
Colony formation assay
Transfected cells were seeded in a 6-well plate and the medium for cultivation was replaced every 3 days. After incubation for 2 weeks, cells of MGC-803 and AGS were first fixed using methanol (Solarbio) and then dyed by crystal violet (Solarbio). Lastly, cells were counted by naked eyes.
TUNEL assay
MGC-803 and AGS cells were plated in 24-well flat-bottomed plates under density of 1 × 105 cells each well, and cells were fixed in paraformaldehyde (Solarbio). TUNEL staining was detected by using the in situ cell death detection kit (Roche), and DAPI was applied to stain the nuclei. The rate of apoptosis cells were recorded via a fluorescence microscope (Nikon).
EdU proliferation assay
The EdU proliferation assay (RiboBio, Guangzhou, China) was used to determine the influence on cell proliferation. Cells were incubated with 50 μm EdU for 2 h and dyed with Apolo and Hoechest 33342 (Beyotime, Shanghai, China), then EdU-positive cells was detected by fluorescence microscopy. The positive rate of EdU was expressed as the ratio of the amount of EdU positive cells to the total DAPI chromogenic cells.
Subcellular fractionation
Cytoplasmic and nuclear RNA were isolated using PARIS™ Kit (Ambion, Austin, TX, USA). The procedures were performed as before [15].
Fluorescence in situ hybridization (FISH)
AGS and MGC-803 cells were washed by PBS and then fixed in 4% formaldehyde. Further, cells were permeabilized with PBS which containing 0.5% Triton X-100, then cleaned with PBS again. After nucleus staining by Hoechst, images were collected via a confocal microscopy (Leica TCS SP8).
In vivo assay
In vivo assay was performed following the laboratory animal guideline for ethical review of animal welfare. MGC-803 and AGS cells were injected into nude mouse. Finally, the mice were killed and their tumors were photographed. The tumor volume and weight were calculated. These experiments gained ethical approval from Laboratory animal welfare committee.
Luciferase reporter assay
The wild-type predictive binding sites and mutant binding sites circATXN7/ENTPD4 with miR-4319 were sub-cloned into pmirGLO dual-luciferase vector to construct circATXN7-WT/ENTPD4-WT and circATXN7-Mut/ENTPD4-Mut. Then the plasmids were co-transfected with miR-4319 mimics into AGS or MGC-803 cells. The luciferase activity was detected after 48 h.
Immunohistochemistry
The steps of assay were conducted the same as before [16].
RIP assay
Based upon the indicated protocols, the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, USA) and the Ago2 antibody (Abcam, Cambridge, UK) were employed to conduct RIP experiment. Co-precipitated RNAs were analyzed by RT-qPCR analysis.
RNA pull-down assay
GC cells were transfected with biotinylated miRNA, and we made cells gather after transfection. M-280 streptavidin magnetic beads (Invitrogen) helped us to cultivate the cell lysates. The purified RNA was detected using RT-qPCR analysis.
Statistical analysis
Data from three independent experiments were denoted as mean ± SD. Statistical analysis was conducted using the SPSS (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5 software (GraphPad Software, San Diego, CA). Significance of the variance was evaluated by Student’s t test or ANOVA. Overall survival of GC patients by Kaplan–Meier analysis and Pearson’s correlation analysis found the correlation. P < 0.05 has statistically significance.
Results
The expression and characteristics of circATXN7 in GC
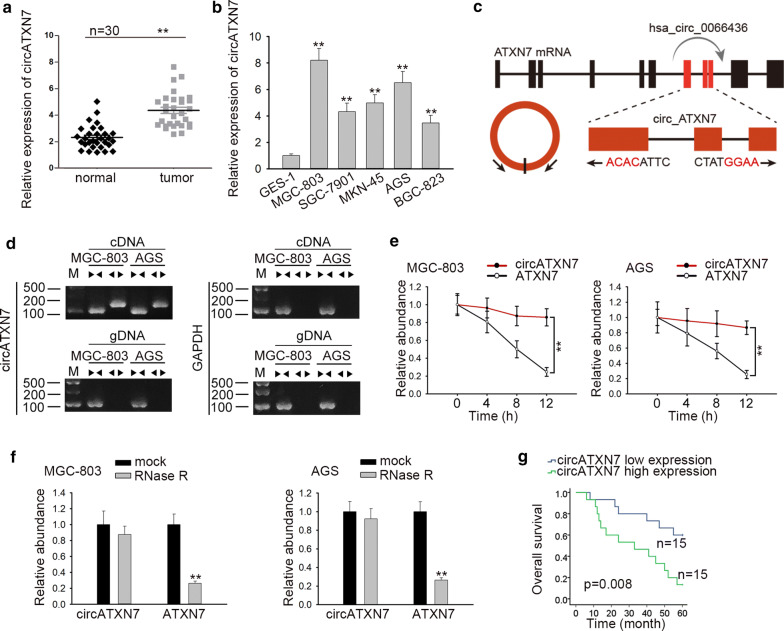
Firstly, the expression of circATXN7 in GC tissues and cell lines (MGC-803, SGC-7901, MKN-45, AGS, BGC-823) was detected. The matched non-tumor tissues and gastric epithelial cell (GSE-1) were used as the control, respectively. Results depicted that whether in GC tissues or in GC cell lines, circATXN7 expression was evidently upregulated (Fig. 1a, b). Later, the genomic loci of the ATXN7 gene and circATXN7 were illustrated in Fig. 1c. Further, agarose gel electrophoresis was used to verify the specificity and accuracy of circATXN7 qRT-PCR products from MGC-803 and AGS cells. As a result, circATXN7 amplified by divergent primers in cDNA but not in gDNA (Fig. 1d). Next, circATXN7 stability was analyzed after MGC-803 and AGS cells were treated with Actinomycin D. The result indicated that circATXN7 was more stable than linear ATXN7 (Fig. 1e). In accordance of the results from qRT-PCR analysis, we found that circATXN7 resisted to digestion after the treatment of RNase R exonuclease in comparison with the linear RNA ATXN7, which further indicated that circATXN7 possessed a loop structure (Fig. 1f). In addition, the survival curve delineated that high circATXN7 expression led to a worse prognosis of patients with GC (Fig. 1g). Further, clinical data uncovered that circATXN7 expression was closely related to TNM stage and pathological type (Table 1). These data revealed the high expression and loop feature of circATXN7 in GC.
Fig. 1.
The expression and characteristics of circATXN7 in GC. a The expression of circATXN7 in GC tissues and matched non-tumor tissues was detected by qRT-PCR. b The expression of circATXN7 in GC cell lines (MGC-803, SGC-7901, MKN-45, AGS, BGC-823) and one gastric epithelial cell line (GES-1) was detected. c The genomic loci of the ATXN7 gene and circATXN7. d Agarose gel electrophoresis was utilized to confirm circATXN7 specificity and accuracy in qRT-PCR products from MGC-803 and AGS cells. e qRT-PCR analysis was applied to assess the expression circATXN7 and ATXN7 RNA after treatment with Actinomycin D. f qRT-PCR analysis was applied to assess the expression of circATXN7 and ATXN7 RNA after treatment with RNase R. g The potential diagnostic value of circATXN7 was evaluated. **P < 0.01
Table 1.
Correlation between circATXN7 expression and clinical features (n = 30)
| Variable | circATXN7 expression | P-value | |
|---|---|---|---|
| Low | High | ||
| Age | |||
| < 60 | 6 | 8 | 0.715 |
| ≥ 60 | 9 | 7 | |
| Gender | |||
| Male | 10 | 8 | 0.710 |
| Female | 5 | 7 | |
| Lymph node metastasis | |||
| Yes | 8 | 6 | 0.715 |
| No | 7 | 9 | |
| TNM stage | |||
| I/II | 11 | 1 | 0.0001*** |
| III/IV | 4 | 14 | |
| Pathological type | |||
| Infiltrative | 12 | 3 | 0.003** |
| Non-infiltrative | 3 | 12 | |
Low/high by the sample median. Pearson χ2 test
***P < 0.001, **P < 0.01 were considered to be statistically significant
CircATXN7 promoted cell proliferation and invasion as well as suppressed apoptosis in GC
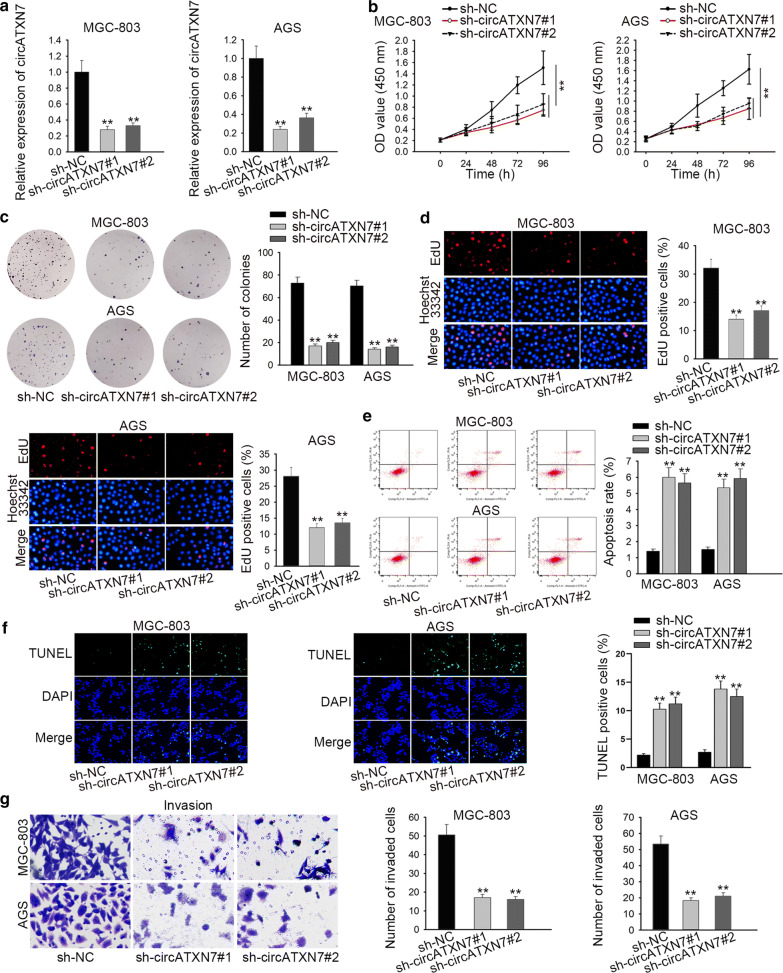
To probe the function of circATXN7 in GC, sh-circATXN7#1 and sh-circATXN7#2 were used to cut down circATXN7 expression (Fig. 2a). Through CCK-8 assay and colony formation assay, in comparison with the control groups, the proliferative ability of MGC-803 and AGS cells was obviously restrained by silenced circATXN7 (Fig. 2b, c). EdU assay was performed to further validate the suppressive role of circATXN7 knockdown in cell proliferation (Fig. 2d). Then, we applied flow cytometry and TUNEL analyses and found that circATXN7 silencing remarkably boosted the apoptosis of MGC-803 and AGS cells (Fig. 2e, f). Moreover, cell invasion capability was markedly weakened in sh-circATXN7#1/2 group compared with that in sh-NC group (Fig. 2g). Taken together, circATXN7 downregulation inhibited the proliferation and migration as well as induced apoptosis in GC.
Fig. 2.
CircATXN7 promoted cell proliferation and migration as well as suppressed apoptosis in GC. a The efficiency of circATXN7 knockdown was tested via RT-qPCR. b–d The effect of circATXN7 knockdown on cell proliferation was evaluated by CCK-8, colony formation and EdU assays. e, f Flow cytometry and TUNEL analyzes were applied to measure the effect of circATXN7 knockdown on cell apoptosis. g Transwell assay was conducted to determine the effect of circATXN7 knockdown on cell invasion. **P < 0.01
MiR-4319 was targeted by circATXN7, and repressed cell proliferation and invasion while boosted cell apoptosis in GC
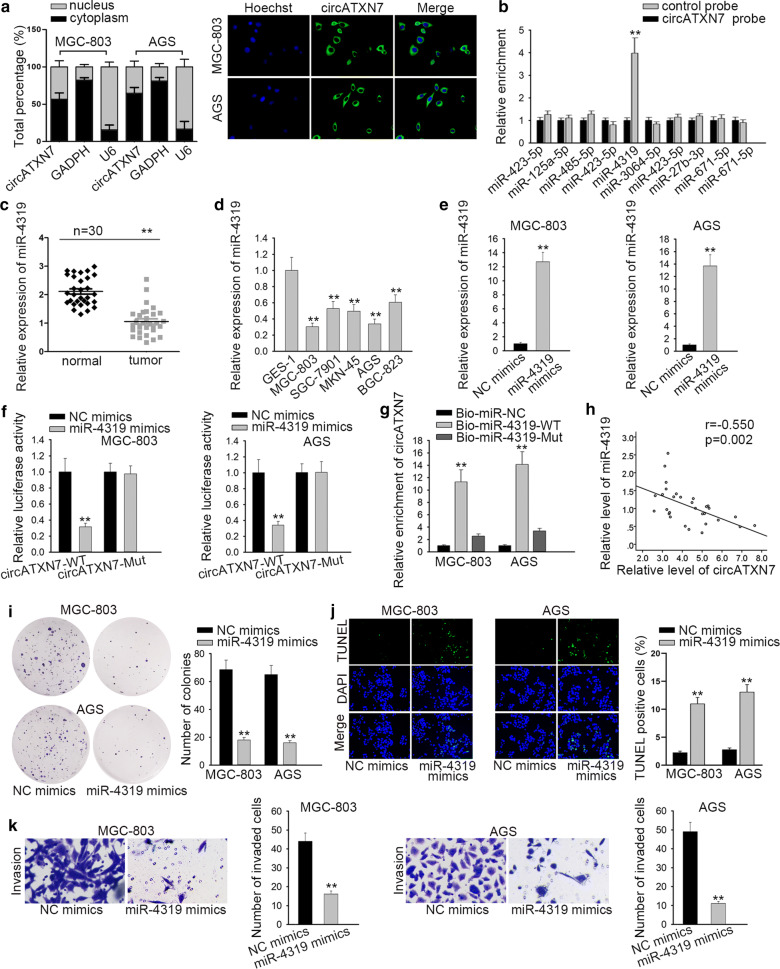
Mounting studies displayed that circRNAs could function as a sponge of miRNAs to regulate gene transcription [17]. Based on nuclear-cytoplasmic fractionation and FISH assay, circATXN7 was mainly discovered in the cytoplasm (Fig. 3a). Hence, we hypothesized that circATXN7 might be a ceRNA in GC. To find potential miRNAs sponged by circATXN7 in GC, starBase (http://starbase.sysu.edu.cn) was applied and 10 miRNAs were speculated. As presented in Fig. 3b, result from RNA pull-down assay suggested that only miR-4319 was dramatically enriched in circATXN7 probe group (Fig. 3b). Therefore, we predicted that miR-4319 might be a downstream target gene of circATXN7 in GC. Later, a low level of miR-4319 was detected in GC tissues and cell lines through qRT-PCR (Fig. 3c, d). We next overexpressed miR-4319 in MGC-803 and AGS cells with the use of miR-4319 mimics to conduct the follow-up experiments (Fig. 3e). Through luciferase reporter assay, with the transfection of miR-4319 mimics, we found that the luciferase activity of wild type circATXN7 was considerably weakened, but that of mutant type circATXN7 showed no notable change (Fig. 3f). Furthermore, RNA pull-down assay verified that the enrichment of circATXN7 demonstrated a terrific increase in Bio-miR-4319-WT group rather than in Bio-miR-4319-Mut group (Fig. 3g). We also found a negative relationship between circATXN7 and miR-4319 in tissues (Fig. 3h). In addition, to explore the role of miR-4319 in GC, colony formation assay was applied to detect the proliferation of GC cells. And the result manifested that miR-4319 overexpression diminished the proliferative cells in MGC-803 and AGS cells (Fig. 3i). And TUNEL assay implied that the promotive effect of miR-4319 mimics on cell apoptosis (Fig. 3j). At last, the number of invaded cells was decreased by overexpression of miR-4319 (Fig. 3k). Conclusively, miR-4319 was targeted by circATXN7, and repressed cell proliferation and invasion while boosted cell apoptosis in GC.
Fig. 3.
MiR-4319 was targeted by circATXN7, and repressed cell proliferation and invasion while boosted cell apoptosis in GC. a The cellular localization of circATXN7 in GC cells was determined by nuclear-cytoplasmic fractionation and FISH assays. b RNA pull-dwon assay was applied to screen out the potential miRNAs for circATXN7. c, d The expression of miR-4319 in GC tissues and cell lines was detected through RT-qPCR. e The efficiency of miR-4319 mimics was tested via RT-qPCR. f, g The interaction between circATXN7 and miR-4319 was verified by luciferase reporter assay and RNA pull-down assay. h The relationship between circATXN7 and miR-4319 levels in GC tissues was revealed via Spearman’s correlation analysis. i The effect of miR-4319 upregulation on cell proliferation was estimated via colony formation assay. j TUNEL assay was employed to measure the effect of miR-4319 mimics on cell apoptosis. k Transwell assay was applied to test the effect of miR-4319 overexpression on cell invasion. **P < 0.01
ENTPD4 was confirmed as a target gene of miR-4319
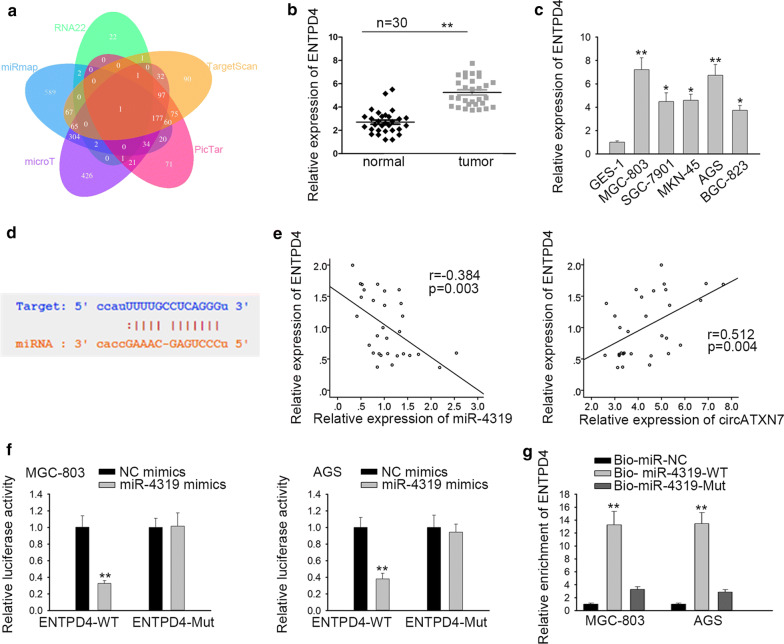
To further support ceRNA hypothesis, here, we explored the target gene of miR-4319. According to these databases (RNA22, TargetScan, PicTar, microT, miRmap), ENTPD4 was screened out (Fig. 4a). By the use of qRT-PCR analysis, we found that ENTPD4 was conspicuously overexpressed in GC tissues and cell lines (Fig. 4b, c). Then, the binding site between miR-4319 and ENTPD4 was predicted by starBase (Fig. 4d). What’s more, it was confirmed that ENTPD4 had a negative relevance to miR-4319 and a positive association with circATXN7 (Fig. 4e). We performed luciferase reporter assay to certificate whether miR-4319 could bind to ENTPD4. Data elucidated that considerable reduction of luciferase activity was observed with co-transfection of miR-4319 mimics and ENTPD4-WT, but no notable difference in ENTPD4-Mut (Fig. 4f). Then, RNA pull-down assay further verified that miR-4319 combined with ENTPD4 (Fig. 4g). In short, ENTPD4 was confirmed as a target gene of miR-4319.
Fig. 4.
ENTPD4 was confirmed as a target gene of miR-4319. a The target gene of miR-4319 was shown via Venn diagram. b, c qRT-PCR assay was conducted to detect the expression of ENTPD4 in GC tissues and cells. d The binding site between miR-4319 and ENTPD4 was presented. e The relationship between ENTPD4 and miR-4319 (or circATXN7) in GC tissues was revealed via Spearman’s correlation analysis. f, g Luciferase reporter and RNA pull-down assays were used to confirm the interaction between ENTPD4 and miR-4319. *P < 0.05, **P < 0.01
The circATXN7/miR-4319/ENTPD4 axis regulated the proliferation, invasion and apoptosis of GC cells
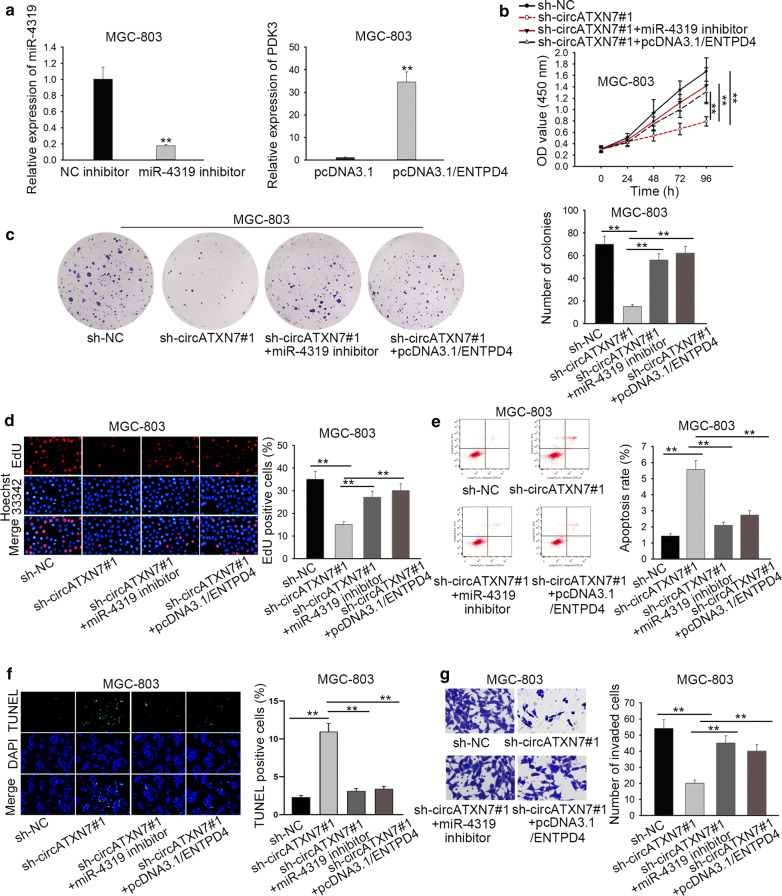
To investigate whether circATXN7 involved in GC progression by sponging miR-4319 and regulating ENTPD4, a series of rescue assays were employed. We firstly reduced the expression of miR-4319 with miR-4319 inhibitor and overexpressed the expression of ENTPD4 with pcDNA3.1/ENTPD4 (Fig. 5a). Then, cell proliferation in GC was examined. As expected, the proliferation of MGC-803 cells transfected with sh-circATXN7#1+miR-4319 inhibitor or sh-circATXN7#1+pcDNA3.1/ENTPD4 was significantly promoted in comparison with those transfected with sh-circATXN7#1 (Fig. 5b–d). The apoptosis of GC cells was facilitated by sh-circATXN7#1, while the transfection of miR-4319 inhibitor or pcDNA3.1/ENTPD4 reversed the effect of sh-circATXN7#1 on cell apoptosis (Fig. 5e, f). At last, it was certificated that miR-4319 repression or ENTPD4 overexpression could counteract the circATXN7 silencing-mediated suppression on cell invasion (Fig. 5g). In a word, the circATXN7/miR-4319/ENTPD4 axis mediated the proliferation, invasion and apoptosis of GC cells.
Fig. 5.
The circATXN7/miR-4319/ENTPD4 axis regulated the proliferation, invasion and apoptosis of GC cells. a The efficiency of miR-4319 inhibitor and pcDNA3.1/ENTPD4 was determined in MGC-803 cells. b–d CCK-8, colony formation and EdU assays were utilized to assess cell proliferation. e, f Flow cytometry and TUNEL assays were performed to test cell apoptosis. g Cell invasion was estimated by transwell assay. **P < 0.01
CircATXN7 contributed to tumor growth of GC in vivo
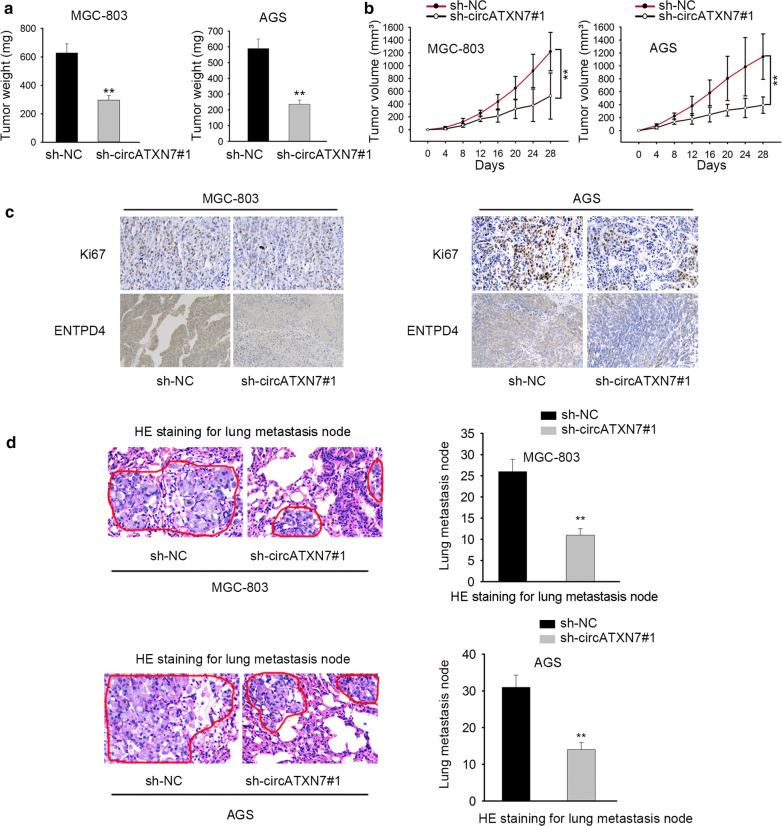
Lastly, the effect of circATXN7 on GC tumor growth in vivo was explored. At first, MGC-803 and AGS cells transfected with sh-circATXN7#1 or sh-NC were subcutaneously injected into nude mice. After inoculation, the mice were killed and then xenograft tumors were extracted. As shown in Fig. 6a, the tumor weight was decreased in sh-circATXN7#1 group compared with that in sh-NC group. In addition, the reductive volume of GC tumors was observed in sh-circATXN7 group in contrast to sh-NC group (Fig. 6b). Besides, the expression of Ki67 and ENTPD4 was decreased in circATXN7‐depleted group (Fig. 6c). Furthermore, knockdown of circATXN7 cut down lung metastasis node, hinting that circATXN7 deficiency could inhibit cell metastasis (Fig. 6d). Besides, compared with that in GC patients with low expression of circATXN7, the expression of Ki67 and ENTPD4 in GC patients with high expression of circATXN7 was elevated (Additional file 1: Figure S1). All the above results disclosed that circATXN7 contributed to the GC tumor growth of GC in vivo.
Fig. 6.
CircATXN7 contributed to tumor growth of GC. a Tumor weight was analyzed at the end point. b The tumor volumes were analyzed at indicative time points post injection. c The expression of Ki67 and ENTPD4 was detected in different groups via immunohistochemistry. d He staining for lung metastasis node was utilized to measure cell metastasis in different groups.**P < 0.01
Discussion
CircRNAs are a type of RNAs which is special for a covalently closed loop and could not code proteins [18]. Up to now, the biological function and mechanism of circRNAs still need to be explored in malignancies progression [19]. Highly conserved sequences and particular covalently closed circular construction formed the feature of circRNAs [20]. It was reported that circRNAs might be the underlying biomarkers to diagnose and cure a series of diseases due to the considerable functions they exerted on the development of diverse diseases, such as cancers [21, 22]. For example, upregulated circRNA hsa_circ_0000069 promotes colorectal cancer cell proliferation and metastasis [23]. What’s more, circular RNA ciRS-7 is involved in the prognosis of lung cancer and drives its development [24]. Although circATXN7 has been confirmed as a tumor facilitator in non-small cell lung cancer [13], its function in GC is still obscure. Here, we found that the expression of circATXN7 was upregulated in GC tissues and cells, and overexpressed circATXN7 resulted in worse prognosis. Additionally, it was discovered that circATXN7 boosted GC cell proliferation, invasion and suppressed cell apoptosis. Hence, we recognized that circATXN7 played oncogenic roles in GC.
MicroRNAs (miRNAs) are a group of small RNAs with 22–24 nucleotides, short of proteins-coding ability and participated in the regulation of biological process [25]. For example, miRNA-135a plays tumor-suppressive role in inhibiting the proliferation of breast cancer cells [26]. MiR-183 is overexpressed in glioblastoma and promotes radioresistance by reducing the expression of LRIG1 [27]. MiR-200a, an upstream gene of FOXA2, promotes EMT process in endometrial cancer [28]. In this study, miR-4319 was predicted to possess binding capacity with circATXN7 through bioinformatics website, and then validated to bind with circATXN7 in GC cells. The suppressive function of miR-4319 on the development of prostate cancer [29] and breast cancer [30] has been uncovered, but the specific role of miR-4319 in GC and the effect that the interaction of circATXN7 with miR-4319 on GC progression remain unclear. In this research, a low level of miR-4319 was detected in GC tissues and cells. Furthermore, miR-4319 was confirmed to be targeted by circATXN7 and negatively associated with circATXN7. In addition, miR-4319 inhibited cell proliferation, invasion and enhanced cell apoptosis in GC. Conclusively, miR-4319 was directly targeted by circATXN7 and acted as a tumor suppressor in GC.
CircRNAs have been acknowledged to act as sponges for miRNAs in order to contest with mRNAs, hence elevating the level of target genes [31]. Disease-related reports have mentioned ENTPD4 in schizophrenia [32] and acquired immune deficiency syndrome [33]. However, ENTPD4 is a novel gene that has not been studied in cancers. Our present study showed that ENTPD4 was overexpressed in GC tissues and cell lines. Besides, ENTPD4 was validated to construct a negative relationship with miR-4319 and a positive relationship with circATXN7. Subsequently, we confirmed that ENTPD4 was a target gene of miR-4319. Through rescue assays, we observed that miR-4319 repression and ENTPD4 overexpression could countervail the suppressive effect of circATXN7 knockdown on GC progression. At last, the facilitating function of circATXN7 on GC tumor growth has been certificated via in vivo assays. All these data revealed the significant effect of the circATXN7/miR-4319/ENTPD4 pathway on regulation of GC progression.
In conclusion, circATXN7 was identified as an oncogene in GC and promotes GC development through regulating miR-4319/ENTPD4 axis, which provided a new therapeutic target for the treatment of GC.
Conclusion
The circATXN7 was identified as an oncogene in GC and promotes GC development through regulating miR-4319/ENTPD4 axis, which provided a new therapeutic target for the patients with GC.
Supplementary information
Additional file 1: Figure S1. The expression of Ki67 and ENTPD4 in GC patients with high or low expression of circATXN7 was detected.
Acknowledgements
We appreciate all the participants who provide supports in this study.
Abbreviations
- circRNAs
circular RNAs
- GC
gastric cancer
- circATXN7
circular RNA ATXN7
- ENTPD4
ectonucleoside triphosphate diphosphohydrolase 4
- ceRNA
competing endogenous RNA
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
fetal bovine serum
- shRNA
short hairpin RNA
- actD
Actinomycin D
- CCK-8
cell counting kit-8
- FISH
fluorescence in situ hybridization
- qRT-PCR
quantitative real-time polymerase chain reaction
Authors’ contributions
ZZ: investigation and preparation for the experiment; HW: conduct the experiment; ZC: collect and collate experimental data; GL: proofread and analyze the experimental data; BL: Figures and tables, complete the manuscript. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Not shared.
Ethics approval and consent to participate
No treatments were performed on patients before this study. Written informed consent was signed by every patient, the study protocol was accepted by the Ethics Committee of the Second Hospital of Shandong University.
Consent for publication
The publication was approved by all the authors.
Competing interests
The authors declare that they have no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1186/s12935-025-03733-x
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/13/2025
This article has been retracted. Please see the Retraction Notice for more detail: 10.1186/s12935-025-03733-x
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12935-020-1106-5.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet (London, England). 2015;385(9972):977–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA (New York, NY). 2014;20(12):1829–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6(5):563–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Huang S. Response to comment on response to “Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer,” Cancer Lett. 2017 Mar 1; 388(2017): 208–219. Cancer Lett. 2017;411:64. [DOI] [PubMed] [Google Scholar]
- 7.Kong Z, Wan X, Zhang Y, Zhang P, Zhang Y, Zhang X, Qi X, Wu H, Huang J, Li Y. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493(3):1217–23. [DOI] [PubMed] [Google Scholar]
- 8.Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ, Xu B. Long noncoding RNA PVT1 facilitates cervical cancer progression via negative regulating of miR-424. Oncol Res. 2017;25(8):1391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Wei Y, Yan Y, Wang H, Yang J, Zheng Z, Zha J, Bo P, Tang Y, Guo X, et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in OSCC. Oncol Rep. 2018;39(3):951–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Q, Wang S, Li X, Yang F, Feng C, Zhong K, Qiu M, Wang J. Circular RNA ATXN7 is upregulated in non-small cell lung cancer and promotes disease progression. Oncol Lett. 2019;17(6):4803–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol Carcinog. 2019;58(4):554–64. [DOI] [PubMed] [Google Scholar]
- 15.Jiang L, Wu Z, Meng X, Chu X, Huang H, Xu C. LncRNA HOXA-AS2 facilitates tumorigenesis and progression of papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 Axis. Hum Gene Ther. 2019;30(5):618–31. [DOI] [PubMed] [Google Scholar]
- 16.Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q, Zhao G, Zhang ZZ. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Wang J, Qiu M, Xu L, Li M, Jiang F, Yin R, Xu L. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumour Biol. 2015;36(3):1643–51. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Sun Q, Chen C, Yin R, Huang X, Wang X, Shi R, Xu L, Ren B. ZYG11A serves as an oncogene in non-small cell lung cancer and influences CCNE1 expression. Oncotarget. 2016;7(7):8029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vannini I, Wise PM, Challagundla KB, Plousiou M, Raffini M, Bandini E, Fanini F, Paliaga G, Crawford M, Ferracin M, et al. Transcribed ultraconserved region 339 promotes carcinogenesis by modulating tumor suppressor microRNAs. Nat Commun. 2017;8(1):1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. [DOI] [PubMed] [Google Scholar]
- 22.Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo JN, Li J, Zhu CL, Feng WT, Shao JX, Wan L, Huang MD, He JD. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. OncoTargets Ther. 2016;9:7451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan B, Zhang W, Mao XW, Jiang LY. Circular RNA ciRS-7 correlates with advance disease and poor prognosis, and its down-regulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(24):8712–21. [DOI] [PubMed] [Google Scholar]
- 25.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92. [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmad A, Zhang W, Wu M, Tan S, Zhu T. Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genom. 2018;40(3):243–51. [DOI] [PubMed] [Google Scholar]
- 27.Fan H, Yuan R, Cheng S, Xiong K, Zhu X, Zhang Y. Overexpressed miR-183 promoted glioblastoma radioresistance via down-regulating LRIG1. Biomed Pharmacother Biomedecine pharmacotherapie. 2018;97:1554–63. [DOI] [PubMed] [Google Scholar]
- 28.Shi W, Wang X, Ruan L, Fu J, Liu F, Qu J. MiR-200a promotes epithelial-mesenchymal transition of endometrial cancer cells by negatively regulating FOXA2 expression. Pharmazie. 2017;72(11):694–9. [DOI] [PubMed] [Google Scholar]
- 29.Lin X, Wang Y. Re-expression of microRNA-4319 inhibits growth of prostate cancer via Her-2 suppression. Clin Transl Oncol. 2018;20(11):1400–7. [DOI] [PubMed] [Google Scholar]
- 30.Chu J, Li Y, Fan X, Ma J, Li J, Lu G, Zhang Y, Huang Y, Li W, Huang X, et al. MiR-4319 suppress the malignancy of triple-negative breast cancer by regulating self-renewal and tumorigenesis of stem cells. Cell Physiol Biochem. 2018;48(2):593–604. [DOI] [PubMed] [Google Scholar]
- 31.Deng N, Li L, Gao J, Zhou J, Wang Y, Wang C, Liu Y. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495(1):189–96. [DOI] [PubMed] [Google Scholar]
- 32.Saito A, Fujikura-Ouchi Y, Ito C, Matsuoka H, Shimoda K, Akiyama K. An association study on polymorphisms in the PEA15, ENTPD4, and GAS2L1 genes and schizophrenia. Psychiatry Res. 2011;185(1–2):9–15. [DOI] [PubMed] [Google Scholar]
- 33.Spadoni JL, Rucart P, Le Clerc S, van Manen D, Coulonges C, Ulveling D, Laville V, Labib T, Taing L, Delaneau O, et al. Identification of genes whose expression profile is associated with non-progression towards AIDS using eQTLs. PLoS ONE. 2015;10(9):e0136989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The expression of Ki67 and ENTPD4 in GC patients with high or low expression of circATXN7 was detected.
Data Availability Statement
Not shared.