Abstract
Purpose:
To investigate two deep learning-based modeling schemes for predicting short-term risk of developing breast cancer using prior normal screening digital mammograms in a case-control setting.
Methods:
We conducted a retrospective Institutional Review Board-approved study on a case-control cohort of 226 patients (including 113 women diagnosed with breast cancer and 113 controls) who underwent general population breast cancer screening. For each patient, a prior normal (i.e., with negative or benign findings) digital mammogram examination (including mediolateral oblique [MLO] view and craniocaudal [CC] view two images) was collected. Thus, a total of 452 normal images (226 MLO view images and 226 CC view images) of this case-control cohort were analyzed to predict the outcome, i.e., developing breast cancer (cancer cases) or remaining breast cancer-free (controls) within the follow-up period. We implemented an end-to-end deep learning model and a GoogLeNet-LDA model and compared their effects in several experimental settings using two mammographic view images and inputting two different sub-regions of the images to the models. The proposed models were also compared to logistic regression modeling of mammographic breast density. Area under the receiver operating characteristic curve (AUC) was used as the model performance metric.
Results:
The highest AUC was 0.73 (95% Confidence Interval [CI]: 0.68 – 0.78; GoogLeNet-LDA model on CC view) when using the whole-breast and was 0.72 (95% CI: 0.67– 0.76; GoogLeNet-LDA model on MLO+CC view) when using the dense tissue, respectively, as the model input. The GoogleNet-LDA model significantly (all p<0.05) outperformed the end-to-end GoogLeNet model in all experiments. CC view was consistently more predictive than MLO view in both deep learning models, regardless the input sub-regions. Both models exhibited superior performance than the percent breast density (AUC=0.54; 95% CI: 0.49–0.59).
Conclusions:
The proposed deep learning modeling approach can predict short-term breast cancer risk using normal screening mammogram images. Larger studies are needed to further reveal the promise of deep learning in enhancing imaging-based breast cancer risk assessment.
Keywords: breast cancer, risk biomarkers, digital mammography, deep learning, breast density
1. Introduction
Digital mammography is a routine screening examination for early detection of breast cancer. Breast density measures the amount of fibroglandular (i.e., dense) tissue imaged on a mammogram, which is mainly assessed in current clinical practice using the qualitative Breast Imaging and Reporting Data System (BI-RADS) density categories [1]. Quantitative breast density measures can also be computed using automated computer programs such as LIBRA [2], Quantra [3], Volpara [4]), etc. Both BI-RADS-based qualitative breast density categories and computer-generated quantitative density measures have been shown to be associated with breast cancer risk [5–8].
In addition to breast density, studies have shown that the mammographic imaging texture features of breast tissue are also related to breast cancer risk [9, 10]. Texture descriptors such as energy, contrast, correlation, etc. compute the local properties at each pixel and derive a set of statistics information from the local properties’ distribution. Hence, texture features capture some of the more subtle and localized micro-structural characteristics of breast tissue that may be associated with breast cancer risk, potentially by a different mechanism from a coarse measure of amount of dense parenchyma. The connection between breast density and texture is still not well understood in regard to breast cancer risk prediction.
Current breast cancer screening guidelines are mainly based on age, a major risk factor for development of breast cancer. However, risk-based screening approaches tailored with regards to an individual’s risk are increasingly being advocated. Such an approach requires accurate risk assessment, taking all known important risk factors into proper consideration. Going beyond the already-known risk markers, in-depth analysis of digital mammogram image contents for unexplored mammographic imaging features potentially associated with breast cancer risk merits further investigation, particularly through use of newly emerged deep learning techniques.
Deep learning is a subset of machine learning and a representative technique of the broader concept of artificial intelligence (AI). Lately, deep learning modeling has shown great promise in many AI applications, including biomedical imaging analysis. Deep convolutional neural networks (CNNs) have been studied for analyzing mammographic images such as breast density category classification [11–15], breast anatomy classification [16], mass detection [17–20], prediction [21], and segmentation [22], etc. [23, 24]. The unique nature of deep learning is that, massive data are fed to the CNN model which then automatically learns/extracts intrinsic imaging traits/features that are associated with the model output (i.e., outcome). This is fundamentally different from existing feature engineering mechanisms that require pre-defined imaging features/descriptors. Feature engineering for clinical tasks can be constrained because the medical domain knowledge is usually abstract, tacit, and hard to describe by exact mathematical descriptors. To date, studies have shown that feature extraction using deep learning models outperformed pre-defined imaging descriptors in many scenarios [25–29]. The purpose of this study was to investigate a deep learning-based CNN modeling approach to predict short-term risk of developing breast cancer using normal screening mammograms of a case-control study cohort.
2. Materials and Methods
2.1. Study cohort and imaging dataset
We performed a retrospective study that was compliant with the Health Insurance Portability and Accountability Act (HIPAA) and received Institutional Review Board (IRB) approval. Informed consent from patients was waived as this was a retrospective study. A case-control cohort of 226 women (1:1 case-control ratio) who underwent general population breast cancer screening in 2013 at our institution were studied. Cases were 113 women diagnosed with breast cancer (including invasive cancers, ductal carcinoma in situ [DCIS], and their mixture). The cancer cases were newly diagnosed unilateral breast cancer confirmed by pathology (interval cancers not included). Asymptomatic cancer-free controls were matched by patient age (±3 years) and year of imaging (±1 year) to the cancer cases. All studied women did not have any prior biopsy or recall on digital mammography. For each patient in the cohort, the most recent (at least 1 year earlier) normal (i.e., BI-RADS 1 or 2) mammogram examination prior to the patient outcome (i.e., cancer vs. breast cancer-free status) was retrospectively identified for analysis: for the 113 cancer cases, we used the normal images of the unilateral breast that later developed breast cancer; for the 113 controls, we used the prior normal images of the side-matched breast to the paired cancer case. Both the mediolateral oblique (MLO) and craniocaudal (CC) view on the processed (i.e., “FOR PRESENTATION”) images were analyzed. In total, we collected 226 normal mammogram examinations and used 452 mammogram images of the 226 unilateral breasts (each with MLO and CC view images). All mammogram examinations were acquired by the Hologic (Marlborough, MA) full-field digital mammography units, with two different models (i.e., Lorad Selenia and Selenia Dimensions) and similar imaging protocol parameters and automatic exposure control settings.
2.2. Deep learning modeling for predicting short-term breast cancer risk
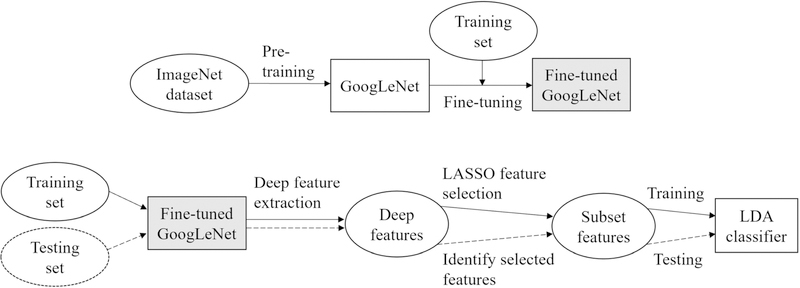
We proposed a two-class deep learning model to classify the prior normal mammogram images of the case-control cohort to predict the outcome of breast cancer vs. breast cancer-free status, which indicates a short-term (i.e., the interval between the acquisition time of normal images and the time of outcome) probability/risk of developing breast cancer. Specifically, our model was implemented in two schemes: one was an end-to-end CNN model using GoogLeNet [30] and the other was in the form of a GoogLeNet combining a linear discriminant analysis (LDA) classifier (denoted as GoogLeNet-LDA). Figure 1 illustrates the flowchart of the proposed modeling schemes.
Figure 1.
The proposed schemes for deep learning-based modeling for short-term breast cancer risk prediction. The top half is an end-to-end prediction model using fine-tuned GoogLeNet, which is also adapted as an offline deep imaging feature extractor in the GoogLeNet-LDA model (bottom half).
The end-to-end model was based on the original structure of the GoogLeNet, where the 2-way softmax function was used to provide normalized probability for binary classification between breast cancer and breast cancer-free outcome. The end-to-end deep learning prediction model does not explicitly extract imaging features for offline analysis. In contrast, the GoogLeNet-LDA model extracts deep imaging features offline using the fine-tuned GoogLeNet model. Note that in this scheme the fine-tuned GoogLeNet was adapted as a feature extractor and deep features were extracted from the layer right before the last fully-connected layer. We compared the performance of the end-to-end model to the GoogLeNet-LDA model. Through this comparison, one expects to gain insights on the effects of breast cancer risk prediction between the end-to-end modeling mechanism and the offline deep learning feature extraction mechanism.
The main component of our schemes was the GoogLeNet model, which was initialized by transfer learning of the pre-trained model on a very large imaging dataset (i.e., ImageNet [29], consisting of more than one million labeled natural images) and then fine-tuned using our own mammogram imaging data. The fine-tuned GoogLeNet directly served as the end-to-end prediction model. In the GoogLeNet-LDA scheme, we re-fed all training samples to the fine-tuned GoogLeNet model to extract 1024 deep imaging features from the layer right before the last fully-connected layer, followed by the least absolute shrinkage and selection operator (LASSO) feature selection and then the LDA classifier training. In the testing phase of the GoogLeNet-LDA model, we used the same fine-tuned GoogLeNet model to extract the 1024 deep imaging features and then identified the same subset features as previously selected in the training phase. The subset features were then fed to the LDA model for testing.
In our schemes we chose to use GoogLeNet, mainly because it has been shown effective in many applications but also has a relatively small scale of parameters (i.e., ~5 million) in comparison to other popular CNN models (for example ~60 million parameters in AlexNet [31]) to potentially reduce chances of overfitting on our dataset. Our model was implemented using MATLAB (R2018b) running on a super computer system with the following specifications [32]: 9 HPE Apollo 6500 servers, each with 8 NVIDIA Tesla V100 Graphics Processing Units (GPUs) and 16GB GPU memory, connected by NVLink 2.0. We employed the parallel GPU programming mechanism to accelerate mode training. A stochastic gradient descent with momentum (SGDM) optimizer was used to find optimal model parameters. We started with a learning rate of 0.01 and dropped the learning rate by factor 0.2 every 10 epochs. Our batch size was 30. For preprocessing, histogram equalization was run to calibrate the intensity contrast across all images, and all images were down-sampled using standard bicubic interpolation to 224×224 pixels from the original resolution in order to accommodate the needs of the pre-trained GoogLeNet model.
CNNs are known as working like black boxes and as an effort to further understand the deep learning modeling for our specific prediction task, we attempted to visualize potential imaging features/regions that are most relevant to the short-term risk prediction. To this send, we used the class activation mapping (CAM) method, where sample mammograms were forward propagated through the fine-tuned GoogLeNet model and the activated feature detectors were projected back into the original image space to visualize the most dominant imaging features/regions. We utilized the deepest convolutional layer in the fine-tuned GoogLeNet (i.e. inception_5b-pool_proj) for CAM feature map visualization.
2.3. Model evaluation and analysis plans
We used patient-wise 10-fold cross-validation to reduce chances of overfitting and to evaluate the general prediction performance of the two deep learning models. The same fold split was applied to the two models for a fair comparison. Area under the receiver operating characteristic (ROC) curve (AUC) [33] was calculated as the model performance metric. The average of the AUCs across the 10-fold validations was reported. We used the bootstrap test method to compare the difference between two AUCs (p<0.05 was considered statistically significant).
In order to evaluate the effects of the two mammographic views, i.e., CC and MLO, we performed experiments by using the CC view images only, MLO view images only, and their combination. The combination mode is implemented in a simple format of placing the two view images in two different channels of the deep learning model input. All these experiments used the same cross-validation and evaluation settings for a fair comparison of the model performance.
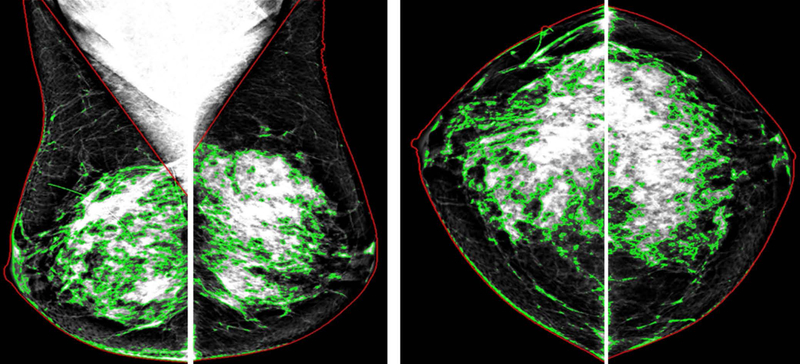
Furthermore, the deep learning models were assessed separately by two different kinds of sub-regional inputs of the mammogram images: 1) the whole-breast region and 2) the dense breast region only. The performance was compared to explore the potential differences on the effects of the imaging features identified over the whole-breast or the dense breast region only, in relation to the prediction of breast cancer risk. To do so, the whole-breast region and dense breast tissue were automatically separated from the non-breast region (i.e., air and chest muscles) and fatty tissue in each image by the previously evaluated LIBRA program [2, 46] (Figure 2).
Figure 2.
Two different kinds of sub-regional inputs for the two deep learning models: One is the whole-breast region (red contours) and the other is the dense breast tissue only (green contours). The two regions were segmented using an automated computer method.
Finally, we also compared effect of the existing imaging marker, namely, mammographic breast density. We employed automated computer methods (LIBRA [2]) to compute the area-based percentage breast density and utilized logistic regression modeling to predict the short-term risk.
3. Results
Table 1 summarizes the key characteristics of the study cohort and imaging parameters. The average age at the mammogram examination was 60.1±10.0 years old for the controls and 61.3±10.3 years old for the cancer cases. Since the acquisition time of the analyzed normal mammogram examinations, the average follow-up length for patient outcome was 1.46 (range 1–3) years for women later diagnosed with breast cancer and 1.48 (range 1–4) years for women who remained breast cancer free by the time of the study. The rate of family history of breast cancer was similar between cancer cases (35%) and controls (31%). Only a small portion of the study cohort was premenopausal (12% for cancer cases and 14% for controls).
Table 1.
Patient and imaging characteristics of the 226 patients including 113 breast cancer cases and 113 matched controls.
| Patient/Imaging characteristics | Cancer cases (N=113) n (%) |
Controls (N=113) n (%) |
|---|---|---|
| Age (years): mean±SD (range) | 61.3±10.3 (41–89) | 60.1±10.0 (41–83) |
| Follow-up length (years): mean±SD (range) | 1.46±0.63 (1–3) | 1.48±0.76 (1– 4) |
| Menopausal status | ||
| Premenopausal | 14 (12%) | 16 (14%) |
| Postmenopausal | 83 (73%) | 82 (73%) |
| Hysterectomy | 9 (8%) | 10 (9%) |
| Sterilization (bilateral oophorectomy & hysterectomy) | 4 (4%) | 4 (4%) |
| Uncertain | 3 (3%) | 1 (1%) |
| Known or test positive pathogenic BRCA1/2 mutation | 0 (0%) | 0 (0%) |
| Personal history of breast cancer | 0 (0%) | 0 (0%) |
| Family history of breast cancer | ||
| No family history | 71 (63%) | 77 (68%) |
| At least 1 1st degree relatives | 24 (21%) | 14 (12%) |
| At least 1 2nd and/or 3rd degree relatives | 16 (14%) | 21 (19%) |
| Unknown | 2 (2%) | 1 (1%) |
| Personal or family history of ovarian cancer | 1 (1%) | 3 (3%) |
| Mammographic density (visual BI-RADS density description) | ||
| Fatty | 6 (5%) | 8 (7%) |
| Scattered fibroglandular tissue | 59 (52%) | 55 (49%) |
| Heterogeneously dense | 46 (41%) | 45 (40%) |
| Extremely dense | 1 (1%) | 5 (4%) |
| Breast thickness and dose | ||
| Breast thickness (cm): mean±SD | 5.95 ± 1.34 | 5.80 ± 1.19 |
| Organ dose (mGy), CC view: mean±SD | 1.72 ± 0.52 | 1.80 ± 0.64 |
| Organ dose (mGy), MLO view: mean±SD | 1.90±0.56 | 1.94±0.59 |
| Tumor size | ||
| ≤2 cm | 69 (61.06%) | - |
| 2–5 cm | 10 (8.85%) | - |
| >5 cm | 3 (2.65%) | - |
| Unknown/missing | 31 (27.43%) | - |
| Cancer type | ||
| Invasive ductal carcinoma (IDC) | 40 (35.4%) | - |
| Invasive lobular carcinoma (ILC) | 8 (7.08%) | - |
| IDC and DCIS | 29 (25.66%) | - |
| DCIS | 36 (31.86%) | - |
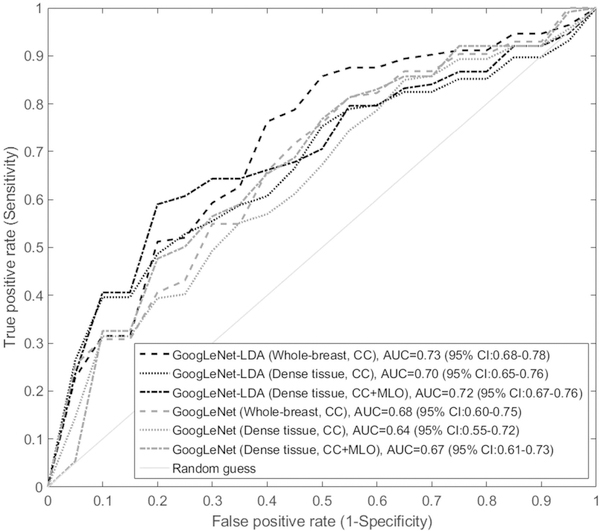
The deep learning model performance AUCs were compared in Table 2 with respect to different experimental settings. The first observation was that the GoogLeNet-LDA model was significantly superior in performance than the end-to-end GoogLeNet model (all p-values<0.05 for corresponding AUC comparisons). In terms of the two views, CC view consistently outperformed MLO view in both the two models, regardless the input sub-region was whole-breast or dense tissue only. When the two view images were combined, the model performance was improved in comparison to using either of the two views alone when the input was the dense tissue: AUC of the GoogLeNet was 0.67 (MLO+CC), significantly higher than 0.64 (CC; p=0.022) and 0.62 (MLO; p=0.002); AUC of the GoogLeNet-LDA was 0.72 (MLO+CC), significantly higher than 0.70 (CC; p=0.021) and 0.67 (MLO; p=0.002). However, the AUCs of MLO+CC view were not increased in comparison to using either of the two views alone when the input was whole-breast, regardless using the GoogLeNet or GoogLeNet-LDA model. Overall, among all the experiments, the highest AUC was 0.73 (GoogLeNet-LDA; CC view) when using the whole-breast and was 0.72 (GoogLeNet-LDA; MLO+CC view) when using the dense tissue, respectively, for predicting the short-term breast cancer risk. The difference of the two AUCs (0.73 vs 0.72) was not statistically significant (p=0.26). Figure 3 shows six selected representative ROC curves: four from all the four experiments using CC view and two from using the MLO+CC view and dense tissue as input.
Table 2.
Comparison of the deep learning model performance for predicting short-term breast cancer risk. The numbers are average AUCs of 10-fold cross-validation with 95% Confidence Interval [CI]. The highest AUC was 0.73 and 0.72 when using the whole-breast and dense tissue region, respectively, both achieved by the GoogLeNet-LDA model.
| Model | CC view | MLO view | MLO + CC view | |||
|---|---|---|---|---|---|---|
| Whole–breast | Dense tissue | Whole–breast | Dense tissue | Whole–breast | Dense tissue | |
| End-to-End GoogLeNet | 0.68(95% CI:0.60 – 0.75) | 0.64(95% CI:0.55–0.72) | 0.60(95% CI:0.55–0.64) | 0.62(95% CI:0.53–0.72) | 0.62(95% CI:0.58–0.66) | 0.67(95% CI:0.61–0.73) |
| GoogLeNet-LDA | 0.73(95% CI:0.68 – 0.78) | 0.70(95% CI:0.65–0.76) | 0.69(95% CI:0.65–0.72) | 0.67(95% CI:0.59–0.75) | 0.64(95% CI:0.58– 0.70) | 0.72(95% CI:0.67– 0.76) |
Figure 3.
Six representative ROC curves for predicting short-term breast cancer risk: four from all the four experiments using CC view and two from using the MLO+CC view and dense tissue as input.
Based on the results shown in Table 2, the AUCs of the two proposed deep learning models were consistently higher than the AUC of 0.54 (95% CI: 0.49–0.59) achieved by the area-based percentage breast density.
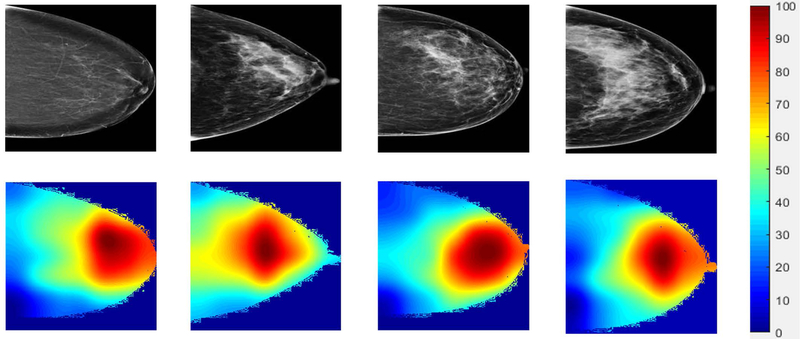
In Figure 4, we showed examples of the CAM-based feature map visualization for several sample images selected from both the case and control groups. The color bar indicates the importance level (highest 100 and lowest 0) of a specific region in the images in predicting the short-term risk. These examples implied that roughly the central image regions behind the nipple were more relevant/predictive for this specific prediction task.
Figure 4.
Feature map visualization for four selected sample images from the control (left two samples) and case (right two samples) groups. The color bar indicates the importance level (highest 100 and lowest 0) of a specific region in the images in predicting the short-term risk.
4. Discussion
In this study, we investigated a deep learning-based approach aiming to predict short-term breast cancer risk on a case-control cohort using normal mammogram images. We proposed an end-to-end deep learning model as well as a GoogLeNet+LDA model coupled with explicit deep feature extraction for offline analysis. We evaluated the effects of the two models in several experimental settings using different mammographic view images and different sub-regions as model input. Our results showed that both the two models can predict short-term risk and outperformed the existing risk factor of mammographic breast density in this specific patient cohort. To the best of our knowledge, this is the first study that investigated and compared two different modeling schemes by deep learning using prior normal screening mammographic images to predict short-term breast cancer risk in a case-control setting.
Deep learning represents a data-driven method to investigate imaging features. It automatically learns and hierarchically organizes essential traits (features), which is fundamentally different from traditional manual feature engineering [34]. Manual feature engineering is about crafting well-defined features (such as shape, contour, texture) to directly describe domain knowledge, such as imaging appearance characteristics [35, 36]. Researchers have crafted hundreds of computer features [9, 35, 37] and tested them in many applications in a trial-and-error manner to identify those that work best for specific tasks [38]. However, it is not always straightforward to translate qualitative and tactic radiological domain knowledge to exact mathematic descriptors to support manual feature engineering. In addition, it becomes even more challenging when the domain knowledge itself is poorly understood and is still under investigation to gain in-depth insights from data (like the problem of risk biomarker identification from normal images). Regarding imaging-based risk biomarker, studies have shown that breast density (both qualitative and quantitative measures) and certain texture measures are associated with breast cancer risk, but the imaging content characteristics in relation to breast cancer risk are still not fully understood, and it is likely that there are forms of potential imaging features/traits that are more relevant or essential to breast cancer risk. This represents the exact motivation of this study, i.e. we seek to implement a deep learning approach to discover potential “new” or more predictive risk markers directly from the mammographic imaging data fed to the CNN models. Our results indicate that both the two deep learning models are able to identify certain deep imaging features for improved breast cancer risk prediction when compared to the simple measure of breast density.
In terms of the two mammographic views, the CC view images showed overall superior performance than the MLO view. The mechanism behind this observation is uncertain, but some previous studies also showed that CC view images exhibited higher performance than MLO view in computer-aided diagnosis tasks [39]. When the two views were combined, one may expect to observe an increase in AUC than using either of the two views alone. Our results showed that it was indeed the case when using the dense breast tissue as model input, but not, when the input was the whole-breast region. We believe further investigation is needed in order to better assess the roles and effects of the two views in this kind of breast cancer risk prediction study.
In terms of the two different sub-regional inputs to the deep learning models, the effects varied with respect to different experimental settings (as seen in Table 2). Simply speaking, the highest AUC on the whole-breast (0.73 on CC view) was very close to the highest AUC on the dense tissue (0.72 on MLO+CC view), where the AUC difference is not statistically significant. This finding may indicate that the most predictive imaging characteristics of breast cancer risk are still within the dense/fibroglandular tissue. Nevertheless, whether a gain can be achieved by analyzing the extended region of whole breast is still worth further study by using large datasets in future work.
Generally speaking, deep learning desires a large number of training samples. Our study included 226 patients and 452 images, which was not considered at the large scale. We would like to point out that our models benefitted from the pre-training on the huge imaging dataset, i.e., ImageNet [31]. Although ImageNet is not a medical imaging dataset, recent studies have shown that pre-training on ImageNet followed by fine-tuning can substantially improve model performance in many medical image-based tasks, such as chest pathology identification [40, 41], lung disease classification [42], and colonoscopy frame classification [28]. Hence, our results may be attributable to the use of the pre-training on ImageNet and fine-tuning by our own data.
The deep imaging features could be developed to breast cancer risk biomarkers after sufficient validation. Those features were identified automatically by deep learning, without any manual feature engineering a priori. Deep learning has been viewed as working like a black-box [4], lacking interpretability of the features. In the attempts of trying to understand/interpret these deep imaging features identified by our deep learning models, we visualized the feature activation maps to highlight the regions of importance/relevance in the images in relation to the specific prediction tasks. As indicated in Figure 4, the central regions behind the nipple may contain the most predictive/relevant deep imaging features for short-term breast cancer risk prediction. While we can visualize these regions in images, it is still difficult though to provide more meaningful interpretation of these regions and deep features in an intuitive or perceptible manner to human radiologists. If we were able to do that, it would provide new insights/knowledge to enhance training/education to radiologists in mammographic image reading. At this time, deep learning feature interpretation is still an active research topic with increasing attention and efforts. We are very positive that further technical advances will contribute to resolve this key issue in future work.
The proposed deep learning modeling has a great potential to improve current breast cancer risk prediction. At the concept level, after thorough validation, the CNN models we built can be used to predict for example a short-term (e.g., 1.5 years in this study) risk for developing breast cancer, and if we were able to use even earlier (e.g., 5 years prior to outcome) normal mammogram images to build these models, it will be possible to estimate a longer (e.g., 5-year) risk. Of course, here the prediction of risk is merely based on mammographic images and this is different from existing risk models such as the Gail model [43, 44], BCSC model [45], etc. It is, however, possible to combine deep learning modeling and other known clinical/personal risk factors to build more powerful breast cancer risk models.
The strengths of our study include: 1) using two different deep learning modeling schemes and compared their effects and 2) analyzing the normal mammogram images prior to outcome in a matched case-control setting for risk prediction by deep learning. Our study has some limitations. Despite using transfer learning, the sample size is still considered relatively small and this is also a single center retrospective study. We only used mammogram images acquired from a single vendor; more extensive testing of our models on other equipment, imaging protocols, and parameter settings is needed. We plant to further evaluate our methods and findings in a larger multi-center mammogram imaging dataset. In addition, our analyses may be enhanced from a second review of the prior normal images to examine the initial assessment. Finally, as mentioned earlier, the technical immaturity of CNN feature interpretation did not enable us to well perceive the essential deep imaging features identified by our deep learning models. We understand that it would not be sufficient to gain clinical trust for a prediction model lacking meaningful feature interpretability. We position this work as a preliminary investigation to demonstrate the feasibility and potential of the proposed deep learning approach for addressing breast cancer risk prediction.
5. Conclusions
In summary, we proposed and evaluated the effects of two deep learning-based models to predict short-term breast cancer risk using prior normal digital mammogram images of a case-control cohort. Our study showed that the GoogLeNet-LDA model outperformed the end-to-end GoogLeNet model, and both the two deep learning models have superior performance than mammographic breast density. This preliminary work demonstrates the feasibility and promise of applying deep learning to enhance breast cancer risk assessment, warranting larger multi-center studies to further evaluate the models and findings.
Acknowledgments
This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) grants (#1R01CA193603, #3R01CA193603–03S1, and #1R01CA218405), a Radiological Society of North America (RSNA) Research Scholar Grant (#RSCH1530), an Amazon AWS Machine Learning Research Award, and a University of Pittsburgh Physicians (UPP) Academic Foundation Award. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges system, which is supported by NSF award number ACI-1445606, at the Pittsburgh Supercomputing Center (PSC).
References
- [1].D’orsi C, Bassett L, Feig S, Breast imaging reporting and data system (BI-RADS), Breast imaging atlas, 4th edn. American College of Radiology, Reston: (1998). [Google Scholar]
- [2].Keller BM, Nathan DL, Wang Y, Zheng Y, Gee JC, Conant EF, Kontos D, Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c‐means clustering and support vector machine segmentation, Medical physics 39(8) (2012) 4903–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ciatto S, Bernardi D, Calabrese M, Durando M, Gentilini MA, Mariscotti G, Monetti F, Moriconi E, Pesce B, Roselli A, A first evaluation of breast radiological density assessment by QUANTRA software as compared to visual classification, The Breast 21(4) (2012) 503–506. [DOI] [PubMed] [Google Scholar]
- [4].Alain G, Bengio Y, Understanding intermediate layers using linear classifier probes, arXiv preprint arXiv:1610.01644 (2016). [Google Scholar]
- [5].Boyd NF, Guo H, Martin LJ, Sun L, Stone J, Fishell E, Jong RA, Hislop G, Chiarelli A, Minkin S, Mammographic density and the risk and detection of breast cancer, New England Journal of Medicine 356(3) (2007) 227–236. [DOI] [PubMed] [Google Scholar]
- [6].McCormack VA, dos Santos Silva I, Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis, Cancer Epidemiology and Prevention Biomarkers 15(6) (2006) 1159–1169. [DOI] [PubMed] [Google Scholar]
- [7].Destounis S, Arieno A, Morgan R, Roberts C, Chan A, Qualitative versus quantitative mammographic breast density assessment: applications for the US and Abroad, Diagnostics 7(2) (2017) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Engmann NJ, Golmakani MK, Miglioretti DL, Sprague BL, Kerlikowske K, Population-attributable risk proportion of clinical risk factors for breast cancer, JAMA oncology 3(9) (2017) 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gastounioti A, Conant EF, Kontos D, Beyond breast density: a review on the advancing role of parenchymal texture analysis in breast cancer risk assessment, Breast Cancer Research 18(1) (2016) 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Keller BM, Chen J, Conant EF, Kontos D, Breast density and parenchymal texture measures as potential risk factors for estrogen-receptor positive breast cancer, Medical Imaging 2014: Computer-Aided Diagnosis, International Society for Optics and Photonics, 2014, p. 90351D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mohamed AA, Luo Y, Peng H, Jankowitz RC, Wu S, Understanding clinical mammographic breast density assessment: a deep learning perspective, Journal of digital imaging 31(4) (2018) 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mohamed AA, Berg WA, Peng H, Luo Y, Jankowitz RC, Wu S, A deep learning method for classifying mammographic breast density categories, Medical physics 45(1) (2018) 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Arefan D, Talebpour A, Ahmadinejhad N, Asl AK, Automatic breast density classification using neural network, Journal of Instrumentation 10(12) (2015) T12002. [Google Scholar]
- [14].Becker AS, Marcon M, Ghafoor S, Wurnig MC, Frauenfelder T, Boss A, Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer, Investigative radiology 52(7) (2017) 434–440. [DOI] [PubMed] [Google Scholar]
- [15].Zhang X, Zhang Y, Han EY, Jacobs N, Han Q, Wang X, Liu J, Classification of whole mammogram and tomosynthesis images using deep convolutional neural networks, IEEE transactions on nanobioscience 17(3) (2018) 237–242. [DOI] [PubMed] [Google Scholar]
- [16].Cheng J-Z, Ni D, Chou Y-H, Qin J, Tiu C-M, Chang Y-C, Huang C-S, Shen D, Chen C-M, Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans, Scientific reports 6 (2016) 24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang J, Yang X, Cai H, Tan W, Jin C, Li L, Discrimination of breast cancer with microcalcifications on mammography by deep learning, Scientific reports 6 (2016) 27327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gao F, Wu T, Li J, Zheng B, Ruan L, Shang D, Patel B, SD-CNN: A shallow-deep CNN for improved breast cancer diagnosis, Computerized Medical Imaging and Graphics 70 (2018) 53–62. [DOI] [PubMed] [Google Scholar]
- [19].Mendel K, Li H, Sheth D, Giger M, Transfer learning from convolutional neural networks for computer-aided diagnosis: a comparison of digital breast tomosynthesis and full-field digital mammography, Academic radiology (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burt JR, Torosdagli N, Khosravan N, RaviPrakash H, Mortazi A, Tissavirasingham F, Hussein S, Bagci U, Deep learning beyond cats and dogs: recent advances in diagnosing breast cancer with deep neural networks, The British journal of radiology 91(1089) (2018) 20170545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Arefan D, Zheng B, Dabbs D, Bhargava R, Wu S, Multi-space-enabled deep learning of breast tumors improves prediction of distant recurrence risk, Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications, International Society for Optics and Photonics, 2019, p. 109540L. [Google Scholar]
- [22].Zhang L, Luo Z, Chai R, Arefan D, Sumkin J, Wu S, Deep-learning method for tumor segmentation in breast DCE-MRI, Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications, International Society for Optics and Photonics, 2019, p. 109540F. [Google Scholar]
- [23].Hamidinekoo A, Denton E, Rampun A, Honnor K, Zwiggelaar R, Deep learning in mammography and breast histology, an overview and future trends, Medical image analysis 47 (2018) 45–67. [DOI] [PubMed] [Google Scholar]
- [24].Aboutalib SS, Mohamed AA, Berg WA, Zuley ML, Sumkin JH, Wu S, Deep Learning to Distinguish Recalled but Benign Mammography Images in Breast Cancer Screening, Clinical Cancer Research 24(23) (2018) 5902–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI, A survey on deep learning in medical image analysis, Medical image analysis 42 (2017) 60–88. [DOI] [PubMed] [Google Scholar]
- [26].Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, Kim L, Summers RM, Improving computer-aided detection using convolutional neural networks and random view aggregation, IEEE transactions on medical imaging 35(5) (2016) 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D, Mok VC, Shi L, Heng P-A, Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks, IEEE transactions on medical imaging 35(5) (2016) 1182–1195. [DOI] [PubMed] [Google Scholar]
- [28].Sirinukunwattana K,Raza S.e.A,Tsang Y-W, Snead DR, Cree IA, Rajpoot NM, Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images, IEEE Trans. Med. Imaging 35(5) (2016) 1196–1206. [DOI] [PubMed] [Google Scholar]
- [29].Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L, Imagenet: A large-scale hierarchical image database, 2009 IEEE conference on computer vision and pattern recognition, Ieee, 2009, pp. 248–255. [Google Scholar]
- [30].Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A, Going deeper with convolutions, Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 1–9. [Google Scholar]
- [31].Krizhevsky A, Sutskever I, Hinton GE, Imagenet classification with deep convolutional neural networks, Advances in neural information processing systems, 2012, pp. 1097–1105. [Google Scholar]
- [32].Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, XSEDE: accelerating scientific discovery, Computing in Science & Engineering 16(5) (2014) 62–74. [Google Scholar]
- [33].Metz CE, ROC methodology in radiologic imaging, Investigative radiology 21(9) (1986) 720–733. [DOI] [PubMed] [Google Scholar]
- [34].Suk H-I, Lee S-W, Shen D, A.s.D.N. Initiative, Latent feature representation with stacked auto-encoder for AD/MCI diagnosis, Brain Structure and Function 220(2) (2015) 841–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Moura DC, López MAG, An evaluation of image descriptors combined with clinical data for breast cancer diagnosis, International journal of computer assisted radiology and surgery 8(4) (2013) 561–574. [DOI] [PubMed] [Google Scholar]
- [36].Pérez NP, López MAG, Silva A, Ramos I, Improving the Mann–Whitney statistical test for feature selection: An approach in breast cancer diagnosis on mammography, Artificial intelligence in medicine 63(1) (2015) 19–31. [DOI] [PubMed] [Google Scholar]
- [37].Tan M, Qian W, Pu J, Liu H, Zheng B, A new approach to develop computer-aided detection schemes of digital mammograms, Physics in Medicine & Biology 60(11) (2015) 4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shao Y-Z, Liu L-Z, Bie M-J, Li C.-c., Wu Y.-p., Xie X.-m., Li L , Characterizing the clustered microcalcifications on mammograms to predict the pathological classification and grading: A mathematical modeling approach, Journal of digital imaging 24(5) (2011) 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Huo Z, Giger ML, Vyborny CJ, Computerized analysis of multiple-mammographic views: Potential usefulness of special view mammograms in computer-aided diagnosis, IEEE Transactions on Medical Imaging 20(12) (2001) 1285–1292. [DOI] [PubMed] [Google Scholar]
- [40].Bar Y, Diamant I, Wolf L, Lieberman S, Konen E, Greenspan H, Chest pathology detection using deep learning with non-medical training, 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), IEEE, 2015, pp. 294–297. [Google Scholar]
- [41].Bar Y, Diamant I, Wolf L, Greenspan H, Deep learning with non-medical training used for chest pathology identification, Medical Imaging 2015: Computer-Aided Diagnosis, International Society for Optics and Photonics, 2015, p. 94140V. [Google Scholar]
- [42].Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM, Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning, IEEE transactions on medical imaging 35(5) (2016) 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, Mulvihill JJ, Projecting individualized probabilities of developing breast cancer for white females who are being examined annually, JNCI: Journal of the National Cancer Institute 81(24) (1989) 1879–1886. [DOI] [PubMed] [Google Scholar]
- [44].Costantino JP, Gail MH, Pee D, Anderson S, Redmond CK, Benichou J, Wieand HS, Validation studies for models projecting the risk of invasive and total breast cancer incidence, Journal of the National Cancer Institute 91(18) (1999) 1541–1548. [DOI] [PubMed] [Google Scholar]
- [45].Ballard-Barbash R, Taplin SH, Yankaskas BC, Ernster VL, Rosenberg RD, Carney PA, Barlow WE, Geller BM, Kerlikowske K, Edwards BK, Breast Cancer Surveillance Consortium: a national mammography screening and outcomes database, AJR. American journal of roentgenology 169(4) (1997) 1001–1008. [DOI] [PubMed] [Google Scholar]
- [46].Keller Brad M., Chen Jinbo, Daye Dania, Conant Emily F., and Kontos Despina, Preliminary evaluation of the publicly available Laboratory for Breast Radiodensity Assessment (LIBRA) software tool: comparison of fully automated area and volumetric density measures in a case–control study with digital mammography, Breast Cancer Research 17(1) (2015), p. 117. [DOI] [PMC free article] [PubMed] [Google Scholar]