Abstract
Objective:
To conduct a genetic and candidate gene association study with samples from phenotype-ascertained dogs to identify putative disease-associated gene/mutation for optic nerve hypoplasia (ONH) in the miniature poodle.
Animals studied:
A total of 43 miniature poodles from the US and Europe, 9 affected bilaterally with ONH, were included in the study. Pedigree information was recorded.
Procedures:
A pedigree including all animals studied was assembled. Twenty-one genes typically expressed in ganglion cells or that are associated with ocular malformations and have a critical function in eye and neural retina development were selected. Exons and exon-intron boundaries of 8 genes were sequenced in four ONH cases and four controls. Furthermore, cases and controls were genotyped with the Illumina CanineHD BeadChip to obtain genotypes for 13 additional candidate genes for haplotype association.
Results:
The assembled pedigree connected all ONH-affected dogs to a possible common founder. Identified variants and haplotypes of the tested candidate genes did not segregate with the phenotype using Identity by Descent approach assuming autosomal recessive inheritance with variable but yet unknown penetrance.
Conclusions:
Pedigree analysis did not reveal the inheritance pattern. There is no evidence of association of the evaluated candidate genes with ONH, therefore, the screened candidate genes can provisionally be ruled out as causally associated with the disease.
Keywords: dog, optic nerve hypoplasia, micropapilla, miniature poodle, candidate genes
Introduction
Canine optic nerve hypoplasia (ONH), characterized by a very small optic nerve head and blindness or severely impaired vision, is a non-progressive congenital defect affecting one or both eyes.[1, 2] The precise pathogenesis is unknown, although it is likely due to a failure of retinal ganglion cell development, or defects in axon guidance to central nervous system targets. Optic nerve hypoplasia and/or aplasia also has been described in humans [3], horses [4], cats [5, 6] and rodents.[7] In man, it is the second leading cause of pediatric blindness, and the most common optic nerve anomaly in the US.[8] Although most cases happen to be sporadic isolates in families, it is now clear that many cases are caused by mutations in genes involved in eye development.[8]
In dogs, the disease has been reported in the miniature poodle breed [9] where it has a particularly high prevalence. Based on records from the Canine Eye Registry Foundation (CERF) summarized by the Genetics Committee of the American College of Veterinary Ophthalmologists for the 2000–2009 time period [10], a total of 34 cases were diagnosed out of 4242 dogs examined. This is statistically significantly higher than for other breeds (e.g. German shepherd: 6/1725; golden retriever: 7/62,695) or for the toy poodle variety (5/3063), examined in the same time period (Fisher’s exact test; p-value < 0.05). With the transfer of record keeping and registry functions from CERF to the Orthopedic Foundation for Animals (OFA), the ability to readily extract the ONH prevalence data for miniature poodles has been lost as all three of the poodle varieties are grouped under “Poodle” in the Blue Book (e.g. [11]).
There are several factors that complicate ONH disease ascertainment, particularly in determining disease incidence and severity. Firstly, the diagnosis in dogs is based strictly on fundus examination, assessment of the pupillary light reflex (PLR), and superficial assessment of vision deficits, and not on a detailed evaluation of visual function, visual fields or in vivo imaging of inner retinal microanatomy by spectral domain optical coherence tomography (sdOCT) like in man.[12] This assessment results in a binary grouping of patients (i.e. ONH affected and normal), and fails to categorize more mildly affected individuals that may represent intermediate phenotypes. Secondly, the normal appearance of the canine optic nerve head is highly variable, and there is no correlation of these variations with visual function (see Supplementary File S1, page 2 for illustration of some variations in optic disc appearance). Thirdly, there is a confounding clinical condition in dogs, particularly miniature poodles, termed micropapilla (Mp) that refers to a very small optic nerve head, which clinically appears similar to ONH, yet the dog has a normal PLR and subjectively ‘normal’ vision. The only histologic study of Mp to date has been done in one beagle dog in which the optic nerve fibers were normal, but myelination of the nerve fibers began behind the lamina cribrosa and not at the optic disc proper.[13] At present, it is not clear whether Mp is a normal variant unrelated to ONH, or is part of the continuum between a normal and a hypoplastic disc.
In spite of ONH being a common finding in dogs, there is no genetic information, and a paucity of clinical literature on the disease. Is ONH inherited, and if yes, how? Is it acquired? If one considers the criteria set out by the ACVO Genetics Committee and included in many editions of the Blue Book (e.g. 5th page in 2010 edition [10], and page 5 in 2014 edition [11]), ONH in miniature poodles fulfills almost all: frequency is higher than in other breeds; frequency is greater in related dogs within a breed; disease has a characteristic appearance, location, age of onset and progression. Only one criterion is missed (looks identical to an entity which has been proven to be inherited in another breed) because no such disorder has been proven to be inherited in another dog breed.
To this end, we have taken an initial step in our ONH studies by first creating a clinical examination form that minimizes examiner bias, and can be used at multiple sites by different clinicians to obtain cases and controls. With the initial samples and pedigree information obtained, we then carried out a screen of candidate genes associated with optic nerve defects with/without ocular malformations in other species for genetic variants, and assessed their segregation with the ONH phenotype. While no positive ‘hits’ were obtained, this initial study informs on future approaches to further investigate the genetic basis of this visually impairing clinical entity.
Materials and Methods
Study samples
The research study was extensively advertised through the web pages of the American and European Colleges of Veterinary Ophthalmologists, through direct contact with members of the Poodle Club of America, publications and newsletters dealing with poodles [14–16] and through the OptiGen, LLC research web pages (http://www.optigen.com/opt9_onh_micropap.html). Blood samples from miniature poodles diagnosed with ONH or Mp, and samples of their normal relatives together with their eye examination clinical records, fundus photographs (if available) and pedigree information, were obtained from veterinary ophthalmologists in the US and different centers in Europe, and submitted through OptiGen, LLC. Samples also were collected at eye clinics organized by dog clubs.
To obtain samples that are clinically characterized in a consistent manner, a specifically developed clinical research form was used (Supplementary File S1). As assessment of normal optic nerve head structure is quite subjective, and often variable, this form provided a consistent means of assessment. The clinical information provided was reviewed by an ACVO board certified veterinary ophthalmologist (GDA) prior to inclusion. For the current study, only dogs that had bilateral ONH on clinical examination, pupillary light responses that were absent or extremely sluggish/incomplete, and blindness were included. Micropapilla samples were used for evaluating family relatedness with ONH cases, but not for the molecular studies. In total, we received nine samples of dogs that were diagnosed as blind due to bilateral ONH, and ten samples of dogs that were diagnosed with Mp (2 unilaterally and 8 bilaterally affected) representing both sexes. Furthermore, we obtained 34 samples of close and distant relatives of affected dogs. Twenty-six out of 34 were examined by veterinary ophthalmologists and had normal optic nerves and vision; the remaining 8 samples are being archived for future use. All procedures were in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Based on the guidelines of the University of Pennsylvania’s Office of Laboratory Animal Welfare, protocol approval by the university IACUC was not required when outside veterinarians submitted blood samples/clinical records for the study.
DNA extraction
Genomic DNA was isolated from EDTA blood samples with the Nucleon Bacc2 kit (GE Healthcare) in accordance with the manufacturer’s instructions. DNA quality was measured with a NanoDrop (http://www.nanodrop.com/). Samples with a ratio of A260/280 between 1.8 and 2 were diluted to a final concentration of approximately 50ng/µl and were used for PCR amplification and genotyping.
Candidate gene analysis
Candidate genes for ONH were chosen based on their reported association with ocular/CNS malformations in other species as well as the knowledge about the gene’s biological function, and are typically expressed in ganglion cells, or are involved in axon guidance and neural cell adhesion. Many of the genes serve as transcription factors, participating in an intricate multistep process critical to eye development and retinal neurogenesis. These include: OPA1, OPA3, OTX2, EFNA5, HESX1, TUBA8, TUBA1A, SIX1, SIX4, SIX6, SOX2, NRPM1, SLIT1, SLIT2, BDNF, EPHB1, EPHB6, NAV2, TMEM126A, GLI3 and NRCAM (Supplementary Table S1 including references). Haplotypes were reconstructed with PHASE software [17] if SNPs were polymorphic and there was more than one genotype available. Fisher’s exact test (two-sided) was used for statistical analysis (p<0.05) of the allelic, genotype and haplotype distribution.
Sequencing
Initially, sequencing of exons and intron-exon boundaries was done for eight candidate genes (OTX2, HESX1, TUBA8, TUBA1A, SIX1, SIX4, SIX6 and SOX2) in four cases that were diagnosed with bilateral ONH and blindness, and four controls with normal optic discs. This was done to identify putative disease-causing mutations, and to identify SNPs to be used in haplotype analysis. To further refine the search for associated variants in non-coding regions of four candidate genes (OTX2, TUBA8, TUBA1A and SOX2), additional SNPs retrieved from the list of canine SNPs published by the Broad Institute of Massachusetts Institute of Technology (http://www.broadinstitute.org/mammals/dog) were sequenced for validation.
Primers for PCR amplification were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and synthesized by Integrated DNA Technologies (Supplementary Table S2). PCR products were visualized on a 1.2% agarose gel and sent for sequencing to the NAPCore facility of the Children’s Hospital of Philadelphia (http://www.research.chop.edu/cores/napcore/).
To increase the number of SNPs and candidate genes for analysis, DNA from the four ONH-affected and three normal control dogs was genotyped with the Illumina CanineHD BeadChip (Illumina, San Diego, CA, USA). The BeadChip contains more than 170,000 evenly spaced markers providing uniform genomic coverage. Genotyping allowed us to obtain genotypes for 13 additional candidate genes (OPA1, OPA3, EFNA5, NRPM1, SLIT1, SLIT2, BDNF, EPHB1, EPHB6, NAV2, TMEM126A, GLI3 and NRCAM).
Pedigree analysis
Pedigree information for 9 bilateral ONH-affected dogs, and 10 dogs with Mp (2 unilateral and 8 bilateral) that spanned 4 to 5 generations was obtained as part of the sample collection process. Prior generations were identified using an online poodle pedigree database (http://www.poodledata.org/). Due to the large number of inbreeding loops, a combined pedigree connecting all affected dogs with an identified single founder was drawn manually using Macromedia FreeHand MX (v11.0). To simplify the pedigree information, the pedigree only shows generations tracing back to that founder. Furthermore, we checked pedigrees spanning ten to twelve generations of 10 randomly selected miniature poodles from the poodle pedigree database unrelated to affected dogs. Those dogs were randomly selected using the command ‘sample’ in the software package R.
Results
Clinical phenotype
The clinical examination form designed specifically for this project allowed the study subjects to be ascertained in a consistent manner. This overcame the variation in optic nerve head anatomy one encounters during routine clinical examinations, and, equally important, avoided subjective assessment (Supplementary Fig. S1). Furthermore, as fundus photographs cannot always be taken during these routine examinations, the forms asked the referring ophthalmologist to “... classify optic discs using the scale (images) above for normal variations, micropapilla, and optic nerve hypoplasia”.
All affected dogs used in the candidate gene association study were examined by ECVO- or ACVO-board certified veterinary ophthalmologists, and had a diagnosis of ONH based on bilateral small optic discs, absent direct and consensual pupillary light responses and blindness. When fundus photographs were provided with the clinical examination record (Fig. 1), the fundus changes were compatible with the diagnosis of ONH provided these findings were accompanied by absent PLR and blindness. If fundus photographs were not provided then the optic nerve findings were scored using the illustration provided in clinical examination form (Supplementary File S1). None of the dogs used in the candidate gene studies had Mp. Control dogs were related to dogs diagnosed with Mp and ONH, but had normal optic nerves and vision based on clinical examination by board certified veterinary ophthalmologists.
Figure 1. Retinal changes in ONH-affected dog.
Fundus photographs of two 8-week old littermate miniature poodles that are normal (A) or affected with ONH (B) and were included in the pedigree and candidate gene analyses; OD-right eye; OS-left eye. Whereas the normal disc is round, pink and has a distinct vascular pattern on the surface at 8 weeks of age (A), the hypoplastic discs (white arrows) are markedly reduced in size. Note that the ONH-affected OS (B) has a pre-retinal hemorrhage on the retinal surface (asterisk). This is a frequent finding in puppies that arises from bleeding from the regressing hyaloid vasculature, and disappears with aging.
Pedigree analysis
Using pedigree information obtained during sample collection and an online poodle pedigree database (http://www.poodledata.org/), we assembled a pedigree connecting miniature poodles diagnosed with ONH as well as all dogs diagnosed with Mp. All affected dogs (ONH and Mp) are related on both the maternal and paternal sides of their respective pedigrees to a single popular sire born more than 40 years ago (Fig. 2). Parents of the affected dogs either had an unknown status, were normal or diagnosed with Mp. To minimize the possibility that the identified founder is connected to all affected dogs by chance, we looked at pedigrees of 10 randomly selected miniature poodles from the poodle pedigree database (http://www.poodledata.org/). The founder was not present in those pedigrees spanning at least 10 generations.
Figure 2. Pedigree connecting Optic Nerve Hypoplasia and Micropapilla affected animals.
All sampled animals are related to a single founder male born more than 40 years ago. For simplicity, the pedigree only shows prior generations tracing back to that founder. No examination records were available for animals with a dot in the center of the symbol. Parents of affected dogs either had an unknown status, were normal or diagnosed with Mp. Dogs sequenced by Sanger sequencing and the Illumina CanineHD BeadChip that were used for candidate gene sequencing and haplotype analysis are marked with an oblique arrow. The control dog that was only sequenced by Sanger sequencing is marked with an asterisk.
Candidate gene analysis
For this preliminary analysis, we hypothesized an autosomal recessive mode of inheritance of unknown penetrance as all ONH and Mp affected dogs share a common ancestor in their maternal and paternal lines. To identify a possible association between the gene responsible for ONH in the established pedigree with a common founder, the ONH affected dogs are expected to be identical by descent for the causative mutation and flanking chromosomal regions. Using a combination of identity by descent and homozygosity mapping we tested 21 candidate genes (Supplementary Table S1) in phenotype-ascertained ONH affected (2 males, 2 females) and normal (1 male, 2–3 females) dogs (Fig. 2, oblique arrows); the number of control females that could be tested depended on the method used.
Sequencing of the exons and intron-exon boundaries determined via alignment with the human genome of 8 candidate genes identified 10 variants (three 5’UTR, three intronic, two exonic and two 3’UTR variants) in OTX2, SIX6, SIX1, SIX4, TUBA1A and SOX2, but not in HESX1 and TUBA8. Four of those variants are not located in dbSNP.[18] No associated changes were found in the coding regions and the exon/intron junctions of the genes analyzed when comparing to the reference sequence. Furthermore, haplotype analysis of OTX2, SIX6, SIX1, SIX4, TUBA1A and SOX2 did not show any haplotypes that segregated with ONH (Fig. 3A).
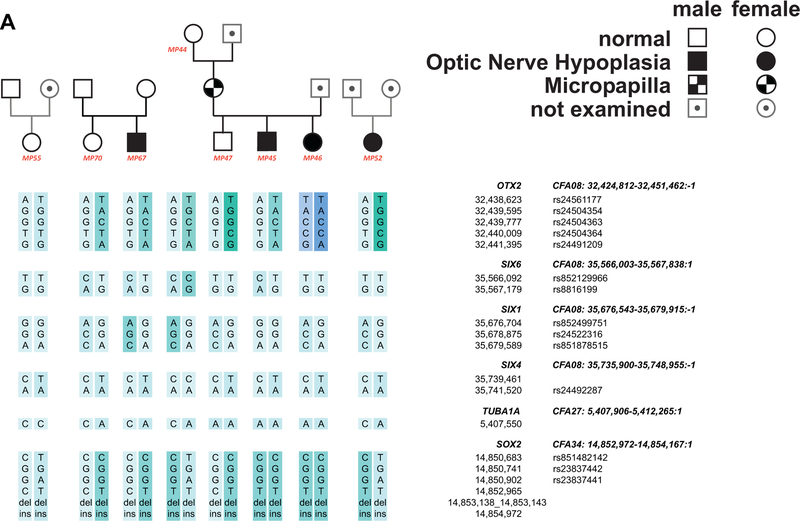
Figure 3. Haplotype analyses of candidate genes in ONH affected dogs and controls.
Haplotypes for the candidate genes studied for ONH affected and control dogs. Note that haplotype analysis was only carried out in normal control and ONH affected dogs, and not in dogs with Mp. Haplotypes for candidate genes sequenced by Sanger sequencing (A), and haplotypes for candidate genes genotyped with Illumina CanineHD BeadChip (B1, B2). Phenotypic status was assigned as detailed in Materials and Methods. Haplotypes for genes with informative SNPs are shown in animals used for association study. Haplotypes of HESX1 and TUBA8 are not shown as all animals shared the same homozygous wildtype haplotype. SNP IDs for SNPs located in dbSNP are given next to the variant base pair position.
For haplotype analysis, we increased the number of genetic variants to seventeen by sequencing additional known SNPs flanking the 5’ and 3’UTR and deep intronic regions of four candidate genes (OTX2, SOX2, TUBA8, TUBA1A). Cases and controls were homozygous wildtype for HESX1 and TUBA8 in the sequenced regions, and could not be evaluated based on homozygosity mapping.
In all ONH cases and three of the controls used for Sanger sequencing, we used the Illumina CanineHD BeadChip and extracted up to 13 genotypes for 13 additional candidate genes (Fig. 3B1, 3B2). For genes with more than one genotype, we reconstructed haplotypes that we used for gene-specific haplotype association (Supplementary Table S3). The number of haplotypes per gene varied from two (SIX4) to thirteen (NAV2). We compared the frequencies of haplotypes and genotypes in affected and unaffected dogs. There were no statistical differences in haplotype and genotype distribution between ONH affected and control dogs and haplotypes did not segregate with the phenotype. Distribution of alleles was also not statistically different between cases and controls. However, TMEM126A and TUBA1A were excluded from haplotype analysis because there was only one genotyped SNP available.
Lastly, because automal recessive inheritance is not definitivefor ONH we also considered the possibility of autosomal dominant inheritance. To this end, we compared haplotype, genotype and allele frequencies between cases and controls, but found no statistical difference between cases and controls.
Discussion
Canine ONH is a rare congenital disorder that may affect one or both eyes, and causes blindness or very severe visual impairment. Based on higher than expected breed prevalence, we have postulated that a genetic basis is likely for the miniature poodle.[10] In dogs, optic nerve head clinical phenotypes are highly variable between breeds and within breeds. Furthermore, disease ascertainment is further complicated in this breed by a clinical condition termed Mp which, in the extreme case, the optic nerve head is clinically indistinguishable from ONH, yet vision is normal, and the retrolaminar optic nerve is presumably unaffected.[13] As visual fields or acuity cannot be reliably assessed in dogs in a standard clinical setting, normal vision in Mp-affected dogs represents a presumptive clinical assessment. It is not clear at this time, whether Mp can be considered a variation of normal and not associated with ONH, or is part of a disease continuum between normal and ONH. Because of this uncertainty, we excluded dogs diagnosed with Mp from the molecular analyses although they were included in the pedigree studies, and samples were actively solicited for future research using the specific clinical research form developed for this study; such an approach ensured consistent phenotyping for all cases and controls enrolled (Supplementary File S1).
Given that all dogs diagnosed with either condition (ONH and Mp) in this pedigree share a common ancestor, for the analyses we posited that ONH has an autosomal recessive inheritance. If this popular sire disseminated the causal variant, subsequent inbreeding among the descendants increased the likelihood of ONH emerging. However, the complex inbreeding loops, and the limited phenotype information for prior generations, make it impossible to determine the precise mode of inheritance. Although a common founder on the maternal and paternal lines suggests an autosomal recessive inheritance pattern, the observation is not conclusive, and cannot exclude an autosomal dominant disorder also with variable penetrance. Furthermore, it is not known if dogs heterozygous for a putative genetic variant causing ONH also express the phenotype, an indication of the penetrance of the disorder. Therefore, this study and the pedigree are only a first step to determine the inheritance of the disorder, and more phenotype-ascertained dogs will be needed to unravel the actual mode of inheritance and direct future molecular studies.
If Mp is part of the ONH disease complex, it is possible that one gene controls the disease predisposition and additional modifying gene(s) influence the expression and severity of the disorder.[19, 20] In such a scenario, all dogs affected with ONH or Mp will have the same genetic variant, while only ONH-affected dogs have sequence changes in the modifier gene(s). At present, this is only speculative as the extant data do not support an additive dominant mode of inheritance, and are insufficient to indicate whether Mp should be considered a ‘milder’ manifestation of ONH or a totally unrelated inconsequential finding.
In humans, ONH can also occur in combination with myriad functional and anatomic anomalies of the central nervous system (CNS) including learning disabilities.[21] In miniature poodles, on the other hand, ONH appears to be a non-syndromic disorder affecting the inner retinal layers and optic nerves as pathologic examinations of ONH-affected miniature poodles did not reveal any gross anatomic anomalies in the brain.[9] Additionally, a necropsy of five ONH affected collie dogs did not reveal gross abnormalities in other organ systems.[22] It is possible, however, that mild CNS abnormalities could be present in affected dogs, but these would be difficult to detect given the difficulties of evaluating cognitive abilities in dogs. For this reason, we included in the analysis candidate genes, which are associated with multiple ocular/CNS syndromes.
Sequenced-based exclusion analysis has been used in multiple studies to eliminate hypothesized candidate genes underlying inherited diseases in dogs.[23–26] We identified no segregating variants in known coding sequences and intron/exon boundaries with ONH. Therefore, variants in the analyzed candidate genes that would lead to an alteration of the translated protein of the candidate genes can be dismissed as causative for the disease. However, we cannot rule out the presence of a pathogenic variant in the promoter region resulting in change of gene expression or large scale genetic structural rearrangements that can result in disease.
Ongoing studies are focused now on obtaining a larger sample set of cases and controls that will permit genome-wide association studies (GWAS). To this end, we urge the veterinary ophthalmology community to participate on these ongoing studies by providing ascertained normal miniature poodles, and those affected with ONH or Mp. To participate, please contact Dina Torjman (dtorjman@vet.upenn.edu) or Lydia Melnyk (lmelnyk@vet.upenn.edu) for the research forms and instructions on shipping blood samples.
Limitations of the study
Optic nerve hypoplasia is a complex disorder that currently has insufficient clinical and no genetic information. Although we posit a genetic cause for the disease based on a statistical higher frequency of disease in miniature poodles, there is no other supporting genetic information for this hypothesis, specifically prospective breeding studies. Although the special research form specifically developed for this study improves clinical disease ascertainment and diagnostic consistency, it does not provide additional in depth clinical assessments that inform on the disease. That would require sdOCT assessment of the inner retina, cSLO assessment of optic disc sizes, and magnetic resonance imaging of the optic nerves in anesthetized dogs that are affected with ONH and Mp. That is planned for the future phases of the study when additional patients are recruited.
Lastly, GWAS has been very successful in uncovering new causative genes for many disorders. The power of GWAS depends not only on sample size and number of markers genotyped, but also on case-to-control ratio, the causative allele frequency, the mode of inheritance, LD between markers and the prevalence and penetrance of the disorder. These factors determine the probability of detecting a putative causative variant.[27] In our study, sample sizes are limited by the number of well characterized cases and/or controls. Furthermore, the mode of inheritance as well as the prevalance of ONH are unknown factors. Therefore, more detailed information about ONH in miniature poodles is needed in order to design a successful GWAS. Whether the disease is inherited as autosomal recessive or dominant, it is likely not fully penetrant and would require a higher sample size to obtain the necessary statistical power.
Supplementary Material
Supplementary Table S1: Candidate genes selected for analysis.
Supplementary Table S2: Primers used for candidate gene sequencing.
Supplementary Table S3: Haplotype distribution in ONH unaffected and affected dogs.
Supplementary File1: Optic Nerve Hypoplasia and Micropapilla Research Form.
Acknowledgements
We are grateful to all veterinarians and veterinary ophthalmologists who provided clinical cases, blood samples and fundus photographs, especially to Nick Whelan, Claus Bundgaard Nielsen and Gerlinde Janssens for contributing detailed phenotypic information and fundus photographs. We also would like to thank the breeders and owners participating in this study. This study was supported by a grant from the Poodle Club of America Foundation, and unrestricted grant from the Van Sloun Fund for Canine Genetic Research, and NIH grant EY-06855.
References
- 1.Saunders LZ, Rubin LF. Ophthalmic Pathology of Animals : an Atlas and Reference Book S. Karger: Basel, 1975. [Google Scholar]
- 2.Kerrison JB, Newman NJ. Genetic causes of blindness. In: Neurology and Clinical Neuroscience Elsevier, 2000; 274–284. [Google Scholar]
- 3.Zion V Optic nerve hypoplasia. Ophthalmic Seminars 1976; 1: 171–196. [PubMed] [Google Scholar]
- 4.Gelatt K, Leipold H, Coffman J. Bilateral Optic Nerve Hypoplasia in a Colt. Journal of the American Veterinary Medical Association 1969; 155: 627–631. [PubMed] [Google Scholar]
- 5.Barnett KC, Grimes TD. Bilateral aplasia of the optic nerve in a cat. British Journal of Ophthalmology 1974; 58: 663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeeman W, Tumbelaka R. Das zentrale und periphere optische System bei einer kongenital blinden Katze. Albrecht von Graefes Archiv für Ophthalmologie 1916; 91: 242–263. [Google Scholar]
- 7.Rubin LF. Atlas of Veterinary Ophhthalmoscopy Lea and Febiger: Philadelphai, 1974. [Google Scholar]
- 8.Chen CA, Yin J, Lewis RA, Schaaf CP. Genetic causes of optic nerve hypoplasia. Journal of Medical Genetics 2017; 54: 441–449. [DOI] [PubMed] [Google Scholar]
- 9.Kern TJ, Riis RC. Optic nerve hypoplasia in three Miniature Poodles. Journal of the American Veterinary Medical Association 1981; 178: 49–54. [PubMed] [Google Scholar]
- 10.ACVO GC. Ocular Disorders Presumed to be Inherited in Purebred Dogs 5th ed., 2010.
- 11.ACVO GC. Ocular Disorders Presumed to be Inherited in Purebred Dogs 7th ed., 2014.
- 12.Pilat A, Sibley D, McLean RJ, Proudlock FA, Gottlob I. High-Resolution Imaging of the Optic Nerve and Retina in Optic Nerve Hypoplasia. Ophthalmology 2015; 122: 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellhorn RW. A survey of ocular findings in eight- to ten-month-old Beagles. Journal of the American Veterinary Medical Association 1974; 164: 1114–1116. [PubMed] [Google Scholar]
- 14.Pearce-Kelling S Why and how of inherited eye diseases in poodles: What breeders need to know. Poodle Variety 2011; Apr-May 76–77.
- 15.Fawver B Researchers seek genetics behind poodle eye diseases. Purina Pro Club Poodle Update 2016; 14: 3–5. [Google Scholar]
- 16.Koolsbergen G Optic nerve hypoplasia. The Poodle Club of Canada: The Poodle Scene 2015; Summer Newsletter: 8–9.
- 17.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics 2001; 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research 2001; 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furlan LL, Marson FA, Ribeiro JD, Bertuzzo CS, Salomao Junior JB, Souza DR. IL8 gene as modifier of cystic fibrosis: unraveling the factors which influence clinical variability. Human Genetics 2016; 135: 881–894. [DOI] [PubMed] [Google Scholar]
- 20.Ivansson EL, Megquier K, Kozyrev SV, Muren E, Korberg IB, Swofford R, Koltookian M, Tonomura N, Zeng R, Kolicheski AL, Hansen L, Katz ML, Johnson GC, Johnson GS, Coates JR, Lindblad-Toh K. Variants within the SP110 nuclear body protein modify risk of canine degenerative myelopathy. Proceedings of the National Academy of Sciences of the United States of America 2016; 113: E3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borchert M, Garcia-Filion P. The syndrome of optic nerve hypoplasia. Current Neurology and Neuroscience Reports 2008; 8: 395–403. [DOI] [PubMed] [Google Scholar]
- 22.Saunders LZ. Congenital optic nerve hypoplasia in collie dogs. Cornell Veterinarian 1952; 42: 67–80. [PubMed] [Google Scholar]
- 23.Short AD, Holder A, Rothwell S, Massey J, Scholey R, Kennedy LJ, Catchpole B, Ollier WE. Searching for “monogenic diabetes” in dogs using a candidate gene approach. Canine Genet Epidemiol 2014; 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiik AC, Ropstad EO, Bjerkas E, Lingaas F. A study of candidate genes for day blindness in the standard wire haired dachshund. BMC Veterinary Research 2008; 4: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet 2002; 3: 391–397. [DOI] [PubMed] [Google Scholar]
- 26.Winkler PA, Bartoe JT, Quinones CR, Venta PJ, Petersen-Jones SM. Exclusion of eleven candidate genes for ocular melanosis in cairn terriers. J Negat Results Biomed 2013; 12: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong EP, Park JW. Sample size and statistical power calculation in genetic association studies. Genomics Inform 2012; 10: 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Candidate genes selected for analysis.
Supplementary Table S2: Primers used for candidate gene sequencing.
Supplementary Table S3: Haplotype distribution in ONH unaffected and affected dogs.
Supplementary File1: Optic Nerve Hypoplasia and Micropapilla Research Form.