Abstract
Permethylation is an essential technique for the detailed structural characterization of glycans by mass spectrometry. However, it requires specialized skills and equipment and is labor-intensive. While this limits glycan analysis to a handful of experts, the increasing awareness of the tremendous importance of glycans in physiological processes of health and disease has drastically raised the demand for detailed structural characterization of glycans. We have developed a simple microplate permethylation method that relies upon solid phase extraction using C18 tips to purify the permethylated glycans. The procedure is easy to perform, making it accessible to non-experts, and fast, promising to accelerate glycan research. A 96-well plate of released glycan samples can be permethylated in less than an hour. The procedure can be carried out without discontinuous steps in an automatic pipette and integrated seamlessly with glycan release and mass spectrometry.
Carbohydrates are an integral part of the functioning of all living organisms. All animal cells are coated with a layer of glycans of specific structure encoding the information required to interact with other cells, either in the context of signaling and molecular recognition or in defense from pathogens.1 Due to the complex structure of carbohydrates, their study has lagged behind that of nucleic acids and proteins, and until recently, only a comparatively small number of researchers have focused on the study of glycan structure and function. Although in the last couple of decades, the glycoscience field has experienced impressive technological advances, these are still not broadly available to the general biomedical science community. The reasons for this lack are that the study of carbohydrates requires highly specialized skill sets and instrumentation and that routine, automated, and high-throughput workflows, common in genomics and proteomics, are not available. As a result, detailed glycan structure analysis is still dependent on a small group of experts, presenting a bottleneck for progress in important biomedical research. The need for new tools to make glycoanalysis more broadly accessible was also recently highlighted in a Nature Methods feature.2 The findings presented in this letter directly respond to this need by introducing a new method for glycan permethylation that is high-throughput, user friendly, and cost-effective, paving the way for automated glycomics and glycoproteomics analysis.
Permethylation consists of replacing all hydrogens attached to oxygen and nitrogen atoms with methyl groups and is a necessary step in the structural characterization of glycans by mass spectrometry (MS). Currently, only permethylation can enable MS to provide sequence, branching, and linkage information for glycans.3 Unfortunately, permethylation is a difficult and time-consuming process that is not easily adapted to large numbers of samples and requires specialized training. Therefore, only a handful of labs are able to perform it consistently. Recently, glycan permethylation has received renewed attention, and new approaches to improve it have been published.4, 5 Attempts have also been made to adapt permethylation for multiple samples,6–8 including a commercially available kit featuring a 96-well plate for high-throughput permethylation of glycans. However, the currently available options are expensive, tedious, and time-consuming. The commercial kit contains one specially made 96-well plate that is sealed under an inert atmosphere to protect the NaOH contained therein from moisture. This entails high cost and lack of flexibility when analyzing less than 96 samples and presents a barrier to widespread use. However, the main disadvantage of the kit is its reliance on liquid-liquid extraction (LLE) for the isolation of the permethylated glycans, as it presents several roadblocks to wider dissemination. LLE is associated with the risk of forming emulsions, difficulty obtaining precise phase separation, as well as large amounts of expensive, highly volatile, and toxic solvents. It requires operations that are disconnected and not easily automated, such as shaking and centrifugation. Furthermore, permethylated glycans bearing polar substituents, such as sulfates, can often not be recovered by LLE.9, 10 Solid-phase extraction (SPE), by contrast, can be performed on smaller volumes, does not require additional solvents, is more efficient and is characterized by higher recovery.11 Most significantly, SPE is more suitable for automation since it can be done in a pipette tip, taking advantage of commercially available C18 pipette tips, and allows sample workup in one continuous flow rather than disconnected steps, as is typical in LLE.
Today, permethylation of glycans is almost always carried out in DMSO with sodium hydroxide as base and iodomethane as methylating agent. The reaction is traditionally performed by adding sodium hydroxide as dry powder.12 Although this method has been widely and successfully applied in various MS based structural studies of complex oligosaccharides, it is a multi-step process involving milliliter scale and multiple LLEs, both of which stand in the way of high throughput and sensitivity. Side reactions, such as oxidative degradation and “peeling”, associated with the high pH conditions of the permethylation are additional limitations that have recently been addressed through methods featuring immobilized NaOH (“solid-phase permethylation”).8, 13 Although the latter are more amenable to larger numbers of samples, these methods require multiple centrifugation steps or several passages through packed capillaries, which pose a hindrance to high-throughput or automated analysis. These obstacles also prevent the widespread use of these methods by non-experts. That goal requires that the permethylation can be performed with ordinary equipment and techniques that are commonly used in biochemistry laboratories. The method we are presenting here can be carried out with regular polypropylene 96-well plates or microcentrifuge tubes and C18 tips, as well as a simple permethylation kit that we plan to make available in the near future.
EXPERIMENTAL SECTION
Permethylation of glycans in 96-well plate.
Lyophilized N-glycans were dissolved in 50 μL of DMSO and transferred to either a microcentrifuge tube or wells of a 96-well plate. To this, 75 μL of the NaOH base in DMSO (see Supplementary Material) was added and drawn up into and expelled from the pipette 3 times for mixing. The samples were treated with 25 μL of MeI, mixed 3 times with the pipette, and incubated for 20 min. The permethylation reaction was quenched by the addition of 100 μL of ddH2O. Excess MeI was evaporated by pipetting 200 μL air into the samples 15 times. The remaining mixtures containing the permethylated N-glycans were drawn up into C18 tips (preconditioned with MeOH and preequilibrated with ddH2O), which retained the permethylated glycans, while the solvents were expelled and discarded. The tips were washed with 5 × 200 μL ddH2O, and the permethylated glycans were eluted with 5 × 30 μL 100% methanol.
RESULTS AND DISCUSSION
The need to repeat the permethylation steps many times in the solid-phase permethylation8, 13 is likely due to the limited contact of the glycan with the surface of the solid NaOH. This is also a detrimental factor in the traditional permethylation with powdered NaOH in DMSO, slowing down the reaction. Anumula and Taylor14 published a modification of the NaOH method that yields a much finer, sol-like dispersion of NaOH in DMSO than the one made from solid, powdered NaOH. This method involves extraction of water with DMSO from a 50% aqueous NaOH solution and gives good permethylation yields with no significant side reactions. This base preparation is stable as long as it is kept away from moisture and is therefore suitable to be included in a potential permethylation kit if it is stored in an appropriate container, such as an ampoule. Because this suspension is finer, we suspected that it would allow faster permethylation reactions. We indeed found that the permethylation is complete in about 15 min, and that no agitation of the reaction mixture is necessary to obtain full permethylation. Thus, this base preparation is ideally suited for an automated process.
The automated procedure is illustrated in Figure 1. It can be performed in a regular automatic 96-channel pipette. Released glycan samples, which themselves can be obtained using automation,15 are dissolved in DMSO in the wells of a polypropylene microplate and treated with the base, followed by iodomethane. The microplate does not need to be covered or agitated but is left for 20 min to ensure complete methylation. The reaction is terminated by addition of water, upon which the excess iodomethane forms a separate lower layer. The iodomethane is removed by repeatedly pushing air through the mixtures by means of the automatic pipette. Then, C18 SPE tips are mounted, and the sample solutions are passed repeatedly through the C18 resin. The resins containing the bound permethylated glycans are washed with water and eluted with a small volume of MeOH. The resulting solution is then mixed with MALDI matrix or ESI infusion buffer for MS. This can also be done automatically by the same pipette.
Figure 1.
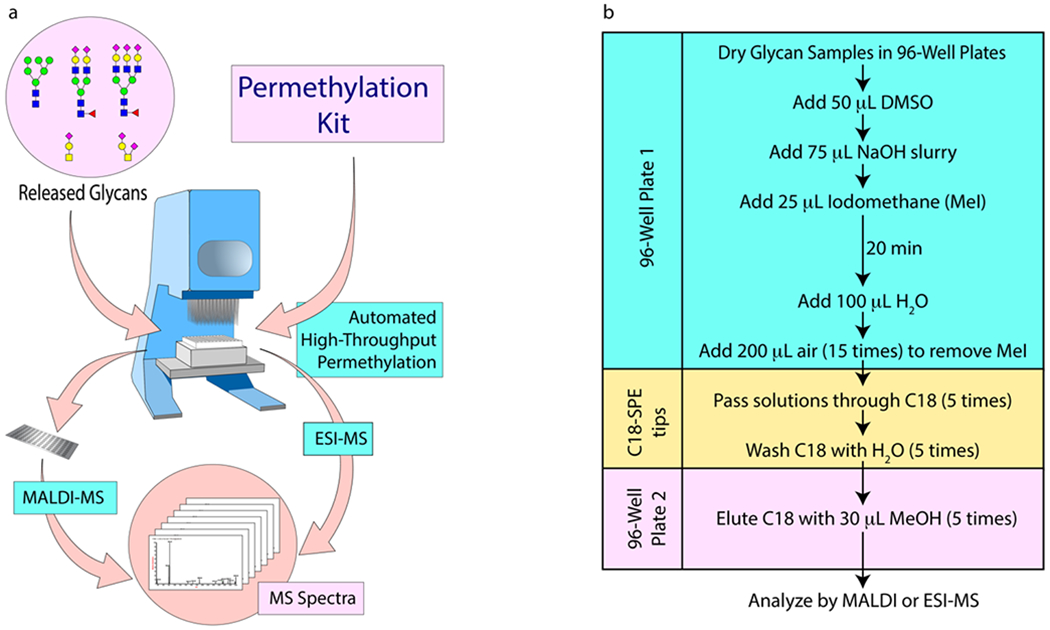
(a) Schematic showing how an automatic 96-well pipette can be used to carry out the permethylation procedure; (b) flow chart, showing the steps involved in the simplified automated permethylation workflow. The permethylated glycans are passed from Plate 1 to the C18 SPE tips, and from there to Plate 2.
The entire micro-permethylation is carried out in conventional microcentrifuge tubes or polypropylene 96-well plates and can be performed manually or by using any degree of automation, from single-channel to multi-channel pipettes to semi-automatic pipettes or fully automatic robots that can process 96 samples at a time. Unlike with the previous approaches to high-throughput or automated glycan permethylation, none of the steps require extensive robotics-based sample handling. The protocol can also be integrated with proteolysis and glycan release. Integrating the protocol with proteolysis and glycan release using rapid trypsin and PNGase F, the whole processing of 96 glycoprotein samples, including acquisition of MALDI-MS or ESI-MSn glycan spectra of all samples can be accomplished in less than one day.
The glycan permethylation method presented here is very sensitive. We were able to detect oligosaccharides in the range of a few femtomoles per injection (Figure 2a) and obtained good spectra of the glycans released from only 1 microgram of glycoprotein (Figure 2b). The resulting mass spectra were equivalent to those obtained from the conventional method performed at milliliter scale. Although the procedure is carried out entirely with plastic supplies, we did not observe any significant contaminating peaks, and the spectra are equivalent to those obtained with the manual method14 performed in glass (Figure 2a,b).
Figure 2.
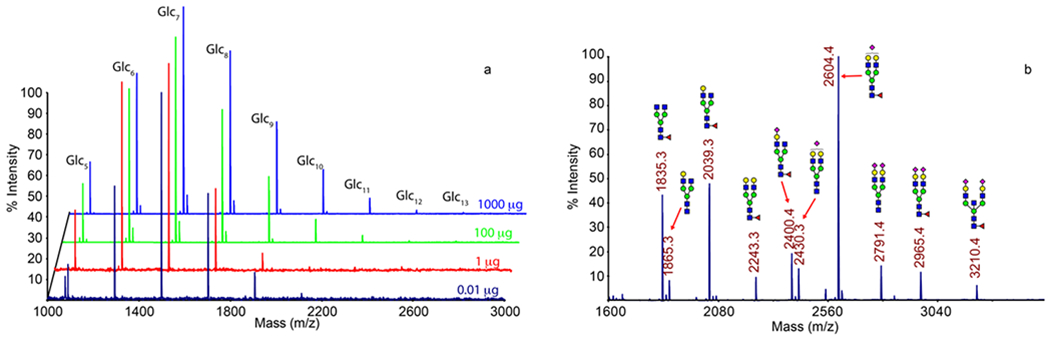
MALDI-TOF MS spectra of oligosaccharides permethylated with the present method. (a) Various amounts of maltooligosaccharides and (b) glycans from 1 μg human serum IgG.
We have employed our micro-permethylation strategy on clinically and therapeutically important glycoproteins such as immunoglobulins (IgG) and transferrin isolated from human serum, along with standard glycoproteins such as bovine fetuin and Ƙ-casein (Figure S1). The glycosylation profiling of IgG and transferrin isolated from human serum are common readouts for aberrant glycosylation, which are hallmarks of various disease conditions such as congenital disorders of glycosylation (CDG), cancer, and many others. 16, 17
The new protocol, along with the permethylation kit, will allow any lab with access to a MALDI or ESI-MS instrument to perform glycomics analysis by MS. Free software (http://www.grits-toolbox.org/?page_id=363) is already available for semi-automated peak assignment of MS spectra. Micro-permethylation of glycopeptides allows convenient, rapid, and reliable high-throughput screening of protein glycosylation. Our strategy will revolutionize the current glycomics analysis approaches and promises the development of a cost-effective kit for the analysis of large sample set of glycoprotein therapeutics, clinical biomarker discoveries and other application where detailed glycan profile is often required.
Supplementary Material
ACKNOWLEDGMENT
Financial support from the US National Institutes of Health (R21GM122633, P41GM10349010, S10OD018530) is gratefully acknowledged.
Footnotes
Competing interests
The authors certify that they have no competing interests.
SUPPORTING INFORMATION
Mass spectra and full experimental details
REFERENCES
- (1).Varki A; Esko JD; Colley KJ Cellular Organization of Glycosylation In Essentials of Glycobiology, 3rd ed.; Varki A; Cummings RD; Esko JD; Stanley P; Hart GW; Aebi M; Darvill AG; Kinoshita T; Packer NH; Prestegard JH; Schnaar RL; Seeberger PH, Eds. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, 2017. [Google Scholar]
- (2).Marx V Metabolism: sweeter paths in glycoscience. Nat. Methods 2017, 14, 667. [DOI] [PubMed] [Google Scholar]
- (3).Ashline DJ; Zhang H; Reinhold VN Isomeric complexity of glycosylation documented by MS(n). Anal. Bioanal. Chem 2017, 409, 439–451. [DOI] [PubMed] [Google Scholar]
- (4).Hu Y; Borges CR A spin column-free approach to sodium hydroxide-based glycan permethylation. Analyst 2017, 142, 2748–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Gao X; Zhang L; Zhang W; Zhao L Design and application of an open tubular capillary reactor for solid-phase permethylation of glycans in glycoprotein. Analyst 2015, 140, 1566–71. [DOI] [PubMed] [Google Scholar]
- (6).Shubhakar A; Kozak RP; Reiding KR; Royle L; Spencer DI; Fernandes DL; Wuhrer M Automated High-Throughput Permethylation for Glycosylation Analysis of Biologics Using MALDI-TOF-MS. Anal. Chem. 2016, 88, 8562–9. [DOI] [PubMed] [Google Scholar]
- (7).Shubhakar A; Pang P-C; Fernandes DL; Dell A; Spencer DIR; Haslam SM Towards automation of glycomic profiling of complex biological materials. Glycoconj. J. 2018, 35, 311–321. [DOI] [PubMed] [Google Scholar]
- (8).Kang P; Mechref Y; Novotny MV High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 721–734. [DOI] [PubMed] [Google Scholar]
- (9).Heiss C; Wang Z; Azadi P Sodium hydroxide permethylation of heparin disaccharides. Rapid Commun. Mass Spectrom. 2011, 25, 774–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yu S-Y; Wu S-W; Hsiao H-H; Khoo K-H Enabling techniques and strategic workflow for sulfoglycomics based on mass spectrometry mapping and sequencing of permethylated sulfated glycans. Glycobiol. 2009, 19, 1136–1149. [DOI] [PubMed] [Google Scholar]
- (11).Ahadi A; Partoazar A; Abedi-Khorasgani M-H; Shetab-Boushehri SV Comparison of liquid-liquid extraction-thin layer chromatography with solid-phase extraction-high-performance thin layer chromatography in detection of urinary morphine. J. Biomed. Res. 2011, 25, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ciucanu I; Kerek F A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984, 131, 209–217. [Google Scholar]
- (13).Kang P; Mechref Y; Klouckova I; Novotny MV Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Commun. Mass Spectrom. 2005, 19, 3421–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Anumula KR; Taylor PB A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal. Biochem. 1992, 203, 101–8. [DOI] [PubMed] [Google Scholar]
- (15).Stöckmann H; Adamczyk B; Hayes J; Rudd PM Automated, High-Throughput IgG-Antibody Glycoprofiling Platform. Anal. Chem. 2013, 85, 8841–8849. [DOI] [PubMed] [Google Scholar]
- (16).Scott K; Gadomski T; Kozicz T; Morava E Congenital disorders of glycosylation: new defects and still counting. J. Inherit. Metab. Dis. 2014, 37, 609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gudelj I; Lauc G; Pezer M Immunoglobulin G glycosylation in aging and diseases. Cellular Immunol. 2018, pii: S0008-8749(18)30325-3 DOI: 10.1016/j.cellimm.2018.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


