Abstract
Effective doctor-patient communication is critical for disease management, especially when considering genetic information. We studied patient-provider communications after implementing a point-of-care pharmacogenomic results delivery system to understand whether pharmacogenomic results are discussed and whether medication recall is impacted. Outpatients undergoing preemptive pharmacogenomic testing (cases), non-genotyped controls, and study providers were surveyed from October 2012-May 2017. Patient responses were compared between visits where pharmacogenomic results guided prescribing versus visits where pharmacogenomics did not guide prescribing. Provider knowledge of pharmacogenomics, before and during study participation, was also analyzed. Both providers and case patients frequently reported discussions of genetic results after visits where pharmacogenomic information guided prescribing. Importantly, medication changes from visits where pharmacogenomics influenced prescribing were more often recalled than non-pharmacogenomic guided medication changes (OR=3.3 [1.6–6.7], p=0.001). Case patients who had separate visits where pharmacogenomics did and did not respectively influence prescribing more often remembered medication changes from visits where genomic-based guidance was used (OR=3.4 [1.2–9.3], p=0.02). Providers also displayed dramatic increases in personal genomic understanding through program participation (94% felt at least somewhat informed about pharmacogenomics post-participation, compared to 61% at baseline, p=0.04). Using genomic information during prescribing increases patient-provider communications, patient medication recall, and provider understanding of genomics, important ancillary benefits to clinical use of pharmacogenomics.
INTRODUCTION
Effective doctor-patient communication is critical for medical management. Successful communication between healthcare providers and patients may facilitate comprehension of information, improve patient satisfaction, and increase likelihood that patients will follow medical advice1–5. Poor doctor-patient communication, however, can be perceived by patients even when providers report adequate communication5–8. Suboptimal discussions surrounding prescription medications are often cited as an area of concern, with studies suggesting that providers should explain prescribing rationales9–11.
Pharmacogenomics, the study of how genes impact drug response, is a key component of precision medicine. As pharmacogenomic results become increasingly available at the point-of-care, it is important to assess patient-provider communications and implications of results delivery, as these considerations will affect widespread adoption12–14. A limited amount of research has suggested that pharmacogenomic results may positively influence doctor-patient communications15, 16. Feelings of shared decision-making between patient and provider in the context of pharmacogenomic results may also positively impact prescribing communications15. Patients may obtain a better understanding of prescribing rationale, potentially even impacting medication adherence17–20.
To our knowledge, no study, to date, has directly assessed patient-provider communications surrounding prescribing when pharmacogenomic results were available. To understand details of patient-provider prescribing communications in the context of pharmacogenomic results availability, we utilized our institutional pharmacogenomics implementation program, for which we previously reported successful and robust adoption of pharmacogenomic results among providers and a positive impact on prescribing in a pattern aimed at reducing patient risk21. We hypothesized that such availability of pharmacogenomic results would facilitate memorable discussions surrounding medication decisions, positively impacting patient medication recommendation recall.
SUBJECTS AND METHODS
Study Design
Preemptive genotyping was offered to case patients through the 1,200 Patients Project, with return of results to enrolled study providers at the point-of-care using our pharmacogenomic decision-support software, the Genomic Prescribing System (GPS)22–25. Non-genotyped controls were recruited from the same clinics. Enrollment continued for the overall 1,200 Patients Project cohort until each provider’s roster was saturated, or until approximately 70–100 case patients were genotyped and approximately 70–100 controls were recruited from each participating provider’s clinic. This sample size was justified based on power estimations for the primary endpoint (previously published)21. As previously described22, cases and controls were subject to the same eligibility criteria. Our prior analyses have shown the cases and controls of the overall cohort to be similar, as no differences were observed between the groups regarding gender, age, and race12. Patients were included in the present study if a survey had been returned for a clinic visit where a medication change took place. All medication change visits were systematically evaluated, including detailed review of electronic medical record decision-making documentation. A formal evaluation process (previously described12) was applied to determine whether pharmacogenomic information influenced each medication change (see details in Supplementary Methods). Overall, survey responses of 245 case patients, 72 control patients, and 18 providers were analyzed in exploratory fashion. The study was approved by the University of Chicago Institutional Review Board and was registered at clinicaltrials.gov (#NCT01280825), and all participants signed written informed consent.
Survey Instruments
To assess communication and understand decision-making, we utilized regularly-administered surveys of patients and providers. Specific patient survey items surrounding medication change recall and discussions of pharmacogenomic results were studied. For each patient, case or control, who was determined to have at least one medication change at a clinic visit, a survey was distributed to the patient either in clinic after the visit, or by mail or email. The same survey was distributed to cases and controls. The provider repeated interval survey assessed provider attitudes toward, knowledge of, and overall use of pharmacogenomics and was distributed to all providers once before study participation and approximately every 3 months during the study period. The provider experience survey studied reasons why pharmacogenomic test results did or did not guide prescribing at individual visits. Additional details, along with the survey instruments, are available in the Supplementary Methods.
Analyses
Demographic statistics were compared between case and control patients using chi-square tests for categorical variables and t-tests for continuous variables. For the primary patient survey analysis, we sought to determine whether survey responses differed between cases and controls. To learn whether providers discussed individual prescribing factors, genetics/DNA, specific genetic test results, and a better prescribing decision due to genetic test results with case patients more often than controls, and whether medication change recall differed between the two groups, linear mixed-effects models were used that took into account two random effects: one for repeated surveys within a patient and another for intra-class correlations within providers. Results were reported as Odds Ratios (OR) [95% Confidence Interval (CI)], with p-values <0.05 considered statistically significant.
For our secondary patient survey analysis, we aimed to study differences between visits where pharmacogenomic information guided prescribing and visits where such information did not influence medication changes. We first used linear mixed-effects models (described above) to assess whether there were differences between case patient visits with only non-pharmacogenomically-influenced medication changes and control visits. These two groups were then combined to form our “traditional medication change visits” group for subsequent analyses. Traditional medication change visits were then compared to all visits with a pharmacogenomically-influenced medication change.
To attempt to control for individual differences that may influence medication change recall, additional patient survey analyses were conducted. Specifically, we aimed to elucidate whether the use of pharmacogenomic information impacted medication change recall independent of any patient-specific, provider-specific, or visit-specific factors. To do so, we studied case patients who had separate pharmacogenomically-influenced medication change and traditional medication change visits. We then compared medication change recall between the two types of visits using a linear mixed-effects model. We also looked at single case patient visits where both a pharmacogenomically-influenced medication change and a traditional medication change took place and assessed medication recall rate for both types of medication changes. Finally, we studied whether or not there was a difference in recall of pharmacogenomically-influenced medication changes based on level of pharmacogenomic risk using Fisher’s exact test.
For the repeated interval survey, responses from each provider’s baseline questionnaire were compared to responses from each provider’s most recently returned survey to directly assess change in knowledge/attitudes/opinions within each provider. P-values were calculated using McNemar’s test, and p<0.05 was considered significant. For some questions on the experience survey, more than one answer could be selected. In these instances, the results were calculated based on how often each response was selected as a percentage of the surveys on which the question was answered.
RESULTS
Participant Demographics
All eighteen invited providers agreed to study participation and represented primary care and subspecialty outpatient clinics. Details of these providers’ practices have been previously published21 (one gastroenterology provider has since joined). Over half of participating providers were male (61%), and the average years in practice at baseline was 20 (range: 3–46). Study providers, on average, saw approximately 9 patients per half-day clinic session (range: 6–16).
Patients were recruited from these providers’ practices. Because enrollment into the genotyped (case) versus non-genotyped (control) cohort was non-randomized, demographics of case and control patients were compared (Table 1). The only studied characteristic that differed between the groups was the average number of evaluable visits (2.8 visits for cases and 2.0 visits for controls [p=0.0001], likely because control enrollment began 1.5 years after case enrollment).
Table 1.
Patient Demographics
| Case Patients | Control Patients | Total Study Population | P-value | |
|---|---|---|---|---|
| Total | 245 | 72 | 317 | |
| Gender | 0.97 | |||
| Male | 123 (50.2) | 37 (51.4) | 160 (50.5) | |
| Age at first study visit | 0.83 | |||
| Mean (SD) | 62.9 (14.6) | 63.3 (13.8) | 63.0 (14.4) | |
| Race/ethnicity | 0.29 | |||
| White | 164 (66.9) | 41 (56.9) | 205 (64.7) | |
| Black | 62 (25.3) | 23 (31.9) | 85 (26.8) | |
| Other | 19 (7.8) | 8 (11.1) | 27 (8.5) | |
| Educational attainment | 0.19 | |||
| <High school or unknown | 8 (3.3) | 6 (8.3) | 14 (4.4) | |
| High school/GED | 39 (15.9) | 6 (8.3) | 45 (14.2) | |
| Some college | 48 (19.6) | 17 (23.6) | 65 (20.5) | |
| College graduate | 64 (26.1) | 20 (27.8) | 84 (26.5) | |
| Graduate school | 86 (35.1) | 23 (31.9) | 109 (34.4) | |
| Number of medications1,mean (SD) | 4.1 (2.3) | 4.6 (2.3) | 4.2 (2.3) | 0.11 |
| Number of medications with known | 1.9 (1.4) | 2.2 (1.4) | 2.0 (1.4) | 0.11 |
| PGx information1, mean (SD) | ||||
| Dates of enrollment | 2/2011–11/2015 | 6/2012–11/2016 | ||
| Number of clinics represented | 18 | 13 | ||
| Average number of evaluable visits during study during study period, mean (SD) | 2.8 (2.4) | 2.0 (1.2) | 2.6 (2.2) | 0.0001 |
| Surveys returned/delivered | 415/781 (53.1) | 95/204 (46.6) | 510/985 (51.8) | 0.38 |
| Median surveys returned per patient (range) | 1 (1–13) | 1 (1–3) | 1 (1–13) | |
| Average number of medication changes per visit (range) | 1.46 (1–5) | 1.37 (1–4) | 1.44 (1–5) | 0.59 |
| Type of medication change | 0.64 | |||
| New medication | 297 (48.9) | 67 (50.0) | 364 (48.9) | |
| Discontinuation | 164 (26.9) | 31 (23.1) | 195 (26.2) | |
| Dose change | 149 (24.4) | 36 (26.9) | 185 (24.9) | |
| Top Medications Changed2 | ||||
| 1. | Hydrochlorothiazide 33 (5.4) | Amlodipine 9 (6.7) | ||
| 2. | Atorvastatin 28 (4.6) | Atorvastatin 9 (6.7) | ||
| 3. | Amlodipine 23 (3.8) | Lisinopril 7 (5.2) | ||
| 4. | Lisinopril 23 (3.8) | Metoprolol 6 (4.5) | ||
| 5. | Omeprazole 19 (3.1) | Omeprazole 5 (3.7) |
Values are represented as no. (%) unless otherwise noted
SD, standard deviation
PGx, pharmacogenomics
at baseline
regardless of pharmacogenomic results availability
On average, case patients had 1.46 medication changes per visit (range: 1–5), while controls had 1.37 (range: 1–4) (p=0.59). The medication change types (new medication, discontinuation, and dose change) were similar between cases and controls. Of the studied case patient visits, 84 (20%) were determined to have ≥1 pharmacogenomically-influenced medication change. The remaining 331 visits (80%) had ≥1 non-pharmacogenomically-influenced, or traditional, medication change.
Communication of Pharmacogenomic Results
Thirteen providers (72%) reported having discussed some genetic result with patients in the six months preceding study participation (Supplementary Table 1). After study initiation, however, providers reported doing so on 93% of surveys, and 100% of providers stated this in their last completed survey (before analysis). This perhaps suggests continually increasing communication with patients about pharmacogenomics during the study.
Case patients also confirmed frequent discussions of genetics (Table 2). Case patients reported a discussion of a specific genetic test result surrounding a medication change at 24% of visits, while no controls reported this over the study period.
Table 2.
Patient-Reported Discussions Surrounding Medication Changes: Case vs. Control
| Survey Question | Case | Control | Odds Ratio (95% CI, p-value) |
|---|---|---|---|
| Did your healthcare provider stop or change one of your medications today or start a new medication? | 280 (69.7) | 66 (71.0) | 1.0 (0.6–1.8, p=0.97) |
| Did your healthcare provider discuss specific factors about you or your personal make-up which would suggest that you were more likely or less likely than other patients to benefit from the medication change or new medication? | 147 (61.5) | 36 (66.7) | 0.7 (0.3–1.8, p=0.51) |
| Did your healthcare provider discuss your genetics or your DNA when talking about the medication change or new medication? | 67 (28.2) | 5 (9.3) | 5.6 (1.7–18.2, p=0.004) |
| Did your healthcare provider discuss a specific genetic test result for you when talking about the medication change or new medication? | 57 (24.1) | 0 (0.0) | p<0.051 |
| Did your healthcare provider say that a specific genetic test result for you helped him or her make a better prescribing decision regarding your medication change or new medication? | 60 (25.5) | 0 (0.0) | p<0.051 |
Values are represented as number of patients who responded “yes” (%)
CI = confidence interval
Statistical modeling was non-estimable due to zero “yes” responses from those in the control group.
Patients with Pharmacogenomically-Influenced Medication Changes Report Discussions Surrounding Pharmacogenomics
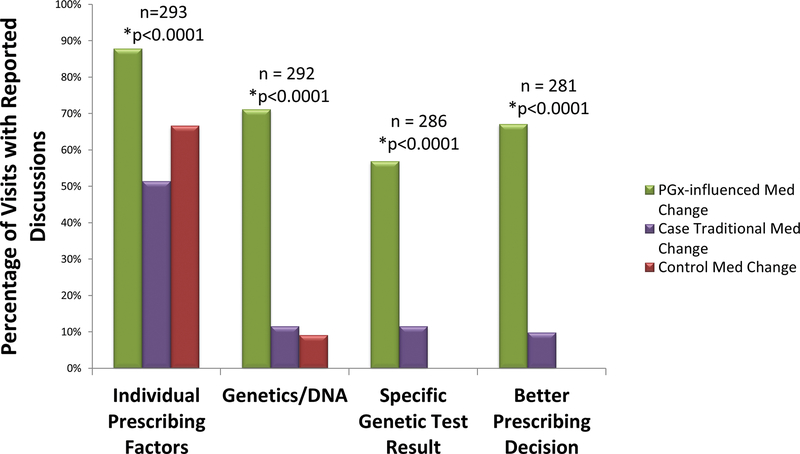
We then specifically sought to assess whether there were differences between visits where patients had medication changes that were influenced by pharmacogenomic information and visits with only non-pharmacogenomically-influenced medication changes. We observed that details of case visits with only non-pharmacogenomically-influenced medication changes were very similar to those of controls (Figure 1). Statistical analysis showed no significant differences between survey responses from these two groups. Hence, case patients with only non-pharmacogenomically-influenced medication changes and control patients were combined to form our “traditional medication change” group in subsequent analyses, for which results are displayed in Supplementary Table 2. Most saliently, patients reported that their provider discussed individual prescribing factors with them at 88% of visits with a pharmacogenomically-influenced medication change, compared to only 55% of traditional medication change visits (OR=10.1 [3.3–31.0], p<0.0001). Genetics or DNA was discussed at 71% of pharmacogenomically-influenced medication change visits compared to only 11% of traditional medication change visits (OR=41.5 [9.5–182.8], p<0.0001).
Figure 1. Patient-Reported Discussions Surrounding Medication Changes: Pharmacogenomically-Influenced Medication Change Visits vs. Traditional Medication Change Visits.
Percentage of medication change visits where patients reported discussions of specific topics, including “individual prescribing factors”, “genetics/DNA”, “specific genetic test results”, and/or a “better prescribing decision” due to use of a specific genetic test result. For statistical comparison, case traditional medication change visits were combined with control medication change visits as there were no statistically significant differences between the two groups. This group was then compared to pharmacogenomically-influenced medication change visits. The “n” value represents the total number of visits analyzed for each question. Patients who had pharmacogenomically-influenced medication changes significantly more often reported discussions of “individual prescribing factors” OR=10.1 [3.3–31.0], p<0.0001; “genetics/DNA” OR=41.5 [9.5–182.8], p<0.0001; “specific genetic test result” OR=30.3 [6.2–148.4], p<0.0001; and “better prescribing decision” OR=27.4 [12.4–60.8], p<0.0001 compared to all non-pharmacogenomically-influenced medication changes (case traditional + control).
Pharmacogenomically-Influenced Medication Changes were More Often Recalled than Non-Pharmacogenomically-Influenced Medication Changes
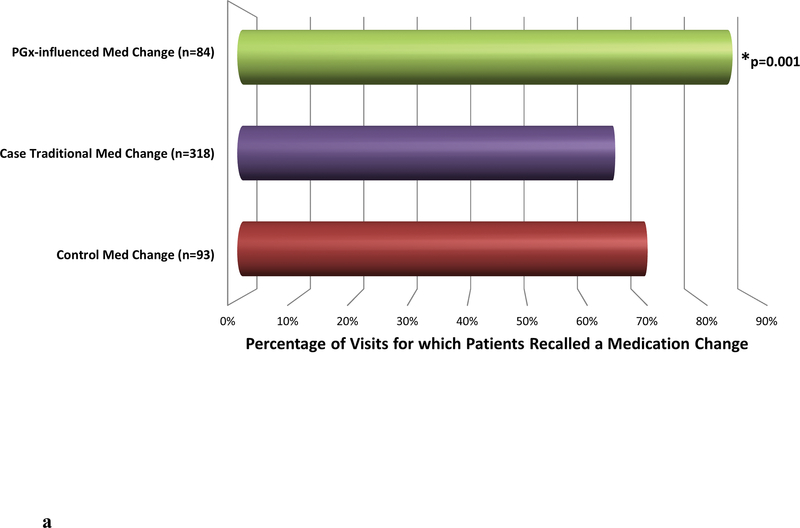
Despite the fact that every patient who received a survey was objectively determined to have had a medication change at the clinic visit in question, not all patients recalled these medication changes. Overall, medication changes were recalled by patients (reported in the surveys) at 70% of the analyzed visits (no significant difference between cases and controls). Yet notably, in the group where pharmacogenomics influenced the medication change, ≥1 medication change was recalled from 86% of visits. On the contrary, medication changes were only recalled at 67% of traditional medication change visits (OR=3.3 [1.6–6.7], p=0.001) (Figure 2a).
Figure 2. Patient Recall of Medication Changes.
a. The graph displays the percentage of visits where at least one medication change was recalled by the patient. Case patient visits with only non-pharmacogenomically-influenced (traditional) medication changes were combined with control medication change visits for statistical analysis. This combined group was then compared to all visits with at least one pharmacogenomically-influenced medication change. The “n” value represents the number of visits analyzed in each group. Pharmacogenomically-influenced medication changes were more often recalled compared to all non-pharmacogenomically-influenced medication changes (case traditional and control medication changes) (OR=3.3 [1.6–6.7], p=0.001).
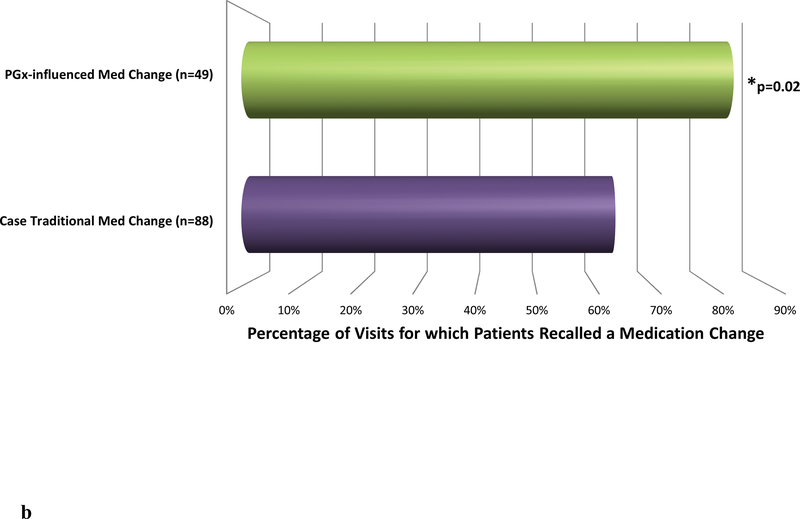
b. A subset of case patients in the study had at least one pharmacogenomically-influenced medication change visit and at least one separate non-pharmacogenomically-influenced (traditional) medication change visit. Medication change recall was examined for these patients’ visits. The “n” value represents the number of visits analyzed in each group. Pharmacogenomically-influenced medication changes were significantly more often recalled compared to non-pharmacogenomically-influenced medication changes (OR=3.4 [1.2–9.3], p=0.02).
To further elucidate whether pharmacogenomics is a key driver of patients recalling medication changes, we next examined case patients who had both a pharmacogenomically-influenced medication change visit and a separate traditional medication change visit. This type of comparison aimed to eliminate possible confounding patient factors—specific to the group of patients who had medication changes influenced by pharmacogenomics—that may have been present when comparing all pharmacogenomically-influenced medication change visits to all traditional medication change visits. There were 42 patients who had both types of visits, comprising 137 visits analyzed. Medication changes were recalled at 84% of pharmacogenomically-influenced medication change visits and only 64% of traditional medication change visits (OR=3.4 [1.2–9.3], p=0.02) (Figure 2b).
Moreover, we investigated single visits where case patients had both ≥1 pharmacogenomically-influenced medication change and ≥1 traditional medication change. This within-case, within-visit analysis aimed to account for the possibility that there could have been specific patient or provider factors that were responsible for the above differences exclusive of the presence or absence of pharmacogenomic information. The number of analyzed visits was 24. The total number of pharmacogenomically-influenced medication changes from these visits was 30, while the total number of traditional medication changes was 26. Importantly, and despite the small number of visits, medication changes were significantly more frequently remembered by patients when providers utilized pharmacogenomic information. Specifically, 67% of pharmacogenomically-influenced medication changes were recalled compared to only 39% of traditional medication changes (OR=3.5 [1.0–11.6], p=0.04). Finally, we assessed whether the degree of pharmacogenomic risk impacted the likelihood of medication change recall. We found that the type of pharmacogenomic information displayed in GPS did not matter – 89%, 91%, and 88% of medication changes for favorable, cautionary, and high-risk pharmacogenomic medications, respectively, were recalled (p=1.0).
Provider-Reported Utility of Pharmacogenomic Results
Prior to pharmacogenomic results availability, two-thirds of providers stated that pharmacogenomic results had never changed their prescribing in the six months before study participation. After study initiation, however, 94% of providers reported that pharmacogenomics had changed their prescribing (p=0.04) (Supplementary Table 1). At individual clinic visits, providers also reported (on the experience survey) how pharmacogenomic information informed prescribing. Providers most frequently stated that pharmacogenomic information helped them make a more informed therapeutic decision (86% of instances where pharmacogenomic results guided prescribing) and simultaneously reported that the given pharmacogenomic results increased the likelihood that their patient would respond favorably to treatment (68% of instances) and helped choose a therapy from multiple options (50%) (Figure 3).
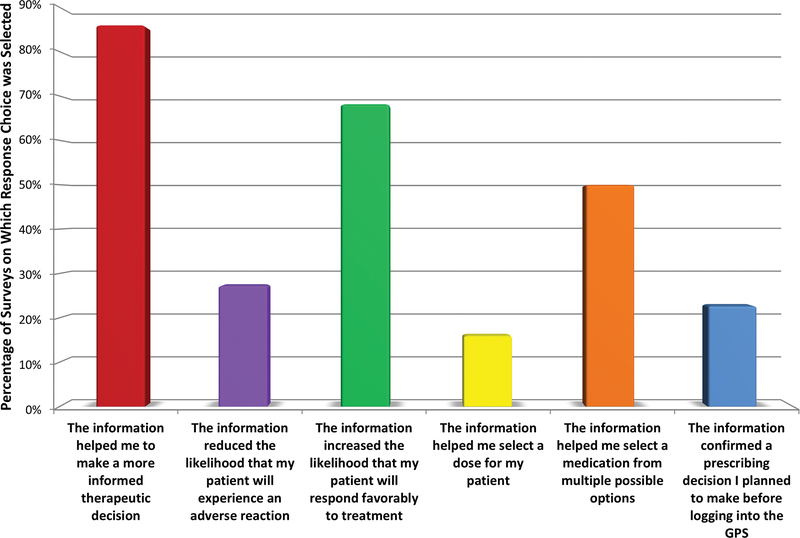
Figure 3. Provider Reported Utility of Pharmacogenomic Information to Guide Prescribing.
At visits where pharmacogenomic information influenced prescribing (as determined by independent assessment12, n=57 clinic visits), providers most frequently stated, on the provider experience survey, that pharmacogenomic information helped them to make a more informed therapeutic decision (cited for 86.0% of instances where a pharmacogenomic result guided prescribing), yet they simultaneously also reported that the given pharmacogenomic results increased the likelihood that their patient would respond favorably to treatment (68.2% of instances), helped choose a therapy from multiple options (50.0%), reduced the likelihood that their patient will experience an adverse reaction (27.3%), reinforced an originally intended prescribing decision (22.7%), and helped select a specific dose (15.9%). Providers could choose more than one response for each visit. Bars represent the number of times the response was chosen as a percentage of the total number of surveys on which the question was answered.
*Out of 761 total experience surveys sent for visits at which pharmacogenomic results were accessed, 395 (51.9%) were returned. Seventeen of the 18 study providers returned ≥1 experience survey (median surveys returned/provider: 15, range 0–77).
Provider Perceived Knowledge of Pharmacogenomics Changes Over Time
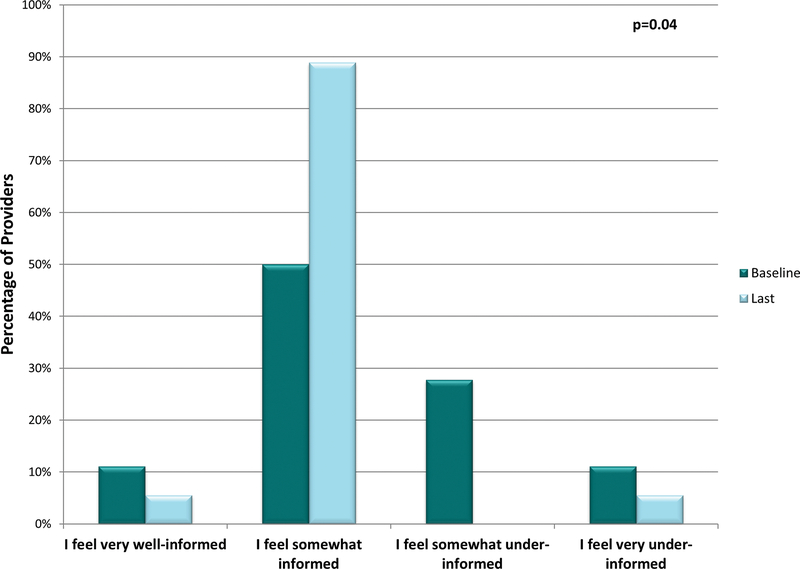
As we discovered that pharmacogenomic result availability was associated with an unexpected benefit to patients (increased medication change recall), we sought to assess personal benefits that implementation may have for providers. Before study participation, only 61% of providers reported feeling informed about pharmacogenomics. When asked post-study participation, 94% of providers felt at least somewhat informed (p=0.04) (Figure 4). This perhaps suggests providers learn about pharmacogenomics simply by being provided patient-specific results and utilizing pharmacogenomics in their clinics. Notably, and likely explaining this finding, providers often stated that their primary source of pharmacogenomic educational information was the provided study materials (47% of the time).
Figure 4. Provider Change in Self-Reported Knowledge of Pharmacogenomics During Implementation.
Two provider repeated interval surveys from each of the study providers were included in this analysis—the baseline survey (prior to availability of the Genomic Prescribing System [GPS] for clinical use) and the last completed survey (after each provider had access to GPS for at least 6 months). At baseline, 61.1% of providers reported feeling at least somewhat informed about pharmacogenomics. When asked the same question post-study participation, 94.4% of providers felt at least somewhat informed (p=0.04). For statistical purposes, “very well-informed” and “somewhat informed” were combined and compared to the combined “somewhat under-informed” and “very under-informed” using McNemar’s test.
*Each provider completed a baseline repeated interval survey and ≥1 post-GPS implementation repeated interval survey. Out of 106 total surveys distributed, 76 (71.7%) were returned (median surveys returned/provider: 4, range 2–8).
DISCUSSION
In this study, we found that when pharmacogenomic results were available via our implementation program, providers reported having frequent discussions of pharmacogenomic test results with patients, which patients almost always recalled. Importantly, communication surrounding pharmacogenomically-informed prescribing remained robust throughout the nearly 5 years of study follow-up, with both patients and providers reporting recurring discussions about pharmacogenomics. In fact, providers increasingly reported discussions as the study progressed, arguing against the idea that initial novelty of pharmacogenomic results was the cause. Moreover, when pharmacogenomic results guided a provider’s prescribing decision, patients were much more likely to recall those medication changes than medication changes at visits where decisions were not guided by pharmacogenomic information, and patients were 10 times more likely at those visits to report discussions of individual prescribing factors. These findings demonstrate that available pharmacogenomic results lead to unique doctor-patient communication about prescription decision-making that results in dramatically strengthened recollections of treatment guidance by the patient. To our knowledge, this is the first study to prospectively assess both patient- and provider-reported communications in the context of broad preemptive pharmacogenomic testing and the first to demonstrate that medication recall rates can be positively influenced by the clinical use of pharmacogenomics.
Previous literature has shown that providers face practical barriers when communicating genomic test results, including varying levels of patient health literacy and suboptimal provider knowledge about genetics15, 26, 27. Providers have, indeed, voiced concerns surrounding the communication of pharmacogenomic results, including debates about which healthcare professionals should ultimately bear the responsibility28, 29. In our study, providers reported feeling significantly more informed about pharmacogenomics simply by being exposed to patient results, suggesting that providers can learn genomics via patient-based exposure. As direct-to-consumer genetic testing is becoming more widespread, our findings support the idea that providers, even without formal genomics training, will learn how to successfully handle such results with increasing exposure. We also anticipate that the role of the pharmacist in pharmacogenomic implementation programs will further positively impact within-provider communication about and understanding of results, and we have recently begun studying this additional question in one of our ongoing implementation projects (clinicaltrials.gov #NCT03225820). From the patients’ perspectives, we found it promising that when pharmacogenomic results influenced medication decision-making, patients significantly more often reported recalling discussions of personalized prescribing, genetics/DNA, and specific genetic test results, suggesting frequent patient-provider communication about these topics during prescribing. As the average age of patient participants in our study neared the Medicare-eligible age, these findings may prove especially important for the medical management of an older population, considering the oftentimes higher number of medications taken by those patients.
We believe these findings could be particularly important in the context of prescribing because a patient’s ability to recall details of medication recommendations is critical for adherence30. Previous research has indeed shown that the likelihood of adhering to medications was higher among patients who recalled a physician recommendation compared to those who did not31. While a limited number of prior studies have suggested that pharmacogenomics can positively impact medication adherence17–20, our study is the first, to our knowledge, to examine direct patient-provider communications surrounding pharmacogenomic results and the resulting impact on medication change recall.
Others have suggested that patients who learn their pharmacogenomic results may have increased perceived efficacy about prescribed medications and/or decreased anxiety surrounding potential side effects17. We believe this to be one potential reason that our patients who had pharmacogenomically-influenced medication changes recalled those medication decisions more often than traditional medication changes, as our providers often directly shared pharmacogenomic results with patients. However, the simple presence of genomic results—results that personify the ideal of “individualized care” that patients seek from providers—may also have facilitated discussions of precision medicine in a way that allowed the patient to become more actively involved in their healthcare. These results are consistent with our prior findings, which showed higher patient-reported scores of their providers surrounding empathy, medical decision-making, and personalized care when pharmacogenomic results were accessed12.
Consideration of genetic results may not need to occur in-person. Charland et al.20 reported increased statin adherence when pharmacogenomic results were delivered to patients via mail, meaning that considerations (and possibly discussions) of results could occur outside of the traditional clinic visit, through other forms of communication. It is even possible that what matters is additional communication (whether about genomics, or not), or more broadly the promotion of resources that increase patient education or patients’ active participation in their healthcare32.
Our study had limitations. This study was completed at one institution and included patients cared for by providers who were actively interested in pharmacogenomics. However, our baseline providers’ knowledge about pharmacogenomics was modest, mirroring the characteristics of >10,000 general U.S. physicians21, 33. The fact that only 18 providers were selected to participate is acknowledged, but provider “spillover” effects in this case patient-control patient study would have tended to reduce detectable between-patient-cohort differences. We also recognize that over one-third of patients in the study completed graduate school (a level of education that is higher than that of the average U.S. population), which may potentially hinder the generalizability of our results. It is acknowledged that cases and controls were not randomized. It therefore remains possible that inherent, undetected differences between the two groups existed. The magnitude of any such effect, however, is likely small, as our within-group analyses showed the same results as the overall cohort comparison. We acknowledge that providers were permitted to order separate pharmacogenomic testing outside of the study, although our data suggest that this occurred very infrequently, only in a small minority of providers, and was restricted to a small list of disease-based specialty tests (e.g., TPMT phenotyping in gastroenterology). Further, medication recall was assessed shortly after (within a week of) the clinic visit in most cases. Assessing recall at a later time point may show different results. We also cannot rule-out the possibility that similar increases in medication change recall would be accomplished through other, non-genomic-focused prescription teaching interventions (i.e., it is possible that beneficial effects on recall might also be achieved by any intervention that focuses doctor-patient discussions on prescribing rationale). Finally, we did not directly measure adherence. Therefore, we can only conclude that discussions of pharmacogenomic results impacted recall of medication changes.
In conclusion, both patients and providers reported that genomic information was often discussed when pharmacogenomic results were available to guide medication decisions. This bidirectional assessment of patient-provider prescribing communications revealed that pharmacogenomically-influenced medication changes were recalled by patients significantly more frequently than medication changes that were not influenced by pharmacogenomic results. Additionally, providers reported increased knowledge of pharmacogenomics simply by incorporating patient-specific results into their decision-making calculus. These findings represent important, previously-unrecognized benefits to the clinical use of pharmacogenomics.
Supplementary Material
Acknowledgments
DISCLOSURE/CONFLICT OF INTEREST
Mr. Danahey, Dr. Ratain, and Dr. O’Donnell are named as co-inventors on a pending patent application for the Genomic Prescribing System. Dr. Ratain is a co-inventor holding patents related to pharmacogenetic diagnostics and receives royalties related to UGT1A1 genotyping, although no royalties were received from the genotyping performed in this study. Dr. Ratain’s work has been funded by the NIH, The Conquer Cancer Foundation of the American Society for Clinical Oncology, and The William F. O’Connor Foundation. Dr. O’Donnell’s work has been funded by the NIH, The University of Chicago Comprehensive Cancer Center, The University of Chicago Bucksbaum Institute for Clinical Excellence, and the Central Society for Clinical and Translational Research.
Footnotes
Ms. Borden, Dr. Lee, Ms. Galecki, Dr. Patrick-Miller, Dr. Siegler, Dr. Sorrentino, Dr. Sacro, Dr. Davis, Dr. Rubin, Dr. Lipstreuer, Dr. Polonsky, Dr. Nanda, Dr. Harper, Dr. Koyner, Dr. Burnet, Dr. Stadler, Dr. Kavitt, and Dr. Meltzer declare no potential conflict of interest.
Supplementary Information is available at The Pharmacogenomics Journal website.
REFERENCES
- 1.Ha JF, Longnecker N. Doctor-patient communication: a review. Ochsner J 2010; 10(1): 38–43. [PMC free article] [PubMed] [Google Scholar]
- 2.Bredart A, Bouleuc C, Dolbeault S. Doctor-patient communication and satisfaction with care in oncology. Curr Opin Oncol 2005; 17(4): 351–354. [DOI] [PubMed] [Google Scholar]
- 3.Herndon JH, Pollick KJ. Continuing concerns, new challenges, and next steps in physician-patient communication. J Bone Joint Surg Am 2002; 84-A(2): 309–315. [DOI] [PubMed] [Google Scholar]
- 4.Zolnierek KB, Dimatteo MR. Physician communication and patient adherence to treatment: a meta-analysis. Med Care 2009; 47(8): 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart MA. Effective physician-patient communication and health outcomes: a review. CMAJ 1995; 152(9): 1423–1433. [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy FD, Gordon GH, Whelan G, Cole-Kelly K, Frankel R, Buffone N, et al. Assessing competence in communication and interpersonal skills: the Kalamazoo II report. Acad Med 2004; 79(6): 495–507. [DOI] [PubMed] [Google Scholar]
- 7.Tongue JR, Epps HR, Forese LL. Communication skills. Instr Course Lect 2005; 54: 3–9. [PubMed] [Google Scholar]
- 8.Taran S An examination of the factors contributing to poor communication outside the physician-patient sphere. Mcgill J Med 2011; 13(1): 86. [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson IB, Schoen C, Neuman P, Strollo MK, Rogers WH, Chang H, et al. Physician-patient communication about prescription medication nonadherence: a 50-state study of America’s seniors. J Gen Intern Med 2007; 22(1): 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarn DM, Heritage J, Paterniti DA, Hays RD, Kravitz RL, Wenger NS. Prescribing new medications: a taxonomy of physician-patient communication. Commun Med 2008; 5(2): 195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora NK. Interacting with cancer patients: the significance of physicians’ communication behavior. Soc Sci Med 2003; 57(5): 791–806. [DOI] [PubMed] [Google Scholar]
- 12.McKillip RP, Borden BA, Galecki P, Ham SA, Patrick-Miller L, Hall JP, et al. Patient Perceptions of Care as Influenced by a Large Institutional Pharmacogenomic Implementation Program. Clin Pharmacol Ther 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinnon RA, Ward MB, Sorich MJ. A critical analysis of barriers to the clinical implementation of pharmacogenomics. Ther Clin Risk Manag 2007; 3(5): 751–759. [PMC free article] [PubMed] [Google Scholar]
- 14.Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013; 15(4): 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haga SB, Mills R, Bosworth H. Striking a balance in communicating pharmacogenetic test results: promoting comprehension and minimizing adverse psychological and behavioral response. Patient Educ Couns 2014; 97(1): 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweet K, Sturm AC, Schmidlen T, Hovick S, Peng J, Manickam K, et al. EMR documentation of physician-patient communication following genomic counseling for actionable complex disease and pharmacogenomic results. Clin Genet 2017; 91(4): 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haga SB, LaPointe NM. The potential impact of pharmacogenetic testing on medication adherence. Pharmacogenomics J 2013; 13(6): 481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant RW, Hivert M, Pandiscio JC, Florez JC, Nathan DM, Meigs JB. The clinical application of genetic testing in type 2 diabetes: a patient and physician survey. Diabetologia 2009; 52(11): 2299–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JH, Joy SV, Haga SB, Orlando LA, Kraus WE, Ginsburg GS, et al. Genetically guided statin therapy on statin perceptions, adherence, and cholesterol lowering: a pilot implementation study in primary care patients. J Pers Med 2014; 4(2): 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charland SL, Agatep BC, Herrera V, Schrader B, Frueh FW, Ryvkin M, et al. Providing patients with pharmacogenetic test results affects adherence to statin therapy: results of the Additional KIF6 Risk Offers Better Adherence to Statins (AKROBATS) trial. Pharmacogenomics J 2014; 14(3): 272–280. [DOI] [PubMed] [Google Scholar]
- 21.O’Donnell PH, Wadhwa N, Danahey K, Borden BA, Lee SM, Hall JP, et al. Pharmacogenomics-Based Point-of-Care Clinical Decision Support Significantly Alters Drug Prescribing. Clin Pharmacol Ther 2017; 102(5): 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Donnell PH, Danahey K, Jacobs M, Wadhwa NR, Yuen S, Bush A, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care--initial results of the University of Chicago “1,200 Patients Project”. Am J Med Genet C Semin Med Genet 2014; 166C(1): 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 2012; 92(4): 446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain S, Kenigsberg BB, Danahey K, Lee YM, Galecki PM, Ratain MJ, et al. Disease-drug database for pharmacogenomic-based prescribing. Clin Pharmacol Ther 2016; 100(2): 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danahey K, Borden BA, Furner B, Yukman P, Hussain S, Saner D, et al. Simplifying the use of pharmacogenomics in clinical practice: Building the genomic prescribing system. J Biomed Inform 2017; 75: 110–121. [DOI] [PubMed] [Google Scholar]
- 26.Burke W, Emery J. Genetics education for primary-care providers. Nat Rev Genet 2002; 3(7): 561–566. [DOI] [PubMed] [Google Scholar]
- 27.Sharp RR, Goldlust ME, Eng C. Addressing gaps in physician education using personal genomic testing. Genet Med 2011; 13(8): 750–751. [DOI] [PubMed] [Google Scholar]
- 28.Unertl KM, Field JR, Price L, Peterson JF. Clinician Perspectives on Using Pharmacogenomics in Clinical Practice. Per Med 2015; 12(4): 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet 2012; 82(4): 388–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin LR, Williams SL, Haskard KB, Dimatteo MR. The challenge of patient adherence. Ther Clin Risk Manag 2005; 1(3): 189–199. [PMC free article] [PubMed] [Google Scholar]
- 31.Kravitz RL, Hays RD, Sherbourne CD, DiMatteo MR, Rogers WH, Ordway L, et al. Recall of recommendations and adherence to advice among patients with chronic medical conditions. Arch Intern Med 1993; 153(16): 1869–1878. [PubMed] [Google Scholar]
- 32.Vahdat S, Hamzehgardeshi L, Hessam S, Hamzehgardeshi Z. Patient involvement in health care decision making: a review. Iran Red Crescent Med J 2014; 16(1): e12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanek EJ, Sanders CL, Taber KA, Khalid M, Patel A, Verbrugge RR, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther 2012; 91(3): 450–458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.