Abstract
We report a simple, one-step enzymatic synthesis of the blue fluorescent noncanonical amino acid β-(1-azulenyl)-l-alanine (AzAla). Using an engineered tryptophan synthase β-subunit (TrpB), stereochemically pure AzAla can be synthesized at scale starting from commercially available azulene and l-serine. Mutation of a universally conserved catalytic glutamate in the active site to glycine has only a modest effect on native activity with indole but abolishes activity on azulene, suggesting that this glutamate activates azulene for nucleophilic attack by stabilization of the aromatic ion.
Keywords: Protein engineering, Amino acids, Biocatalysis, Sustainable chemistry, Aromatic ions
Graphical Abstract
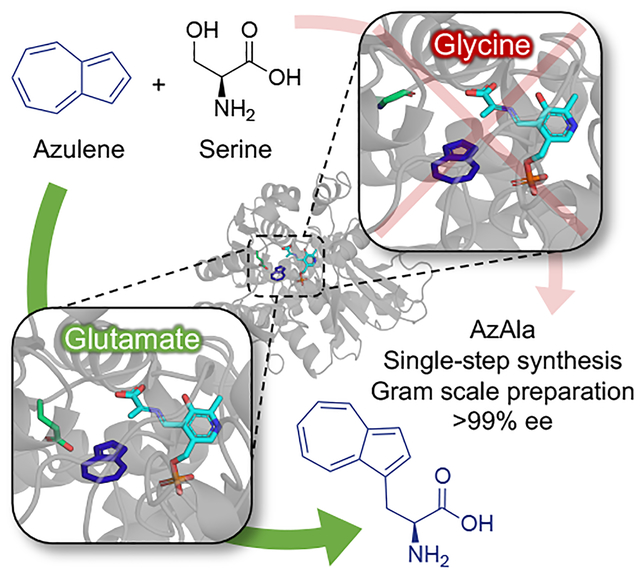
Into the wild blue yonder:
Tryptophan synthase β-subunit substrate scope is expanded to accept the non-indole nucleophile azulene to synthesize the blue fluorescent noncanonical amino acid β-(1-azulenyl)-l-alanine (AzAla). Ina one-step enzymatic synthesis, AzAla is synthesized on gram scale.
Proteins and peptides can be imbued with new chemical and physical properties via the inclusion of noncanonical amino acids (ncAAs). These molecules resemble the natural building blocks of proteins but contain distinct structures and functional groups. When incorporated into proteins, ncAAs can serve as handles for chemical reactions or as spectroscopic probes to examine protein function, including reactivity, localization, and interaction with other biomolecules.[1–3] Unfortunately, applications of many potentially useful ncAAs are limited owing to their high cost and lack of availability. The paucity of available ncAAs also hinders engineering of aminoacyl tRNA synthetases (aaRS) necessary for site-specific, in vivo ncAA incorporation into proteins.
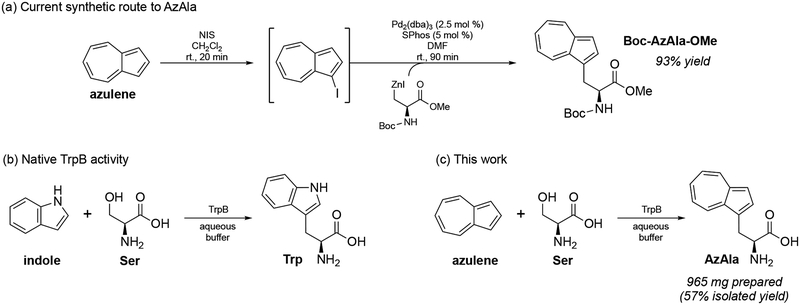
The ncAA β-(1-azulenyl)-l-alanine (AzAla, Scheme 1) is a tryptophan (Trp) isostere with unique fluorescent properties that make it a useful spectroscopic probe for investigating protein dynamics and protein-protein interactions. It can be incorporated into proteins in place of Trp without significantly disturbing tertiary structure or function.[4–8] In contrast to Trp, its spectroscopic properties are insensitive to the environment, making it ideal in contexts where local conditions and quenchers (e.g., methionine, histidine) could complicate analysis of fluorescent signals.[4] Its qualities have been leveraged for Förster resonance energy transfer (FRET) experiments to elucidate protein-protein interactions as well as vibrational energy transfer (VET) studies to probe anisotropic energy flow within proteins.[9,10] Recently, a method for synthesis of AzAla via Negishi cross-coupling was described (Scheme 1a).[11] Although proceeding with good yields on gram scale, the multi-step process is highly time-sensitive and uses precious metal catalysts and organic solvents. A simpler route to AzAla would expand its applications in biochemical studies.
Scheme 1.
(a) Current synthetic route to Boc-protected AzAla (b) TrpB natively catalyzes the condensation of indole and serine to form tryptophan. (c) Single-step biocatalytic synthesis of AzAla from azulene and serine described in this work.
Researchers have begun to look to enzymes as complementary or alternative approaches for the synthesis of enantiomerically pure ncAAs.[12] Enzymes can perform enantio- and regioselective chemistry in the presence of reactive moieties such as primary amines, obviating the need for expensive and intricate chiral catalysts, chiral separations, and protecting groups. To this end, our lab previously reported the directed evolution of the tryptophan synthase β-subunit (TrpB) as a stand-alone biocatalytic platform for the synthesis of diverse tryptophan analogues (Scheme 1b).[13,14] Here we report a simple, efficient route for synthesis of AzAla from stable, commercially available starting materials using an engineered TrpB (Scheme 1c).
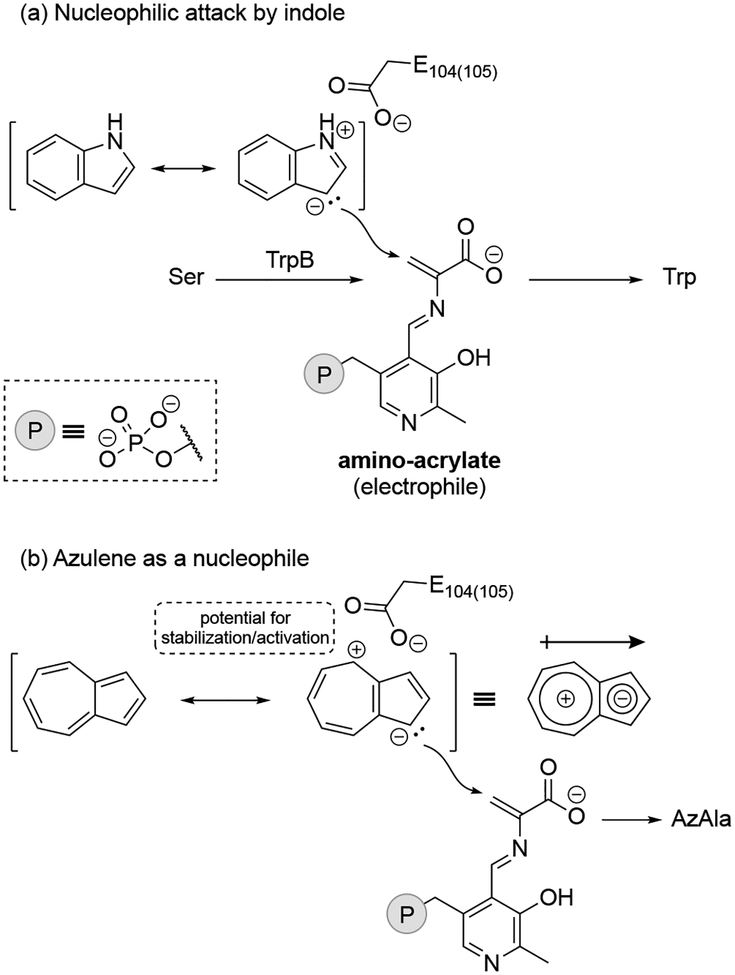
TrpB is a Type-II pyridoxal phosphate (PLP)-dependent enzyme that natively performs a conjugate addition reaction between indole and L-serine (Ser) to make Trp. During the catalytic cycle, Ser binds the PLP cofactor, and subsequent β-elimination and release of water forms an electrophilic amino-acrylate species (Scheme 2a). A highly conserved active site glutamate stabilizes the accumulation of positive charge on the pyrrole ring of indole and helps facilitate the nucleophilic attack on the amino-acrylate to form a new C–C bond that produces Trp (Figure S1). The similarity of AzAla to Trp prompted us to investigate whether TrpB could accept azulene as a nucleophile in the place of indole. Unlike indole, azulene lacks heteroatoms that can help stabilize the accumulation of charge during nucleophilic attack. Despite this, azulene has a permanent dipole exemplified by its resonance structure of a cycloheptatrienyl cation (tropylium) fused to a cyclopentadienyl anion (Cp−). We hypothesized that, analogous to indole, the buildup of electron density on the Cp− could promote nucleophilic attack by azulene in the TrpB catalytic cycle, while the tropylium system could stabilize the resulting positive charge (Scheme 2b). Aromatic ions are common moieties in synthetic chemistry[15] and as enzyme inhibitors,[16] but there are few reports of enzymes that can interact productively with aromatic ions in their catalytic cycles. We were therefore unsure if the active site of TrpB could accommodate or activate such a substrate, or if the reactivity of azulene would be sufficient for nucleophilic attack.
Scheme 2.
Parallels between indole and azulene in the TrpB reaction.
We first examined the conversion of azulene and Ser to AzAla using a small panel of previously engineered TrpB variants from the thermophilic organisms Pyrococcus furiosus (PfTrpB) and Thermotoga maritima (TmTrpB). The variants were selected to provide an efficient sampling of the engineered TrpB evolutionary lineage beginning from wild-type TrpS and ending with stand-alone TrpB variants evolved for activity with different indole and serine analogues. Nearly every enzyme we tested demonstrated significant activity for this reaction, the exception being variants in which the highly conserved glutamate mentioned above was mutated to glycine.
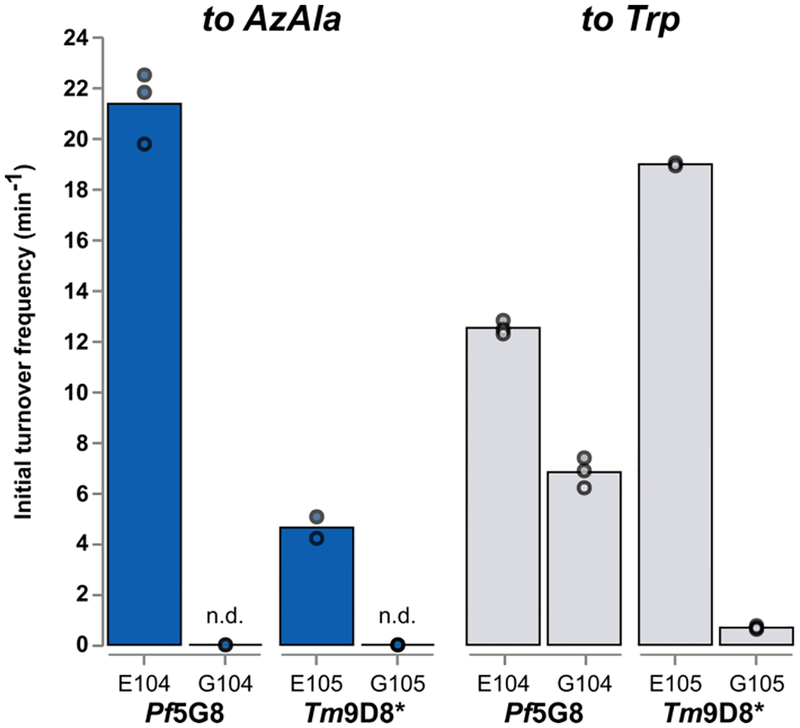
The significant effect of the E104(105)G mutation suggested that this conserved catalytic residue may be playing an important role in the non-native azulene reaction. We explored this possibility by examining two engineered variants with and without the E104(105)G mutation: Pf5G8, which exhibits optimal activity at 75 °C,[13] and Tm9D8*, which exhibits optimal activity at lower temperatures such as 37 °C.[18] Challenging the enzymes with indole demonstrated that this mutation only modestly decreases the rate of Trp formation (Figure 1), with an additional slight decrease in the chemoselectivity of the reaction that leads to formation of trace amounts of isoTrp (a product of the N-alkylation of indole, shown in Figure S2 and described previously[13]). In contrast, the E104(105)G mutation exerts a profound effect on AzAla production, practically abolishing all activity with azulene. Product was only detected in low amounts when the reaction was performed with high catalyst loadings overnight (0.1 mol %, 16 h; Figure S3). We speculate that the glutamate residue stabilizes the tropylium cation to facilitate nucleophilic attack from Cp−. However, further mechanistic studies are required to elucidate the role of this mutation.
Figure 1.
Mutation of a conserved glutamate residue affects the rate of AzAla production more significantly than native Trp production. Bars represent the average of two to three replicates, with replicates being shown as individual points. Reaction conditions can be found in Table S1; n.d. = not detected by LC-MS.
Next, we wished to develop a biocatalytic method for the production of AzAla at scale. Although Pf5G8 catalyzed the reaction roughly 4.5-fold faster at its optimum temperature of 75 °C than Tm9D8* at 37 °C, azulene readily sublimes at high temperatures, making its containment at 75 °C difficult. We thus opted to use directed evolution to improve the activity of Tm9D8* at 37 °C to create a stand-alone biocatalyst for the production of AzAla in vivo and in vitro.
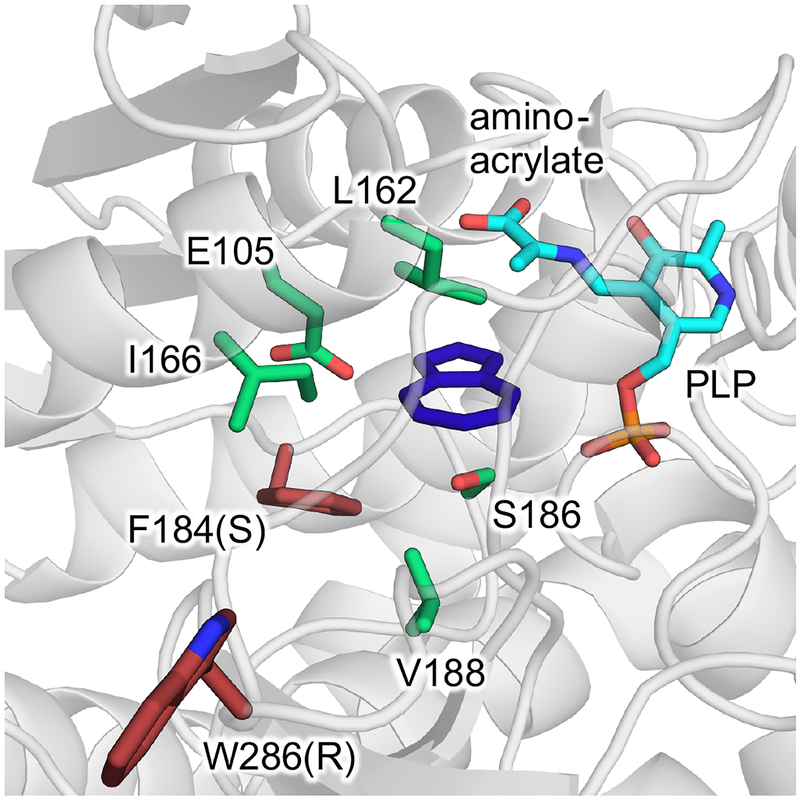
A single round of random mutagenesis and screening identified two variants containing the mutations F184S and W286R. These mutations were combined to yield the final variant TmAzul, which had a three-fold improved rate of AzAla formation compared to Tm9D8* (14.0 turnovers per minute; Table S1). As we have not been successful in obtaining a crystal structure of TmTrpS and its variants, we constructed a homology model of Tm9D8* to try to understand the locations of each mutation and their possible effects (Figure 2). The W286R mutation sits on a flexible loop, where its role in catalysis is difficult to infer. F184S sits directly in the active site and is one of only a handful of residues whose side chains are in close proximity to the azulene substrate. This residue may interact directly with the substrate during catalysis or adjust the active site to be more accommodating for AzAla synthesis.
Figure 2.
Homology model of Tm9D8* with azulene in the active site. Azulene (deep blue) is shown in a putative productive binding mode within a model of Tm9D8* with the amino-acrylate intermediate (cyan) formed in the active site (see SI Section 3.11 for model construction). Active-site residues in the native TmTrpB enzyme (green) and the mutations found in this study (red, mutation in parentheses) are shown as sticks.
Because the Escherichia coli expression cultures were heat treated at 75 °C for three hours prior to screening, TmAzul retains high thermostability, and highly pure enzyme can be obtained simply by heating the E. coli expression host at 75 °C for >1 h, pelleting the denatured E. coli proteins by centrifugation, and collecting the enzyme-bearing supernatant. To demonstrate scalability of this method, we synthesized AzAla on a gram scale using heat-treated lysate from a 1-L culture expressing the evolved TmAzul variant. We found that increasing the concentration of DMSO in the reaction increased the reaction rate at this scale, presumably by keeping azulene from forming insoluble crystals in the reaction mixture due to its sparing solubility in aqueous buffer. We thus used 20% DMSO cosolvent, and the reaction progress was monitored by taking small aliquots of the reaction mixture and combining with an equal volume of ethyl acetate to observe the relative ratio of azulene to AzAla in the organic and aqueous layers, respectively. After 48 hours, the product was purified by removing any remaining azulene by extraction with ethyl acetate, removing the aqueous solvent in vacuo, and precipitating AzAla from the remaining DMSO cosolvent by the addition of excess ethyl acetate. The crude precipitate was collected by filtration and then further purified by reverse-phased column chromatography to afford 965 mg of pure AzAla (57% isolated yield). Crude azulene can be recovered from ethyl acetate by gently evaporating off the solvent under a constant stream of nitrogen and reused. The enantiopurity of the isolated AzAla product is >99% ee.
In conclusion, we have described a new-to-nature reaction catalyzed by tryptophan synthase and identified a conserved residue that is critical for this non-native reaction. Based on comparisons to the native reaction, we suggest that E104(105) stabilizes the aromatic ions in azulene to facilitate nucleophilic attack from Cp−. To improve access to this useful ncAA, we have engineered a highly active enzyme catalyst that synthesizes enantiomerically pure AzAla from commercially available starting materials in a single step. Given that an engineered synthetase/tRNA pair has been reported for this ncAA, TmAzul also closes the gap for in vivo synthesis and incorporation of AzAla.[10]
Supplementary Material
Acknowledgments
We thank Giovanni Tomaleri for his experimental assistance, Nathaniel Goldberg and Dr. Ben Levin for insightful discussions, and Dr. Tina Boville and Dr. David Romney for valuable contributions to experimental design. We thank Dr. Scott Virgil and the Center for Catalysis and Synthesis. This work was supported by the Rothenberg Innovation Initiative and by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM125887. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted after peer review and appears as an Accepted Article online prior to editing, proofing, and formal publication of the final Version of Record (VoR). This work is currently citable by using the Digital Object Identifier (DOI) given below. The VoR will be published online in Early View as soon as possible and may be different to this Accepted Article as a result of editing. Readers should obtain the VoR from the journal website shown below when it is published to ensure accuracy of information. The authors are responsible for the content of this Accepted Article.
Supporting information for this article is given via a link at the end of the document.
Experimental Section
Experimental details can be found in the Supporting Information.
Conflict of Interest
Enzyme variants in this manuscript and enzymatic synthesis of AzAla are the subject of a patent application (P.J.A. inventor).
References
- [1].Liu CC, Schultz PG, Annu. Rev. Biochem 2010, 79, 413–444. [DOI] [PubMed] [Google Scholar]
- [2].Neumann H, FEBS Lett 2012, 586, 2057–2064. [DOI] [PubMed] [Google Scholar]
- [3].Agostini F, Völler JS, Koksch B, Acevedo-Rocha CG, Kubyshkin V, Budisa N, Angew. Chemie - Int. Ed 2017, 56, 9680–9703. [DOI] [PubMed] [Google Scholar]
- [4].Gosavi PM, Moroz YS, Korendovych IV, Chem. Commun 2015, 51, 5347–5350. [DOI] [PubMed] [Google Scholar]
- [5].Loidl G, Musiol HJ, Budisa N, Huber R, Poirot S, Fourmy D, Moroder L, J. Pept. Sci 2000, 6, 139–144. [DOI] [PubMed] [Google Scholar]
- [6].Sartori E, Toffoletti A, Corvaja C, Moroder L, Formaggio F, Toniolo C, Chem. Phys. Lett 2004, 385, 362–367. [Google Scholar]
- [7].Venanzi M, Valeri A, Palleschi A, Stella L, Moroder L, Formaggio F, Toniolo C, Pispisa B, Biopolymers 2004, 75, 128–139. [DOI] [PubMed] [Google Scholar]
- [8].Mazzuca C, Stella L, Venanzi M, Formaggio F, Toniolo C, Pispisa B, Biophys. J 2005, 88, 3411–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Moroz YS, Binder W, Nygren P, Caputo GA, Korendovych IV, Chem. Commun 2013, 49, 490–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baumann T, Hauf M, Schildhauer F, Eberl KB, Durkin PM, Deniz E, Löffler JG, Acevedo-Rocha CG, Jaric J, Martins BM, et al. , Angew. Chemie - Int. Ed 2019, 58, 2899–2903. [DOI] [PubMed] [Google Scholar]
- [11].Stempel E, Kaml RFX, Budisa N, Kalesse M, Bioorganic Med. Chem 2018, 26, 5259–5269. [DOI] [PubMed] [Google Scholar]
- [12].Almhjell PJ, Boville CE, Arnold FH, Chem. Soc. Rev 2018, 47, 8980–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Romney DK, Murciano-Calles J, Wehrmüller J, Arnold FH, J. Am. Chem. Soc 2017, 139, 10769–10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Buller AR, Brinkmann-Chen S, Romney DK, Herger M, Murciano-Calles J, Arnold FH, Proc. Natl. Acad. Sci 2015, 112, 14599–14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Komatsu K, Kitagawa T, Chem. Rev 2003, 103, 1371–1427. [DOI] [PubMed] [Google Scholar]
- [16].Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark AD, Ryan K, Hickey MJ, Love RA, et al. , Structure 2009, 17, 1625–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Murciano-Calles J, Romney DK, Brinkmann-Chen S, Buller AR, Arnold FH, Angew. Chemie - Int. Ed 2016, 55, 11577–11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Boville CE, Romney DK, Almhjell PJ, Sieben M, Arnold FH, J. Org. Chem 2018, 83, 7447–7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.