Abstract
Here we report the design, synthesis and characterization of bifunctional chemical ligands that induce the association of Ras with ubiquitously expressed immunophilin proteins such as FKBP12 and cyclophilin A. We show this approach is applicable to two distinct Ras ligand scaffolds, and that both the identity of the immunophilin ligand and the linker chemistry affect compound efficacy in biochemical and cellular contexts. These ligands bind to Ras in an immunophilin-dependent fashion and mediate the formation of tripartite complexes of Ras, immunophilin and the ligand. The recruitment of cyclophilin A to GTP-bound Ras blocks its interaction with B-Raf in biochemical assays. Our study demonstrates the feasibility of ligand-induced association of Ras with intracellular proteins and suggests it as a promising therapeutic strategy for Ras-driven cancers.
Keywords: Ras, cancer, drug design, immunophilin, dimerization
Graphical Abstract
Bifunctional ligands are designed and synthesized that induce the association of K-Ras and immunophilin proteins including FKBP12 and cyclophilin A. The recruitment of cyclophilin A to GTP-bound Ras blocks its interaction with B-Raf.

Introduction
Mutation of the RAS genes (K-, N-, and H-RAS) is the most common oncogenic lesion in cancer, occurring in more than 16% of all human cancers[1]. Absent deep accessible pockets, Ras proteins are often considered difficult to target directly by small molecule agents. Recent efforts have led to the identification of both macromolecular[2–9] and small-molecule[10–17] direct binders of Ras. Targets of these binders include the nucleotide binding pocket, shallow hydrophobic patches, and dynamic pockets near the Switch-II region revealed only upon ligand binding (namely, the Switch-II pocket (S-IIP) and Switch-II groove (S-IIG)). While efficacious, the Switch-II pocket ligands face a few limitations: they depend on the presence of a nucleophilic Cys residue at position 12 and also require Ras to be in a GDP-bound (inactive) state[14,18]. Despite these limitations, several Switch-II pocket ligands have demonstrated potent cellular[19], animal[20] Ras inhibition and anti-tumor effects, and three compounds (AMG510[21], MRTX849[22], JNJ-74699157[23]) have entered clinical trials in patients with K-Ras(G12C) mutant tumors. The S-IIG binding ligands, which target an unnatural M72C mutant, are at an earlier stage of development. They exhibit the striking feature of recognizing both the GDP and GTP states and surprisingly block the association of PI3K but not Raf with drug-bound GTP-K-Ras (M72C)[16]. The limitations of these two classes of ligands bound to Switch II confound their use against several of the most frequent oncogenic mutations (e.g., G12D, G12V and Q61L), which compromise GTP hydrolysis and render Ras constitutively GTP-bound, and lack available cysteine residues. We wondered whether we could overcome these limitations by recruiting an endogenous protein to Ras with small molecule ligands. In so doing, we hoped to exploit the large surface area of the recruited protein to achieve high-affinity binding that might allow reversible ligand recognition of K-Ras and also introduce steric hinderance to block the interaction between Ras and its effectors.
Equipping an intracellular protein with a small molecule ligand to confer neo-functions is a concept employed by a number of natural and artificial systems. For example, rapamycin forms a complex with the PPIase FKBP12 and this complex binds the protein kinase mTOR to sterically block the access of its substrates[24,25], while thalidomide induces the association of the E3 ligase Cereblon and the transcription factor Ikaros to promote the proteolytic degradation of the latter[26,27]. We envisioned that a bifunctional molecule could be constructed by chemically linking a Ras ligand and a high-affinity ligand of an abundant intracellular protein such as FKBP12 or cyclophilin A (CypA). FKBP12 has been reported to bind palmitoylated H-Ras and N-Ras[28]. Here we report the design and synthesis of such bifunctional ligands and their successful induction of the association of Ras-FKBP12 or Ras-CypA.
Results and Discussion
To design bifunctional molecules that recruit an intracellular protein to Ras, we first considered the identity of the intracellular protein. We chose two immunophilins, FKBP12 and CypA, as candidates because they are ubiquitously expressed chaperone proteins across all tissue types, and nature has repeatedly utilized them to target flat surfaces upon binding of a small molecule agent (e.g., FK506[29], rapamycin[24,25], cyclosporin A[29], and sanglifehrin A[30]). Several high-affinity ligands of FKBP12 and CypA have been reported (Fig. 1) and they each contain a solvent-exposed (when bound to FKBP12 or CypA) functional group handle that allows facile chemical derivatization. Next, we selected two chemical scaffolds as the Ras-binding component: S-IIP ligands (e.g., ARS1620 (A)) represent a family of covalent ligands that selectively binds the oncogenic Ras mutant G12C in the GDP-state[14,19,20], whereas S-IIG ligands (e.g., B) represent another class of covalent ligands that irreversibly reacts with a non-naturally occurring Ras mutant, M72C, regardless of the nucleotide binding state[16]. By analyzing co-crystal structures of these ligands bound to Ras, FKBP12 or CypA, we identified respective solvent-exposed sites for linker attachment that would likely preserve critical binding interactions. Finally, we devised a modular synthetic strategy that allowed us to independently vary three integral components (Ras ligand, FKBP12/CypA ligand and linker) to assemble the bifunctional ligands.
Figure 1.
Small molecule ligands of Ras and immunophilins. Blue shades and dashed circles indicate the solvent-exposed sites of the ligands. Red arrows indicate the predicted trajectory of substituents attached at these sites. a) Chemical ligands of two distinct scaffolds that bind near the Switch II region of Ras. b) High-affinity ligands of FKBP12 (SLF and FK506) and of CypA (Cyclosporin A). PDB entries of the structures shown (from left to right): 5V9U, 5VBE, 1FKG, 1FKJ, 1CWA.
An initial series of compounds focused on the S-IIP scaffold were synthesized with various linker lengths and recruiter ligands including SLF, FK506 and cyclosporin A (Fig. 2a). All of these compounds maintained potent binding to FKBP12 or CypA (Kd < 200 nM, Supplementary Fig. 1). We measured the rate of their covalent labeling reaction with K-Ras G12C CysLight (1–169, C51S/C80L/C118S, GDP-bound) by intact protein mass spectrometry. Although the SLF-based compounds (1–3) failed to show substantial reactivity, both FK506- and cyclosporin-derived compounds (4–6 and 7–10, respectively) labeled K-Ras G12C CysLight at appreciable rates. Interestingly, including an excess (10 μM) of recombinant FKBP12 or CypA significantly accelerated the reactions of FK506-derived and cyclosporin A-derived compounds (Fig. 2b-c), respectively. By contrast, no change in reaction rates was observed with mismatched protein/compound combinations.
Figure 2.
Bifunctional Ras(G12C) ligand created by linking a ligand of Ras and a ligand of FKBP12 or CypA. a) Modular design of bifunctional Ras ligands. b) Covalent modification of K-Ras CysLight(G12C) (GDP-state) after treatment with 10 μM compound for 24 h at 23 °C. c) Immunophilin proteins accelerate the covalent labeling of K-Ras CysLight(G12C) by their cognate bifunctional molecules. All measurements were the average of three independent replicates. CL, CysLight.
In both FK506- and cyclosporin-derived series, compounds with a 1,6-diaminohexyl linker (6 and 10) demonstrated the slowest covalent adduct formation with K-Ras G12C CysLight. A plausible explanation is that long linear linkers impart more conformational flexibility and incur higher entropic cost upon compound binding [31].
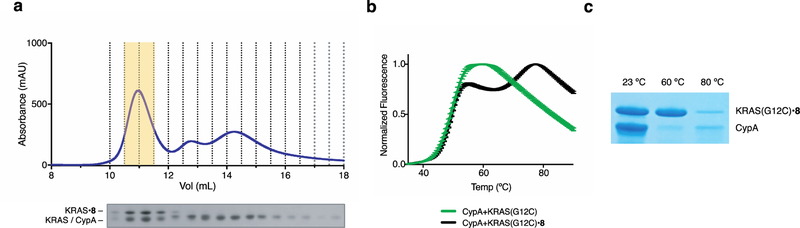
To assess compound-induced association of Ras and immunophilins, we incubated a mixture of K-Ras G12C CysLight (4 μM), CypA (10 μM) and 8 (10 μM) at 23 °C for 24 h and confirmed >95% labeling of K-Ras by intact protein mass spectrometry. Size exclusion chromatography of the reaction mixture yielded a distinct major peak with a retention volume (Kav = 0.23) consistent with a ternary complex of the two proteins and the ligand at 1:1:1 stoichiometry (40 kDa), along with two smaller unresolved peaks corresponding to unreacted K-Ras and excess CypA (Fig. 3a). Differential scanning fluorimetry revealed two-stage thermal denaturation of the protein complex at 48.9 °C and 71.0 °C (Fig. 3b). By analysis of a protein sample heated at 60 °C for 10 min we confirmed that the first melting event corresponds to the denaturation of CypA (Fig. 3c), suggesting that the ligand, while greatly stabilizing K-Ras (Tm = 50.9 °C, Supplementary Fig. 2), did not increase the thermal stability of CypA (Tm = 49.1 °C, Supplementary Fig. 2).
Figure 3.
K-Ras(G12C) labeled by compound 8 forms a stable complex with CypA. a) Size exclusion chromatography of a reaction mixture consisting of K-Ras(G12C), 8 and cyclophilin A allows isolation of a 40-kDa protein complex. b) The K-Ras•8•CypA complex exhibits a two-step thermal denaturation profile. c) K-Ras(G12C)•8•CypA complex was heated at 23 °C, 60 °C or 80 °C for 10 min and the soluble fraction was analyzed by SDS-PAGE.
The cellular efficacy of the S-IIP bifunctional ligands was tested in NCI-H358, a cell line that harbors a heterozygous G12C mutation at the KRAS locus and has been shown to be sensitive to the G12C-selective covalent inhibitor ARS1620[20]. Partial covalent modification of Ras, revealed by the extra higher molecular weight bands in the immunoblot analysis with a pan-Ras antibody, was achieved by compounds 5 and 9 after 24-h incubation at 10 μM (Fig. 4a). The identity of these bands was confirmed by their selective pulldown using GST-FKBP12 or GST-CypA protein captured with glutathione agarose beads. Compounds 5 and 9, both bearing a piperazine linker, led to the highest level of cellular target engagement in their respective series. Nevertheless, partial engagement appeared insufficient to impair the MAPK signaling pathways downstream of Ras, and the p-ERK levels remained unchanged, although we noticed a reduction of p-S6 level in 7- and 9-treated cells (Supplementary Fig. 3). Questioning whether the intracellular concentrations of CypA and FKBP12 were too low in this cell line to permit efficient recruitment by the compounds, we transiently transfected H358 cells with plasmids encoding Human influenza hemagglutinin (HA)-tagged FKBP12 or CypA proteins and treated the cells with 5 or 9 (Fig. 4b). We observed similar degrees of Ras modification and no change in p-ERK levels. We next considered that actively signaling Ras proteins are membrane-anchored and that both FKBP12 and CypA are cytosolic proteins, and hence investigated whether this discrepant sub-cellular distribution limited compound efficacy. With ectopic expression of plasma-membrane localized FKBP12 or CypA (each has a 11-mer N-terminal sequence derived from Lyn to direct plasma-membrane localization[32]), we observed compound-dependent attenuation of p-ERK signal (Fig. 4b & Supplementary Figure 9), despite that the level of Ras modification was comparable to that in mock-transfected cells. Together, these results confirmed that a high level of target engagement of membrane-bound K-Ras G12C is essential to impede its signaling output through the MAPK pathway, and suggest that further optimization on these compounds is necessary to improve their cellular activity.
Figure 4.
Bifunctional K-Ras(G12C) ligands have limited efficacy against H358, a cell line harboring a G12C mutation in one KRAS allele. a) H358 cells were treated with DMSO or compounds for 24 h and analyzed by immunoblotting. b) HA-tagged FKBP12, Lyn-FKBP12, CypA, or Lyn-CypA was ectopically expressed in H358 cells by transient transfection, then the cells were treated with DMSO or indicated compounds for 24 h and analyzed by immunoblotting. The presented data is representative of three independent replicates. Annotations in FKBP12 and CypA Blots: 1 – endogenous FKBP12/CypA; 2 – FKBP12-HA/CypA-HA; 3 – Lyn-FKBP12-HA/Lyn-CypA-HA.
In order to access the GTP-bound state of Ras, we explored a different scaffold (S-IIG ligands) that has been demonstrated to bind both the GDP- and GTP-bound states of Ras harboring a non-natural mutation M72C (Fig. 1a)[16]. When covalently bound to Ras, compounds derived from this scaffold inhibit the interaction of Ras with its nucleotide exchange factor Sos and its downstream signal transducer PI3K, but the structural changes induced by compound binding were insufficient to block the binding of Raf. To enable recruitment of FKBP12 or CypA to GTP-bound Ras, we synthesized FK506-based compound 11 and cyclosporin A-based compound 12 using a similar synthetic route (Fig. 5a). While the M72C mutation was initially introduced to guide site-directed ligand discovery, it also served as a useful tool to evaluate compound binding. Analogous to S-IIP bifunctional ligands, 11 and 12 displayed FKBP- and CypA-accelerated labeling of K-Ras M72C CysLight (GDP-bound or GppNHp-bound), and the cyclosporin-derived compound 12 appeared superior to the FK506-derived 11 in its rate of covalent labeling (Fig. 5b). Ternary complexes consisting of K-Ras M72C CysLight (GDP- or GppNHp-bound), CypA and 12 were purified (Supplementary Fig. 4) and used to study whether the induced association of CypA and Ras is able to block the binding of Ras effectors such as Sos and Raf.
Figure 5.
Bifunctional Ras(M72C) ligands and their biochemical characterization. a) Chemical structures of two bifunctional Ras(M72C) ligands. b) Compounds 11 and 12 covalently react with K-Ras CysLight(M72C) (GDP-state) in an FKBP12/CypA-dependent fashion. CL, CysLight. c) The K-Ras(M72C)•12•CypA complex is resistant to Sos-catalyzed but not EDTA-mediated nucleotide exchange reaction. d) The K-Ras•12•CypA complex retains the ability to bind Raf-RBD but not full length BRAF protein. All measurements are the average of three independent replicates.
Similar to the adduct formed between K-Ras M72C and S-IIG ligand B[16], the K-Ras M72C•12•CypA complex was insensitive to Sos-catalyzed nucleotide exchange but susceptible to the EDTA-mediated reaction (Fig. 5c). To investigate the effect of the associated CypA on Raf binding, we employed a solution-based time-resolved fluorescence resonance energy transfer (TR-FRET) assay that allows the quantitation of the Ras•Raf complex formation. We compared K-Ras M72C, K-Ras M72C•B and K-Ras M72C•12•CypA in both GDP- and GppNHp-bound forms in terms of their ability to form a complex with the GST-tagged Ras-binding domain of Raf1 (Raf1-RBD) or full length Raf[33,34]. We observed a compound-dependent effect on Raf binding: compound B-labeled K-Ras showed weakened association with full length B-Raf, whereas the K-Ras•12•CypA complex completely lost this interaction (Fig. 5d). This decrease in affinity between the ternary complex and B-Raf supports our hypothesis that CypA can act as a steric blocker of Ras-effector communication.
Conclusion
Recent discoveries of direct small molecule ligands of Ras have transformed the therapeutic prospects of Ras-driven cancer [4,6,10–13,17]. Switch-II pocket and Switch-II groove binders reveal new opportunities to inhibit Ras, and the former have demonstrated efficacy in xenograft models for K-RAS(G12C)-driven cancer and are currently in clinical trials[20]. Meanwhile, three key challenges remain: 1) the ability to access the GTP-bound state of Ras is essential to inhibiting many oncogenic mutants of Ras that are constitutively GTP-bound; 2) ligands must be able to block the interaction between Ras•GTP and multiple effector proteins including Raf and PI3K to have therapeutic effect; and 3) the ability to discover non-cysteine dependent ligands to target K-Ras. Neither current S-IIP nor S-IIG ligands meet all of these criteria. We sought to draw inspiration from the unique mode of action of drugs that induce the formation of ternary protein-drug-protein complexes[35,36] and to develop small molecules that recruit intracellular proteins to block Ras-effector interactions.
Our study showed that small molecule-induced association of K-Ras and immunophilins is possible even though these proteins do not have measurable intrinsic affinity for each other. The complete blockade of Ras-Raf and Ras-Sos interaction by the combination of compound 12 and CypA illustrates the power of steric hinderance at insulating Ras from its effectors. We acknowledge that the compounds discussed in the present work do not possess the necessary efficacy for detailed cellular studies, and that the S-IIG ligands are yet dependent on their covalent reactivity with a non-natural cysteine (M72C). Further chemical optimization will be necessary to improve the affinity of current ligand scaffolds for Ras. Perhaps new ligands for Ras directed at other surface pockets and grooves when they are reported will be attractive directions.
Supplementary Material
Acknowledgements ((optional))
The authors would like to thank Dr. Roger Briesewitz for his invaluable advice and encouragement throughout all stages of the project, Dr. Ming Sun and Dr. Adam Frost for their help with structural work. ZZ is a Damon Runyon HHMI Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2281-17). KMS would like to acknowledge NIH (R01 CA 190408) and the Samuel Waxman Cancer Research Foundation.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Competing financial interests
KMS is a stockholder and consultant to Araxes Pharma, Erasca, Inc., Revolution Medicines, and a consultant to Janssen Research & Development.
References
- [1].Prior IA, Lewis PD, Mattos C, Cancer Res. 2012, 72, 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nickerson S, Joy ST, Arora PS, Bar-Sagi D, Nat. Chem. Biol. 2011, 7, 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hons M, Niebel B, Karnowski N, Weiche B, Famulok M, ChemBioChem 2012, 13, 1433–1437. [DOI] [PubMed] [Google Scholar]
- [4].Trinh TB, Upadhyaya P, Qian Z, Pei D, ACS Comb. Sci. 2016, 18, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, et al. , Nat. Chem. Biol. 2017, 13, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sogabe S, Kamada Y, Miwa M, Niida A, Sameshima T, Kamaura M, Yonemori K, Sasaki S, Sakamoto J, Sakamoto K, ACS Med. Chem. Lett. 2017, 8, 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Martín-Gago P, Fansa EK, Klein CH, Murarka S, Janning P, Schürmann M, Metz M, Ismail S, Schultz-Fademrecht C, Baumann M, et al. , Angew. Chemie - Int. Ed. 2017, 56, 2423–2428. [DOI] [PubMed] [Google Scholar]
- [8].Guillard S, Kolasinska-Zwierz P, Debreczeni J, Breed J, Zhang J, Bery N, Marwood R, Tart J, Overman R, Stocki P, et al. , Nat. Commun. 2017, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McGee JH, Shim SY, Lee SJ, Swanson PK, Jiang SY, Durney MA, Verdine GL, J. Biol. Chem. 2018, 293, 3265–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Taveras AG, Remiszewski SW, Doll RJ, Cesarz D, Huang EC, Kirschmeier P, Pramanik BN, Snow ME, Wang YS, Del Rosario JD, et al. , Bioorg. Med. Chem. 1997, 5, 125–133. [DOI] [PubMed] [Google Scholar]
- [11].Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, et al. , Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 5299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW, Angew. Chemie - Int. Ed. 2012, 51, 6140–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, et al. , Proc. Natl. Acad. Sci. 2013, 110, 8182–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM, Nature 2013, 503, 548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, Carrasco M, Hunter J, Kim ND, Xie T, et al. , Angew. Chemie - Int. Ed. 2014, 53, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gentile DR, Rathinaswamy MK, Jenkins ML, Moss SM, Siempelkamp BD, Renslo AR, Burke JE, Shokat KM, Cell Chem. Biol. 2017, 24, 1455–1466.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, Zhang Y, Sanchez-Martin M, Badgley MA, Huang CS, et al. , Cell 2017, 168, 878–889.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hansen R, Peters U, Babbar A, Chen Y, Feng J, Janes MR, Li LS, Ren P, Liu Y, Zarrinkar PP, Nat. Struct. Mol. Biol. 2018, 25, 454–462. [DOI] [PubMed] [Google Scholar]
- [19].Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler VL, Chen Y, Kucharski JM, Feng J, Ely T, et al. , Cancer Discov. 2016, 6, 316–329. [DOI] [PubMed] [Google Scholar]
- [20].Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, et al. , Cell 2018, 172, 578–589. [DOI] [PubMed] [Google Scholar]
- [21].Fakih M, O’Neil B, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Rasmussen E, Morrow PKH, Ngang J, et al. , J. Clin. Oncol. 2019, 37, 3003. [Google Scholar]
- [22].Papadopoulos KP, Ou S-HI, Johnson ML, Christensen J, Velastegui K, Potvin D, Faltaos D, Chao RC, J. Clin. Oncol. 2019, 37, TPS3161–TPS3161. [Google Scholar]
- [23].First-in-Human Study of JNJ-74699157 in Participants With Tumors Harboring the KRAS G12C Mutation, https://clinicaltrials.gov/ct2/show/NCT04006301, accessed July 27, 2019.
- [24].Brown EJ, Albers MW, Shin Bum T., Ichikawa K, Keith CT, Lane WS, Schreiber SL, Nature 1994, 369, 756–758. [DOI] [PubMed] [Google Scholar]
- [25].Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH, Cell 1994, 78, 35–43. [DOI] [PubMed] [Google Scholar]
- [26].Krönke J, Udeshi ND, Narla A, Grauman P, Hurst SN, Mcconkey M, Svinkina T, Heckl D, Comer E, Li X, et al. , Science 2014, 343, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li YJ, Wang HX, Wu HF, Science 2014, 343, 305–309.24292623 [Google Scholar]
- [28].Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, Fehrenbacher N, Linder ME, Philips MR, Mol. Cell 2011, 41, 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu J, Farmer JD, Lane WS, Friedman J, Weissman I, Schreiber SL, Cell 1991, 66, 807–815. [DOI] [PubMed] [Google Scholar]
- [30].Pua KH, Stiles DT, Sowa ME, Verdine GL, Cell Rep. 2017, 18, 432–442. [DOI] [PubMed] [Google Scholar]
- [31].Lai AC, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, Hines J, Crews CM, Angew. Chemie - Int. Ed. 2016, 55, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Inoue T, Do Heo W, Grimley JS, Wandless TJ, Meyer T, Nat. Methods 2005, 2, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bondeva T, Balla A, Várnai P, Balla T, Mol. Biol. Cell 2002, 13, 2323–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Brtva TR, Drugan JK, Ghosh S, Terrell RS, Campbell-Burk S, Bell RM, Der CJ, J. Biol. Chem. 1995, 270, 9809–9812. [DOI] [PubMed] [Google Scholar]
- [35].Corson TW, Aberle N, Crews CM, ACS Chem. Biol. 2010, 5, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schreiber SL, Isr. J. Chem. 2018, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.